Abstract
The interaction of Fas (CD95), a member of the tumor necrosis factor receptor (TNFR) family, and its ligand (FasL) triggers programmed cell death (apoptosis) and is involved in the regulation of immune responses. Although the Fas–FasL interaction is conserved across species barriers, little is currently known about the molecular details of this interaction. Our aim was to identify residues in Fas that are important for ligand binding. With the aid of a Fas molecular model, candidate amino acid residues were selected in the Fas extracellular domain 2 (D2) and D3 and subjected to serine-scanning mutagenesis to produce mutant Fas molecules in the form of Ig fusion proteins. The effects of these mutations on FasL binding was examined by measuring the ability of these proteins to inhibit FasL-mediated apoptosis of Jurkat cells and bind FasL in ELISA and BIAcore™ assays. Mutation of two amino acids, R86 and R87 (D2), to serine totally abolished the ability of Fas to interact with its ligand, whereas mutants K84S, L90S, E93S (D2), or H126S (D3) showed reduced binding compared with wild-type Fas. Two mutants (K78S and H95S) bound FasL comparably to wild type. Therefore, the binding of FasL involves residues in two domains that correspond to positions critical for ligand binding in other family members (TNFR and CD40) but are conserved between murine and human Fas.
Programmed cell death (apoptosis) mediated by the Fas– FasL system is a mechanism used to control immune responses. The Fas (CD95) antigen, a 45-kD protein of the TNF receptor (TNFR) family, is widely expressed and binds a TNF-like ligand (FasL) (1). Perturbations of the Fas–FasL interaction have drastic functional consequences in lpr and gld mice, leading to lymphadenopathy and severe immune disregulation (2, 3). A human lymphoproliferative disorder, the Canale–Smith syndrome, appears to be due to mutation of the signal transduction domain of Fas (4). Although FasL is expressed as a cell surface molecule, it is also released after cleavage by metalloproteinases (5), enabling FasL to act as a soluble mediator of cell death. Fas-mediated cell death is thought to be involved in the pathology of a number of disease states, including fulminant hepatitis and chronic liver disease (6, 7), multiple sclerosis (8), and it may also have a role in neutrophil-mediated tissue destruction (9). In addition, some tumors are able to escape immune surveillance by releasing FasL, which kills activated T cells infiltrating the tumor (10, 11).
Molecular details of the Fas–FasL interaction have yet to be determined. Fas is a type I membrane protein, consisting of three TNFR-like extracellular domains (D1, D2, and D3), a hydrophobic transmembrane region, and a cytoplasmic tail containing a death domain. The death domain binds Fas death domain–binding protein (FADD, MORT1), which links Fas to a cascade of IL-1β-like proteolytic enzymes known as caspases (12). Recently, the three dimensional structure of the Fas death domain was solved using NMR spectroscopy, and was shown to consist of six antiparallel, amphipathic α helices arranged in a novel fold (13). Fas binds to FasL across the murine and human species barrier, suggesting the conservation of amino acid residues important for binding. We wished to investigate the structural basis for the Fas–FasL interaction. Using the TNFR three-dimensional structure as a template, we were able to generate a model of the Fas extracellular domains (14). On this model, we were able to map residues conserved between human and murine Fas, and positions implicated in the interaction of TNFR with TNF (15), and/or positions implicated by mutagenesis analysis in the interaction of another family member, CD40, with the CD40L (16,17). A surface was identified on the extracellular D2 of Fas and a part of D3, which consists of residues conserved in murine and human Fas, but not conserved between Fas and TNFR or CD40. Residues in this region were considered potential candidates for FasL binding and were subjected to serine-scanning mutagenesis. We found that binding of FasL is centered on D2 of Fas and involves a region that corresponds to the ligand binding sites in TNFR and CD40.
Materials and Methods
Monoclonal Antibodies and Fusion Proteins.
Soluble FasL was produced in a manner similar to that described for a closely related TNF family member, gp39, the ligand for CD40 (18), by fusing the extracellular domain of FasL to the extracellular domain of murine CD8. cDNA encoding for the extracellular domain of human FasL (amino acids 105–281) was amplified by PCR from monocyte cDNA using the primers CGC CGC GGA TCC CTT CCA CCT ACA GAA GGA GCT G (forward primer containing a BamHI site) and GGC TGC TCT AGA CCC AAA GTG CTT CTC TTA GAG CTT ATA TAA GCC (reverse primer containing a XbaI restriction enzyme site). Amplified cDNA was digested with BamHI and XbaI, gel purified, and ligated into the pCDM7− vector containing cDNA encoding for murine CD8. CD8–FasL was produced in COS cells following transfection by the DEAE–Dextran chloroquine method. Supernatants containing CD8–FasL fusion proteins were harvested and passed through a 0.22-μm filter. CD8–FasL was affinity purified on an anti-CD8 (53-6) column as previously described for CD8–CD40L (19).
A soluble protein (FasthrRγ1), consisting of the extracellular domain of Fas fused to the hinge, CH2, and CH3 regions of human IgG1 containing a thrombin cleavage site, was constructed. In brief, cDNA was amplified by PCR using oligonucleotide primers 5′-CGC CCC AAG CTT CGG AGG ATT GCT CAA CAA CC (containing a HindIII site) and 3′-CGC CGC GGA TCC CCC AAG TTA GAC CTG GAC CCT TCC TC (naturally occurring BamHI and BglII sites were removed and a BamHI site was added). Purified PCR products were digested with HindIII and BamHI restriction enzymes, gel purified, and ligated into CDM7− containing a thr–human IgG1 (thrRγ1) cassette (20). Fasthr Rγ1 supernatants were produced as described above for CD8–FasL, affinity purified on protein A–Sepharose, eluted with ImmunoPure elution buffer (Pierce, Rockford, IL), dialyzed in PBS, and concentrated. The concentration of each protein was determined using an anti-human IgG binding ELISA. Preparation of murine CD6 SRCR–D1thrRγ1 (mCD6D1thrRγ1) used as a negative control protein has been previously described (21).
Three anti-Fas mAbs were used to confirm the structural integrity of the mutant Fas fusion proteins. SM1/1 (IgG2a), SM1/17 (IgG1), and DX-2 (IgG1) were purchased from Biosource International (Camarillo, CA) for ELISA. These were unable to immunoblot reduced FasthrRγ1, an indication that they recognize conformation-dependent epitopes on Fas.
Site-directed Mutagenesis.
Eight amino acid residues in the extracellular domain of Fas were selected for site-directed mutagenesis to serine based on the molecular model of the Fas extracellular region (14). FasthrRγ1 mutants were produced by encoding the desired mutation in overlapping oligonucleotide primers, and using FasthrRγ1 cDNA as a template, the mutants were generated by PCR. The 5′ and 3′ oligonucleotide primers described for the production of FasthrRγ1 were also used for the generation of mutants. PCR products were digested with HindIII and BamHI and ligated into the CDM7−–thrRγ1 cassette as described above. Mutant proteins were produced as described above for FasthrRγ1. Each cDNA construct was sequenced to confirm insertion of the appropriate mutation and to ensure that the PCR had not produced unwanted mutation.
Stimulation of Jurkat Cell Death.
Jurkat cells that were susceptible to FasL-mediated apoptosis were added to the wells of 96well microtiter plates at a final concentration of 1 × 105 cells/ well in a total volume of 200 μl with 25 μl of a CD8–FasL containing supernatant and various concentrations of FasthrRγ1 or mutant FasthrRγ1. Microtiter plates were incubated overnight at 37°C, 5% CO2 before the addition of 25 μl/well of Alamar Blue (BioSource International, Camarillo, TX). Plates were incubated for a further 8–12 h at 37°C before measuring the OD (A570nm − A595nm).
Enzyme Immunoassays.
The gross structural integrity of the fusion proteins was confirmed using the panel of mAbs described above in an ELISA assay. Fusion proteins (300 ng/ml in carbonate/bicarbonate buffer, pH 9.6) were immobilized on Immunolon II (Dynatech Laboratories, Inc., Alexandria, VA) plates overnight at 4°C. Plates were blocked with 3% BSA–PBS, followed by addition of mAb at various concentrations. Plates were washed, followed by 1:5,000 dilution of HRP-conjugated goat anti–mouse IgG (Jackson ImmunoResearch Labs., Inc., West Grove, PA). HRP substrate (GIBCO BRL, Gaithersberg, MD) was added, the color reaction stopped with H2SO4, and OD measured (A450nm − A725nm).
Binding of FasthrRγ1 mutants to FasL was measured by a capture ELISA. Various amounts of FasthrRγ1 or FasthrRγ1 mutants were captured on anti-human IgG (100 μl of 5 μg/ml; Jackson ImmunoResearch)–coated Immunolon II plates that had been blocked with 3% BSA–PBS. Plates were washed with 0.05% Tween-20 (Sigma Chem. Co., St. Louis, MO) in PBS, followed by the addition of a fixed, saturating concentration of CD8–FasL containing supernatant. Bound CD8–FasL was detected by the addition of 1 μg/ml anti-FasL mAb (NOK-2, PharMingen, San Diego, CA) and subsequently 1:5,000 dilution of HRP-conjugated anti-mouse IgG (Jackson ImmunoResearch). HRP substrate was added, the color reaction stopped with H2SO4, and absorbances were measured as described above. To confirm that equivalent amounts of FasthrRγ1 and FasthRγ1 mutants were captured on the plates in these assays, duplicate plates were set up. HRPconjugated anti-human IgG (1:2,000; Amersham Corp., Arlington Heights, IL) was added to these plates after the addition of the FasthRγ1 proteins, and the binding quantified as described above.
Surface Plasmon Resonance Analysis.
All experiments were run on a BIAcore™ 1000 instrument (Pharmacia Biosensor, Uppsala, Sweden) at 25°C using PBS, pH 7.4, containing 0.005% surfactant P20 (Pharmacia Biosensor) as the running buffer. CD8–FasL was immobilized on research-grade CM5 sensor chips (Pharmacia Biosensor) using standard N-ethyl-N-dimethylaminopropyl carbodiimid/N-hydroxysuccinimide coupling. Purified CD8–FasL protein (15 μg/ml) was diluted in 10 mM sodium formate buffer, pH 4.0, and incubated with activated sensor chips for 3 min. After coupling, excess N-hydroxysuccinimide groups were inactivated with ethanolamine. Immobilization of ∼6,000 RU of FasL was achieved.
Apparent association and dissociation rates for wild-type FasthrRγ1 were determined using concentrations ranging from 10 to 200 nM. After the injection, flow of running buffer alone was established to allow observation of the dissociation of bound protein. Dissociation to baseline was obtained in the allotted time, so it was not necessary to inject any reagent to regenerate the ligand surface. Apparent association and dissociation rates were determined by curve-fitting using BIAevaluation 2.1 (Pharmacia Biosensor).
Results and Discussion
Generation of FasthrRγ1 Mutants.
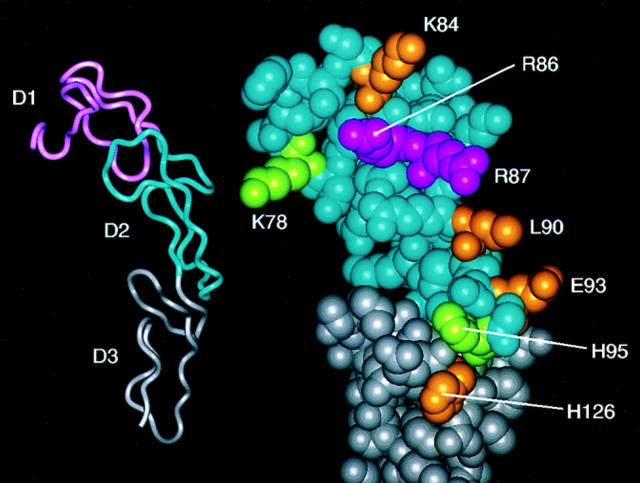
Fas belongs to the TNFR superfamily and its extracellular region includes three domains (D1–D3) with distinct sequence homology to TNFR (1). On this basis, a detailed three-dimensional model was generated (14) using the TNFR x-ray structure (15) as a template. Eight residues that were spatially adjacent in the model (K78, K84, R86, R87, L90, E93, H95 in D2, and H126 in D3) were selected as candidates for mutagenesis. These eight residues are conserved in murine and human Fas, but they are not conserved between family members (14). Six of these positions (all except R86 and H95) correspond to residues that are implicated in the TNFR and/or CD40–ligand interactions (14, 16, 17). The molecular model and mutated residues are shown in Fig. 1. Color-coding of this model is based upon the results of the mutagenesis studies described below and discussed at the end of this report. Mutant proteins were generated, expressed as Ig fusion proteins (FasthrRγ1), and purified for use in FasL binding and functional assays.
Figure 1.
Mapping of residues important for ligand binding to Fas. On the left, a ribbon representation of the Fas molecular model is shown, which illustrates its extracellular domain (D1, D2, D3) organization. On the right, a close-up view of D2 (blue) and D3 (gray) is shown in similar orientation. Residues subjected to mutagenesis were mapped on the model and color-coded according to their importance for binding (magenta, critical for binding; gold, support binding; green, not important for binding).
Binding of Mutant Proteins to mAb.
The binding of each mutant protein to a panel of anti-Fas mAbs, which recognize three epitopes based on the following (a) their ability to block CD8–FasL-mediated apoptosis (DX-2); (b) their ability to induce apoptosis (SW1/1); or (c) their inability to block or induce apoptosis (SW1/17) in our system (data not shown), was examined. Each mAb was unable to immunoblot FasthrRγ1 when the protein was reduced before loading on the gel, indicating that the epitopes the mAb recognize are conformationally sensitive. Therefore, these mAbs are suitable to monitor the overall structural integrity of expressed mutant proteins. Each mAb bound to each mutant and to wild-type FasthrRγ1 equivalently as determined by ELISA (Fig. 2), indicating that the overall structural integrity of the proteins was not significantly compromised as a consequence of the mutations.
Figure 2.
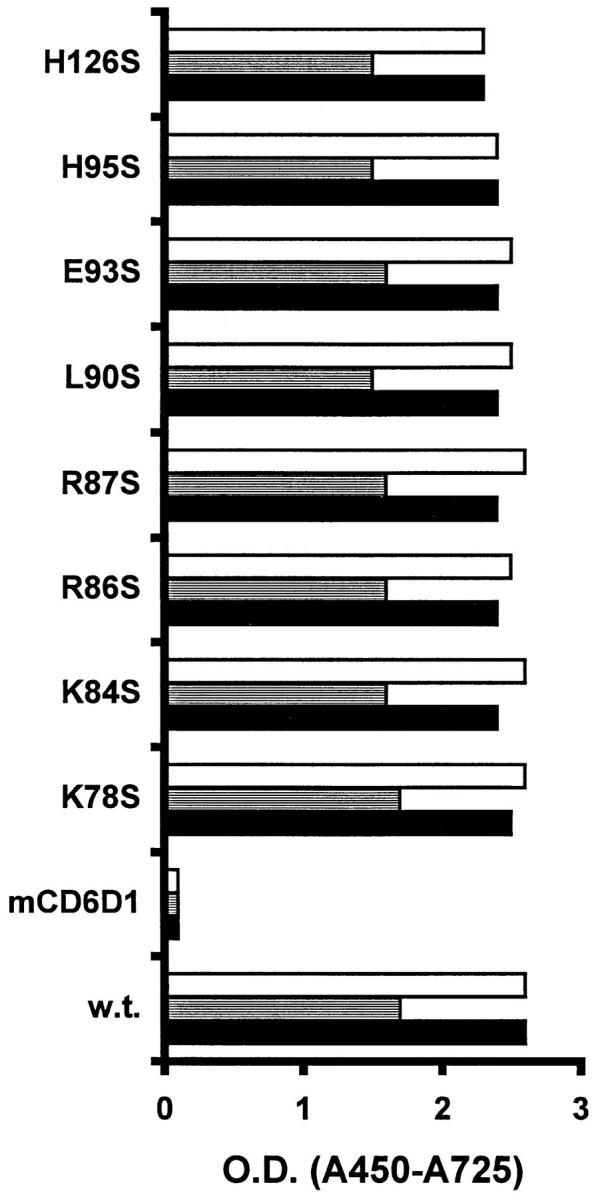
A panel of anti-Fas mAbs bind FasthrRγ1 and FasthrRγ1 mutants. Fusion proteins were immobilized, and binding was detected by anti-Fas mAbs at various concentrations. Data is shown for mAb at 1 μg/ml, but similar relative binding was observed at lower concentrations of mAb. Open bar, SW1/17; shaded bar, SW1/1; closed bar, DX-2. Negative control mAb OD values for each fusion protein were ∼0.1.
FasL Binding Capabilities of FasthrRγ1 Mutants.
The functional capabilities and binding characteristics of FasthRγ1 mutants were compared with wild-type FasthRγ1 in vitro.
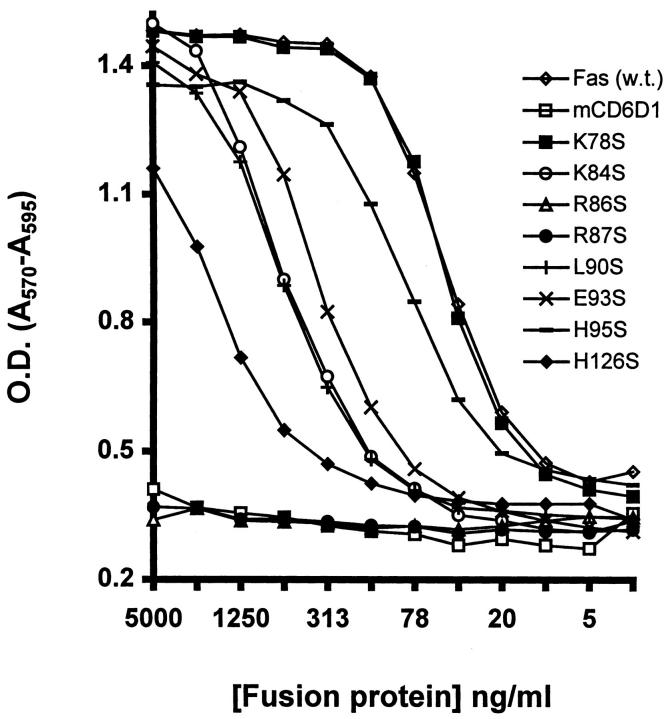
Supernatants of CD8–FasL were able to kill Jurkat cells in a dose-dependent manner (data not shown), and the killing could be inhibited by wild-type FasthrRγ1. FasthrRγ1 wild-type and mutants were titered at a constant concentration of CD8–FasL, and the viability of the Jurkat cells was determined by measuring the change in color of Alamar Blue added to the microtiter wells. In this assay, FasthrRγ1 inhibited apoptosis in a concentration-dependent manner, whereas mutant proteins showed a range of inhibitory activities (Fig. 3). Mutation of residues K78 and H95 to serine had little effect on the ability of the fusion proteins to inhibit killing, whereas mutation of residues, K84, L90, E93, and H126 to serine markedly reduced the ability of FasthrRγ1 to inhibit the apoptotic activity of CD8–FasL. Mutation to serine at positions R86 and R87 completely abolished the inhibitory effect of the fusion proteins, suggesting they are critical for the Fas–FasL interaction. A control fusion protein, mCD6D1thrRγ1, had no protective effect in these assays.
Figure 3.
Mutation of FasthrRγ1 at positions R86 and R87 abrogates its ability to inhibit FasL-mediated killing of Jurkat cells. Jurkat cells were incubated with a 1:8 dilution of CD8–FasL-containing supernatant in the presence of wild-type and mutant FasthrRγ1 in triplicate wells for each concentration of fusion protein. Increased cell death is represented by low OD values.
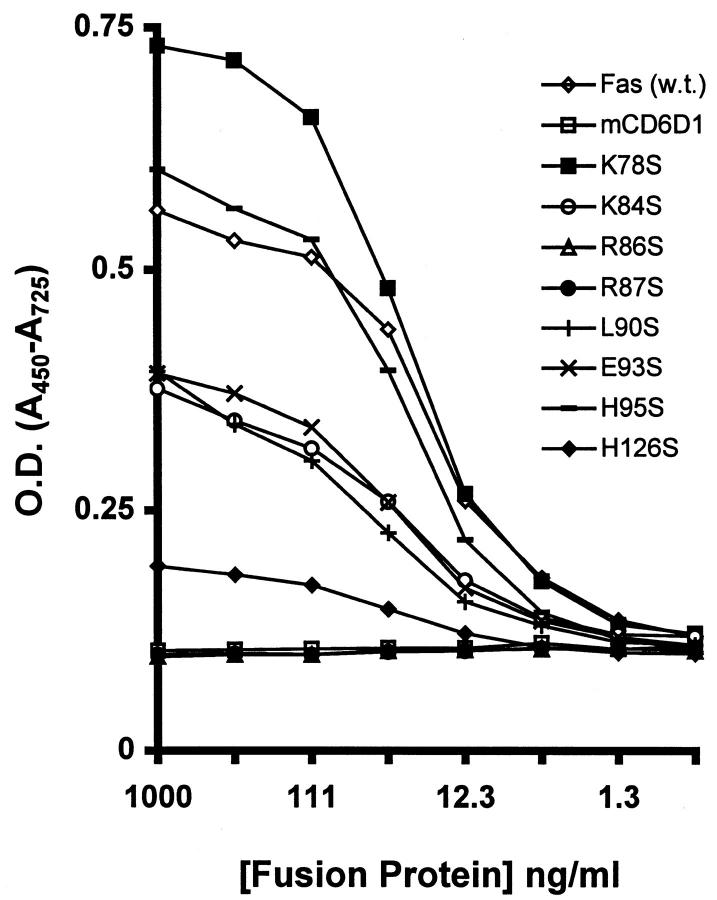
To measure directly the binding of FasthrRγ1 mutants to FasL, an ELISA assay was developed. FasthrRγ1 and mutants were captured on anti-human Ig-coated plates, followed by addition of CD8–FasL-containing supernatant. Bound FasL was detected using an anti-FasL mAb, NOK-2, and subsequently anti-mouse IgG. The results parallel those obtained in the biological assay (Fig. 4). K78S and H95S, which showed wild-type inhibitory activity of FasL-mediated killing bound at similar levels to wild type. Although saturation of binding by K78S was higher than wild type, the linear portions of the binding curves were similar. Mutants R86S and R87S, which were unable to protect Jurkat cells from FasL-mediated lysis, were likewise unable to bind FasL in the ELISA. Their binding characteristics were identical to the mCD6D1thrRγ1 control fusion protein, indicating that positions 86 and 87 are critical for FasL binding. Mutants with intermediate inhibitory activity in the viability assay also showed intermediate binding to FasL, thus indicating a strong correlation of binding and functional characteristics. Of these intermediate mutants, H126S showed the least binding, whereas the binding levels of K84S, L90S, and E93S were roughly equivalent but ∼10-fold reduced compared with wild-type FasthrRγ1. To confirm that the hierarchy of mutant to binding FasL was not a consequence of differential capture of the mutants, duplicate plates were analyzed for amount of proteins captured by detecting the Fc portion of the fusion proteins with an anti-human IgG reagent. Equivalent binding profiles were obtained for each fusion protein (data not shown), confirming that the differential FasL binding observed was due to the mutation in the Fas region and not due to differences in protein concentration or ability to bind anti-human Ig.
Figure 4.
Binding of FasL to immobilized FasthrRγ1 mutants in ELISA. Fusion proteins were captured on ELISA plates using donkey anti– human IgG. CD8–FasL was added to plates, and binding of FasL was detected using the NOK-2 mAb and HRP-conjugated anti-mouse IgG.
Surface Plasmon Resonance Analysis of FasthrRγ1 and Mutant Binding to FasL.
Surface plasmon resonance analysis of the Fas–FasL interaction was undertaken by BIAcore™ analysis to obtain an approximation of the binding constant for this interaction. Purified CD8–FasL was immobilized on a sensor chip and various concentrations of FasthrRγ1 in fluid phase was passed over the chip. Association and dissociation rates were calculated, and a Kd of value of ∼7 × 10−8 M was obtained from the ratio of dissociation/association. This is a relatively weak interaction compared with Kd values obtained for the TNFR(p55)Ig–TNF interaction (6.5 × 10−11 M for TNF-α and 6.4 × 10−10 M for TNFβ) obtained by Scatchard analysis (22). The Kd of the FasthrRγ1–CD8–FasL interaction is also considerably weaker than that reported for CD40–CD40L (Kd, ∼5 × 10−10 M [23]) but similar to another TNFR–TNF family member, 4-1BB/4-1BBL (Kd of low affinity sites using recombinant ligand ∼7 × 10−8 M and Kd of high affinity sites ∼3 × 10−10 M [24]), each Kd value obtained by binding Ig fusion proteins to ligand expressed on cells.
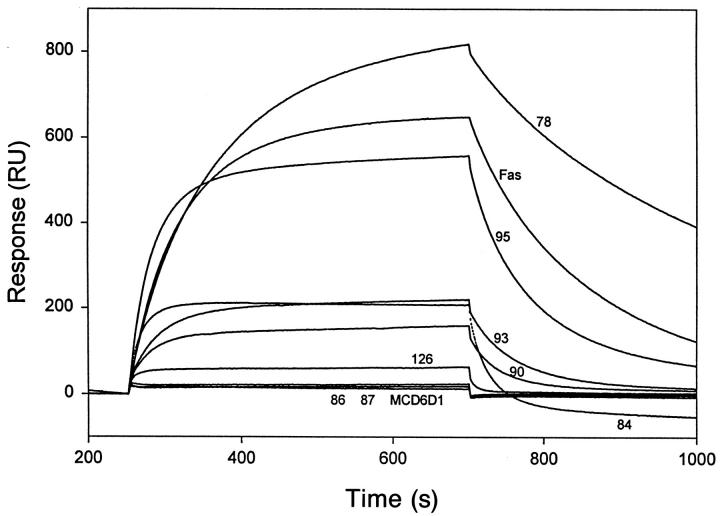
Equilibrium binding curves (Fig. 5) showed results equivalent to the ELISA. K78S saturation binding was higher than wild type due to a slower off rate from FasL. H95S binding was similar to wild type; however, the curves were steeper on each side of the equilibrium plateau, suggesting faster on and off rates. R86S and R87S showed no binding to the chip, whereas mutants K84S, L90S, E93S, and H126S showed reduced binding to the chip-bound FasL. Again, the hierarchy of the intermediate mutants was consistent between different assays, with K84S, L90S, E93S having roughly equivalent RU values, whereas binding H126S was markedly lower than the other three mutants.
Figure 5.
Equilibrium binding of FasthrRγ1 and FasthrRγ1 mutants to immobilized CD8–FasL. Fusion proteins were passed over a BIAcore™ sensor chip containing immobilized CD8–FasL protein and equilibrium binding curves obtained. Curves represented by binding of mutant proteins are labeled with the numerical position of the amino acid residue mutated to serine. The curve labeled Fas represents wild-type FasthrRγ1 binding.
Outline of the Fas Ligand-binding Site.
Mutants with deleterious effects on ligand binding were mapped with the aid of the three-dimensional Fas model (see Fig. 1). Two adjacent arginine residues (R86 and R87; colored magenta in Fig. 1) are critical for binding. Four other residues support binding (K84, L90, E93, H126; colored in gold in Fig. 1) but are not critical as mutation of these residues does not obliterate binding to ligand. In the model, these six residues form a surface that is likely to constitute a center of Fas– FasL interactions. Therefore, binding of ligand is centered on extracellular D2, but residues in D3 may support ligand binding. Mutation of residues K78 and H95 have no deleterious effects on ligand binding and have been colored in green in the model (see Fig. 1). Positions equivalent to H95 have not been implicated in the ligand binding of TNFR or CD40. Residues critical for ligand binding in Fas (R86 and R87) are adjacent to a disulfide bond, which is conserved in TNFR and CD40 (14); however, positions equivalent to only one of these residues (R87) are implicated in ligand binding for the other family members (15). Although the surface of ligand binding for TNFR, CD40, and Fas are located in a similar location, the amino acid composition of the binding surfaces differ. Different residues at equivalent positions alter the ligand binding surface and therefore determine the ligand binding specificity. At present, it is unclear whether all of the identified residues are directly involved in FasL recognition, or whether some mutations introduce local conformational changes sufficient to compromise binding to ligand but not to mAb. Additionally, other residues may be expected to contribute to the interaction.
In summary, individual residues in Fas have been identified as critical for ligand binding and an important role of charged residues in mediating Fas–FasL interactions has been demonstrated. Residues important for binding are conserved in murine and human Fas, which provides a rationale for the observed cross-species Fas–ligand interactions. However, these residues are not conserved in TNFR or CD40, thus explaining the specificity of the Fas receptor–ligand interaction. On the basis of our study, Fas D2 includes two residues that are critical for ligand binding and three residues that support ligand binding. Four of these residues (except R86) correspond to residues that contribute to CD40 (17) and TNFR ligand binding (15). This suggests that although specific residue contributions differ, equivalent regions are utilized by these receptors for mediating different ligand binding specificities.
Acknowledgments
We thank G. Whitney for mCD6D1thrRγ1, R. Peach for helpful discussions, and T. Nelson for assistance in preparation of the manuscript.
References
- 1.Nagata S, Golstein P. The Fas death factor. Science (Wash DC) 1995;267:1449–1456. doi: 10.1126/science.7533326. [DOI] [PubMed] [Google Scholar]
- 2.Watanabe-Fukunaga R, Brannan CI, Copeland NG, Jenkins NA, Nagata S. Lymphoproliferation disorder in mice explained by defects in Fas antigen that mediates apoptosis. Nature (Lond) 1992;356:314–317. doi: 10.1038/356314a0. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi T, Tanaka M, Brannan CI, Jenkins NA, Copeland NG, Suda T, Nagata S. Generalized lymphoproliferative disease in mice, caused by a point mutation in the Fas ligand. Cell. 1994;76:969–976. doi: 10.1016/0092-8674(94)90375-1. [DOI] [PubMed] [Google Scholar]
- 4.Drappa J, Vaishnaw AK, Sullivan KE, Chu J-L, Elkon KB. Fas gene mutations in the Canale–Smith syndrome, an inherited lymphoproliferative disorder associated with autoimmunity. N Engl J Med. 1996;335:1643–1649. doi: 10.1056/NEJM199611283352204. [DOI] [PubMed] [Google Scholar]
- 5.Kayagaki N, Kawasaki A, Ebata T, Ohmoto H, Ikeda S, Inoue S, Yoshino K, Okumura K, Yagita H. Metalloproteinase-mediated release of human Fas ligand. J Exp Med. 1995;182:1777–1783. doi: 10.1084/jem.182.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogasawara J, Watanabe-Fukunaga R, Adachi M, Matsuzawa A, Kasugai T, Kitamura Y, Itoh N, Suda T, Nagata S. Lethal effect of the anti-Fas antibody in mice. Nature (Lond) 1993;364:806–809. doi: 10.1038/364806a0. [DOI] [PubMed] [Google Scholar]
- 7.Galle PR, Hofmann WJ, Walczak H, Schaller H, Otto G, Stremmel W, Krammer PH, Runkel L. Involvement of the CD95 (APO-1/Fas) receptor and ligand in liver damage. J Exp Med. 1995;182:1223–1230. doi: 10.1084/jem.182.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dowling P, Shang G, Raval S, Menonna J, Cook S, Husar W. Involvement of the CD95 (APO-1/Fas) receptor/ligand system in multiple sclerosis brain. J Exp Med. 1996;184:1513–1518. doi: 10.1084/jem.184.4.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liles WC, Kiener PA, Ledbetter JA, Aruffo A, Klebanoff SJ. Differential expression of Fas (CD95) and Fas ligand on normal human phagocytes: implications for the regulation of apoptosis in neutrophils. J Exp Med. 1996;184:429–440. doi: 10.1084/jem.184.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Connell J, O'Sullivan GC, Collins JK, Shanahan F. The Fas counterattack: Fas-mediated T cell killing by colon cancer cells expressing Fas ligand. J Exp Med. 1996;184:1075–1082. doi: 10.1084/jem.184.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hahne M, Rimoldi D, Schroter M, Romero P, Schreier M, French LE, Schneider P, Bornand T, Fontana A, Lienard D, et al. Melanoma cell expression of Fas (Apo-1/CD95) ligand: implications for tumor immune escape. Science (Wash DC) 1996;274:1363–1366. doi: 10.1126/science.274.5291.1363. [DOI] [PubMed] [Google Scholar]
- 12.Nagata S. Apoptosis: telling cells their time is up. Curr Biol. 1996;6:1241–1243. doi: 10.1016/s0960-9822(02)70706-9. [DOI] [PubMed] [Google Scholar]
- 13.Huang B, Eberstadt M, Olejniczak ET, Meadows RP, Fesik SW. NMR structure and mutagenesis of the Fas (APO-1/CD95) death domain. Nature (Lond) 1996;384:638–641. doi: 10.1038/384638a0. [DOI] [PubMed] [Google Scholar]
- 14.Bajorath J, Aruffo A. Prediction of the threedimensional structure of the human Fas receptor by comparative molecular modeling. J Computer-Aided Mol Design. 1997;11:3–8. doi: 10.1023/a:1008011024584. [DOI] [PubMed] [Google Scholar]
- 15.Banner DW, D'Arcy A, Janes W, Gentz R, Schoenfeld H-J, Broger C, Loetscher H, Lesslauer W. Crystal structure of the soluble human 55 kd TNF receptor–human TNFβ complex: implications for TNF receptor activation. Cell. 1993;73:431–445. doi: 10.1016/0092-8674(93)90132-a. [DOI] [PubMed] [Google Scholar]
- 16.Bajorath J, Chalupny NJ, Marken JS, Siadak AW, Skonier J, Gordon M, Hollenbaugh D, Noelle RJ, Ochs HD, Aruffo A. Identification of residues on CD40 and its ligand which are critical for the receptor–ligand interaction. Biochemistry. 1995;34:1833–1840. doi: 10.1021/bi00006a003. [DOI] [PubMed] [Google Scholar]
- 17.Bajorath J, Marken JS, Chalupny NJ, Spoon TL, Siadak AW, Gordon M, Noelle RJ, Hollenbaugh D, Aruffo A. Analysis of gp39/CD40 interactions using molecular models and site-directed mutagenesis. Biochemistry. 1995;34:9884–9892. doi: 10.1021/bi00031a009. [DOI] [PubMed] [Google Scholar]
- 18.Hollenbaugh D, Grosmaire LS, Kullas CD, Chalupny NJ, Braesch-Andersen S, Noelle RJ, Stamenkovic I, Ledbetter JA, Aruffo A. The human T cell antigen gp39, a member of the TNF gene family, is a ligand for the CD40 receptor: expression of a soluble form of gp39 with B cell co-stimulatory activity. EMBO (Eur Mol Biol Organ) J. 1992;11:4313–4321. doi: 10.1002/j.1460-2075.1992.tb05530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malik N, Greenfield BW, Wahl AF, Kiener PA. Activation of human monocytes through CD40 induces matrix metalloproteinases. J Immunol. 1996;156:3952–3960. [PubMed] [Google Scholar]
- 20.Hollenbaugh D, Douthwright J, McDonald V, Aruffo A. Cleavable CD40Ig fusion proteins and the binding to sgp39. J Immunol Methods. 1995;188:1–7. doi: 10.1016/0022-1759(95)00194-8. [DOI] [PubMed] [Google Scholar]
- 21.Whitney GS, Starling GC, Bowen MA, Modrell B, Siadak AW, Aruffo A. The membrane-proximal scavenger receptor cysteine rich domain of CD6 contains the activated leukocyte cell adhesion molecule binding site. J Biol Chem. 1995;270:18187–18190. doi: 10.1074/jbc.270.31.18187. [DOI] [PubMed] [Google Scholar]
- 22.Marsters SA, Frutkin AD, Simpson NJ, Fendly BM, Ashkenazi A. Identification of cysteine-rich domains of the type 1 tumor necrosis factor receptor involved in ligand binding. J Biol Chem. 1992;267:5747–5750. [PubMed] [Google Scholar]
- 23.Armitage RJ, Sato TA, Macduff BM, Clifford KN, Alpert AR, Smith CA, Fanslow WC. Identification of a source of biologically active CD40 ligand. Eur J Immunol. 1992;22:2071–2076. doi: 10.1002/eji.1830220817. [DOI] [PubMed] [Google Scholar]
- 24.Alderson MR, Smith CA, Tough TW, Davis-Smith T, Armitage RJ, Falk B, Roux E, Baker E, Sutherland GR, Din WS, et al. Molecular and biological characterization of human 4-1BB and its ligand. Eur J Immunol. 1994;24:2219–2227. doi: 10.1002/eji.1830240943. [DOI] [PubMed] [Google Scholar]