Abstract
Natural killer (NK) cells in mice and humans express a number of structurally diverse receptors that inhibit target cell lysis upon recognition of major histocompatibility complex (MHC) class I molecules expressed on targets. The contribution of peptide to the structural features of class I required for NK cell inhibition appears to vary depending on the type of receptor engaged. Thus, while there is no peptide specificity in NK inhibition mediated by Ly-49A in the mouse, human histocompatibility antigen (HLA)-B*2705–specific NK clones displayed selectivity for peptides. In this report, we examine the role of peptide in the recognition of HLA-C by the defined killer cell inhibitory receptor (KIR) cl42 with established specificity for HLA-Cw4. Binding of soluble KIR cl42 molecules to HLA-Cw4 expressed on transporter associated with antigen presentation (TAP)-deficient RMA-S cells occurred only upon exogenous peptide loading. Moreover, there was peptide selectivity in that certain substitutions at positions 7 and 8 of the nonamer peptide QYDDAVYKL abolished Cw4 interaction with KIR cl42 despite similar surface expression of HLA-C. The specificity of this direct interaction between peptideloaded HLA-Cw4 on RMA-S cells and soluble KIR cl42 correlated with recognition by NK clones in that they were inhibited only by HLA-Cw4 loaded with the appropriate peptides.
NK cells lyse cellular targets such as certain tumor cells or virus-infected cells. NK-mediated lysis of normal cells is prevented by inhibitory receptors on NK cells that recognize MHC class I molecules on target cells (1, 2). MHC class I molecules consist of a trimolecular complex of a polymorphic heavy chain, β2-microglobulin (β2m), and an intracellularly derived short peptide. MHC class I recognition is mediated by a number of structurally diverse inhibitory receptors present on NK cells in mice and humans. The effect of peptide binding on the structural nature of the class I molecule recognized by the different receptors on NK cells is not well defined. Does peptide fulfill only a generic role in providing proper conformation and adequate cell surface expression of MHC molecules or do receptors on NK cells bind to specific conformational determinants on MHC class I induced only by certain peptides?
Mouse NK cells express inhibitory type II transmembrane glycoproteins called Ly-49 that bind H-2 ligands (1). Studies on the role of peptide in the recognition of H-2Dd by Ly-49A+ NK cells revealed that H-2Dd expressed on transporter associated with antigen presentation (TAP)-deficient cells confers protection from NK-mediated lysis only after loading with peptide but regardless of the sequence of the bound peptide (3, 4). Thus, there is no apparent peptide specificity for the mouse inhibitory receptor Ly-49A and the contribution of the peptide bound to H-2Dd is to ensure proper folding and reasonable cell surface expression.
Human NK cells express at least two different types of inhibitory receptors that are involved in the specific recognition of HLA-A, -B, and -C allotypes on target cells. One type belongs to the C-type lectin family and consists of the type II glycoprotein CD94 covalently associated with structurally heterogeneous members of the NKG2 receptor family (5, 6). The HLA ligands for CD94–NKG2 are not clearly established but appear to include polymorphic HLA-A, -B, and -C molecules (5, 7). There is no information on the role of peptide in the recognition of HLA ligands by the CD94–NKG2 complex. The second type of NK receptors is the p58/p70 family of receptors, called killer cell inhibitory receptors (KIR) that are type I transmembrane glycoproteins of the immunoglobulin superfamily (8–10). Whereas the p70 receptors are specific for certain HLA-A and -B alleles (11–15), members of the p58 group discriminate among defined HLA-C allotypes (allotypes with N77K80, including Cw2, Cw4, Cw5, Cw6 versus allotypes with S77N80, such as Cw1, Cw3, Cw7, Cw8) (13, 16, 17).
In contrast with Ly-49 on mouse NK cells, there is some degree of peptide selectivity in the recognition of HLAB*2705 by NK clones expressing p70 (18, 19). This peptide discrimination can be mediated by a single p70 KIR, cl11 (20). Single–amino acid substitutions along a protective peptide identified residues at positions P7 and P8 as critical for NK recognition of HLA-B*2705 (19). However, there is no data on whether these substitutions affected direct binding of the p70 KIR to HLA-B*2705. Recognition of empty HLA-C molecules expressed by TAP-deficient cells indicated that bound peptide was not required for the inhibition of CTL or NK-mediated lysis (21, 22) by HLA-C. However, the identity of the inhibitory receptor(s) (e.g., KIR, CD94–NKG2) was not established in these studies.
In this study, we investigated the role of peptide in the recognition of HLA-C by KIR cl42, a receptor previously shown to be specific for HLA-Cw4 by binding studies and by functional reconstitution experiments (13). We used a direct in vitro binding assay to show that the recognition of HLA-Cw4 by KIR cl42 is dependent on the presence of peptide in the groove. Moreover, there is peptide selectivity, because certain peptides that stabilized HLA-Cw4 surface expression on the TAP-deficient RMA-S cells did not render these molecules competent to interact with KIR cl42.
Materials and Methods
NK Clones and Target Cells.
CD3−CD56+ NK cells were obtained from PBMC, cloned by limiting dilution, and maintained as previously described (20, 23). The TAP-deficient cell line RMA-S transfected with the human β2m gene (referred to as RMA-S below) was obtained from P. Cresswell (Yale University, New Haven, CT) (24). This line was transfected with the cDNA constructs RSV.5gpt-Cw4 and RSV.5gpt-Cw8. A SalI–HpaI fragment containing either the HLA-Cw*0401 cDNA or the HLA-Cw*0802 cDNA (obtained from R. Biassoni, Istituto Nazionale per la Ricerca sul Cancro, Genova, Italy) was cloned into the SalI–HpaI sites of vector RSV.5gpt (25). Cells were electroporated at 250 V, 960 μF with 20 μg of these constructs. 48 h later, cells were selected with 10 μg/ml mycophenolic acid and 10 μg/ml xanthine and plated in V-bottomed 96-well plates at 103 cells/well. Cells were screened by flow cytometry using the mAb F4/326 and subcloned. Clones with the highest class I expression were used for the experiments described here.
Peptides, Antibodies, and Immunostaining.
Peptides were synthesized and purified as described (26). Purity was confirmed by analytical reverse phase HPLC and by mass spectrometry. QYDDAVYKL is a consensus peptide that binds Cw*0401 with an IC50 value of 18 nM (27). It contains residues present in the endogenous peptides bound to Cw*0401, as determined by pool sequencing (28). The anchor motif for peptides bound to Cw*0401 was proposed to be Y,P at position 2, V,I,L at position 6 and L,F,M at position 9 (27, 28). The following mAbs were used: F4/326 (IgG2a), a gift from S.Y. Yang (Memorial Sloan-Kettering Cancer Center, New York); HP3E4 (IgM), a gift from M. Lopez-Botet (Hospital de la Princesa, Madrid, Spain); DX22 (IgG1), a gift from L. Lanier (DNAX, Palo Alto, CA). 1 × 105 cells were incubated for 30 min on ice with primary mAb, washed in PBS containing 2% FCS, and stained with FITC-conjugated goat anti–mouse IgG+IgM (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA). Cells were washed twice and analyzed on a FACScan® (Becton Dickinson, Mountain View, CA). To test the binding of cl42–Ig (13) to RMA-S transfectants in the absence or presence of peptides, cells were incubated with 100 μg/ml of KIR cl42–Ig (or control cl43–Ig) for 30 min on ice, followed by staining with a fluoresceinated goat anti– human Fc-specific reagent as previously described (13). Cells were then washed and analyzed by flow cytometry.
Peptide Binding Assay for Stabilization of HLA-C Surface Expression.
RMA-S transfectants were plated at 2.5 × 105 cells/well in a 48-well plate in a final volume of 0.5 ml. Cells were incubated for 24 h at 25°C. Peptide (100 μM final concentration) was added at the onset of culture and again 8 h later. Cells were harvested, washed in PBS containing 2% FCS, and divided into three aliquots. One aliquot was used to measure the surface levels of class I molecules on peptide-loaded RMA-S transfectants as described above. The second aliquot was used to test for binding of KIR cl42–Ig to peptide-loaded RMA-S as described above. The third aliquot was used as targets in the cytotoxicity assay described below.
Cytotoxicity Assay.
HP3E4+, DX22+ NK clones were tested for killing at several effector to target ratios in duplicate wells. NK clones were resuspended at the desired concentration in a medium consisting of Iscove's modified essential medium with 10% FCS, 2 mM glutamine, and 50 U/ml rIL-2 (Hoffman-La Roche) and plated in V-bottomed 96-well plates in a final volume of 100 or 50 μl in the presence of mAb. Target cells were incubated with peptide as described above and labeled overnight during the incubation at 25°C with sodium 51Cr (50 μCi/well; Amersham, Arlington Heights, IL). Targets were washed and resuspended at a final concentration of 2.5 × 104 cells/ml and 100 μl were added to the NK clones. The assay was incubated at 37°C for 3 h and 51Cr release was measured as previously described (23). mAbs were added at the onset of the assay and were present throughout the assay.
Results and Discussion
Cell Surface Expression of HLA-Cw4 and HLA-Cw8 in RMA-S Cells.
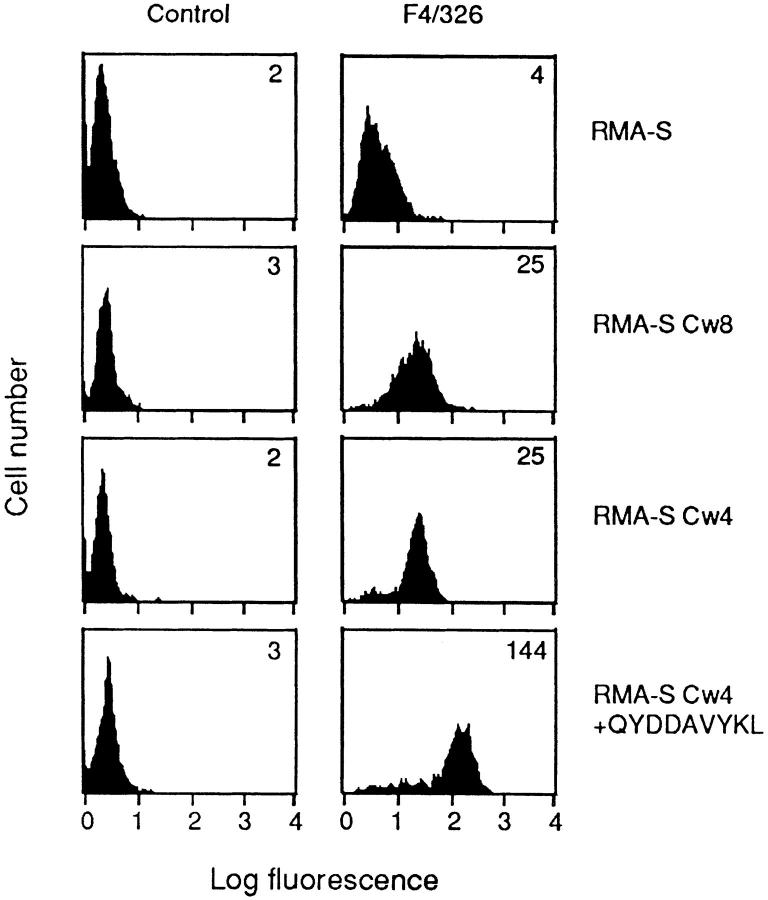
TAP-deficient RMA-S cells previously transfected with human β2m (24) (referred to in this report as RMA-S) were used for transfections with HLA-Cw*0401 or HLA-Cw*0802 cDNA. HLA-Cw*0401 belongs to the group of HLA-C allotypes with amino acids N77K80, and HLA-Cw*0802 belongs to the group with amino acids S77N80. Transfectants were incubated at 25°C to faciliate surface expression of empty HLA-C molecules, screened by flow cytometry using the HLA-C-reactive mAb F4/326, and subcloned. One clone each, expressing high cell surface levels of either HLA-Cw4 or HLA-Cw8 at 25°C, was chosen for further study (Fig. 1). RMA-S-Cw4 transfectants were incubated with the peptide QYDDAVYKL, a high affinity consensus peptide that bound Cw*0401 (27). Peptide loading resulted in a dramatic increase in cell surface expression of HLA-Cw4 as depicted by the greater than fivefold increase in fluorescence intensity. This peptide did not affect the cell surface expression of HLA-Cw8 (see Fig. 2).
Figure 1.
Surface HLA-C expression on RMA-S transfectants. RMA-S cells and RMA-S transfected with HLA-Cw4 and HLA-Cw8 were incubated at 25°C for 24 h and stained with HLA-C–reactive mAb F4/326. The peptide QYDDAVYKL was added at 100 μM to RMA-S-Cw4 (bottom). The mean fluorescence intensity for each profile is given in the top right corner.
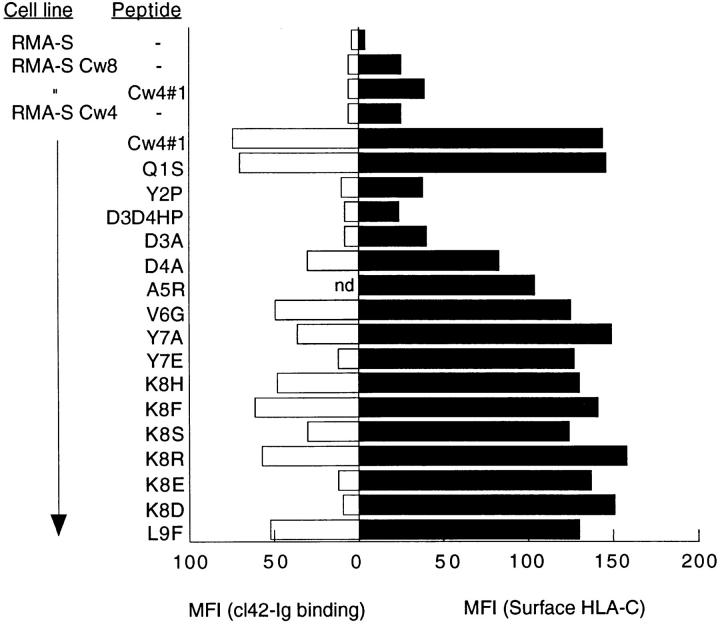
Figure 2.
Interaction of peptide-stabilized surface HLA-C with cl42– Ig. Cells were incubated with peptides (100 μM) for 24 h at 25°C and tested for induction of HLA-C surface expression (closed bars) or KIR cl42–Ig binding (open bars). The data are represented as mean fluorescence intensity. The cl42–Ig binding to peptide Cw4#1 A5R was not done in this particular experiment. In other experiments, binding was comparable to that seen with peptide Cw4#1. Three other experiments gave similar results.
A panel of 16 different amino acid substitutions in the QYDDAVYKL peptide were synthesized and tested for their capacity to bind to Cw4 molecules on the surface of RMA-S-Cw4 cells as assessed by flow cytometry using mAb F4/326 (Table 1; Fig. 2). Each residue in the peptide was substituted for other amino acids that had been identified as being either dominant or strong signals by pool sequencing (28). Additional substitutions of amino acids at P7 and P8 for negatively charged residues were chosen to test whether P7 and P8 were important for NK recognition, as had been observed in the recognition of HLAB*2705 by NK cells (19). Most of the substitutions in the peptide sequence did not affect binding to HLA-Cw4 molecules on RMA-S-Cw4, as judged by the increase in HLA-Cw4 at the cell surface. Peptides with substitutions that failed to increase the cell surface level of HLA-Cw4 included proline for tyrosine at P2. This was surprising, because proline at P2 was proposed to be an auxiliary anchor for the Cw4 binding motif (28). However, it is possible that B*3503-bound peptides contributed to the proline signal at P2, because C1R cells expressing low levels of B*3503 were used as a source of HLA-Cw4 for pool sequencing (28). Substitution of aspartic acid at P3 and P4 with histidine and proline also abolished binding. Substitutions D3A and D4A pinpointed the importance of aspartic acid at position 3 for binding.
Table 1.
Stabilization of HLA-Cw4 Molecules on RMA-S Cells
| Peptide | Sequence | Relative surface levels* | ||||
|---|---|---|---|---|---|---|
| Cw4#1 | QYDDAVYKL | 1 | ||||
| Cw4#1 | Q1S | SYDDAVYKL | 1.01 | |||
| Cw4#1 | Y2P | QPDDAVYKL | 0.11 | |||
| Cw4#1 | D3D4HP | QYHPAVYKL | 0 | |||
| Cw4#1 | D3A | QYADAVYKL | 0.13 | |||
| Cw4#1 | D4A | QYDAAVYKL | 0.49 | |||
| Cw4#1 | A5R | QYDDRVYKL | 0.75 | |||
| Cw4#1 | V6G | QYDDAGYKL | 0.84 | |||
| Cw4#1 | Y7A | QYDDAVAKL | 1.04 | |||
| Cw4#1 | Y7E | QYDDAVEKL | 0.86 | |||
| Cw4#1 | K8H | QYDDAVYHL | 0.88 | |||
| Cw4#1 | K8F | QYDDAVYFL | 0.97 | |||
| Cw4#1 | K8S | QYDDAVYSL | 0.83 | |||
| Cw4#1 | K8R | QYDDAVYRL | 1.11 | |||
| Cw4#1 | K8E | QYDDAVYEL | 0.94 | |||
| Cw4#1 | K8D | QYDDAVYDL | 1.05 | |||
| Cw4#1 | L9F | QYDDAVYKF | 0.77 | |||
| Cw4#2 | SFNCGGEFF | 1.09 |
Surface levels of HLA-Cw4 are expressed relative to that induced by peptide Cw4#1. It is calculated as follows: 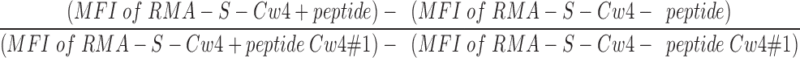 .
.
Interaction of Peptide-Stabilized Surface HLA-C with the Soluble KIR cl42–Ig.
To assess the role of peptide in the direct recognition of HLA-C by p58 KIR receptors, we examined the interaction between peptide-loaded RMA-S-Cw4 and cl42–Ig, a soluble KIR that was previously shown to bind HLA-Cw4 expressed by 721.221 cells (13). Peptideloaded RMA-S-Cw4 cells were incubated with cl42–Ig and cl43–Ig, specific for HLA-Cw3 (29), and binding was detected by flow cytometry. The first striking observation was that cl42–Ig did not bind to HLA-Cw4 molecules in the absence of peptide (Fig. 2). However, there was good binding of cl42–Ig to RMA-S-Cw4 cells loaded with the consensus peptide QYDDAVYKL. As expected, there was no binding of cl43–Ig to peptide-loaded RMA-S-Cw4 (data not shown). Most of the peptides that stabilized surface expression of HLA-Cw4 on RMA-S-Cw4 cells also permitted interaction with cl42–Ig. However, peptides Y7E, K8E, and K8D were not recognized by cl42–Ig even though they stabilized surface expression of HLA-Cw4 as well as the consensus peptide, QYDDAVYKL. This second striking result highlights the importance of residues at P7 and P8 of the Cw4-bound peptide in the interaction between KIR-cl42 and HLA-Cw4. Importantly, this result suggests that the lack of binding to empty HLA-C on RMA-S-Cw4 cells was not simply due to the lower surface expression of HLA-C on these cells but to a requirement for peptide in the recognition by KIR. The HLA-Cw4– restricted T cell epitope peptide SFNCGGEFF (30) that bound RMAS-Cw4 as well as QYDDAVYKL (Table 1) was also tested. Cl42–Ig did not bind RMA-S-Cw4 molecules loaded with this peptide (data not shown). The negatively charged E at P7 may be responsible for the lack of recognition by KIR cl42. Thus, as was seen in studies with HLA-B*2705, side chains P7 and P8 were identified as being important in the recognition of HLA-Cw4 by KIR cl42. Side chain P8 of nonamer peptides bound to HLAB*2705 contacts residues 76, 77, and 80 of the class I heavy chain (19). It is likely that side chain P8 of HLA-Cw4– bound peptides is also in proximity to these residues, two of which form the basis for the distinction among HLA-C allotypes by NK cells (17, 22, 31).
Our data show that peptide is not required merely to ensure adequate levels of surface HLA-Cw4 but that the interaction with KIR cl42 is influenced by the composition of the peptide in the groove. This contrasts the situation with Ly-49 on mouse NK cells where any peptide that stabilized class I cell surface expression induced protection from NK-mediated lysis. mAb blocking studies suggest that Ly-49A interacts with the amino-terminal portion of the α2 domain of H-2Dd, away from the peptide-binding groove (4). In contrast, the site of interaction between KIR and class I includes the carboxy-terminal half of the α1 domain such that side chains P7 and P8 in the bound peptide could influence KIR binding.
Recognition of Peptide-stabilized HLA-Cw4 by NK Clones.
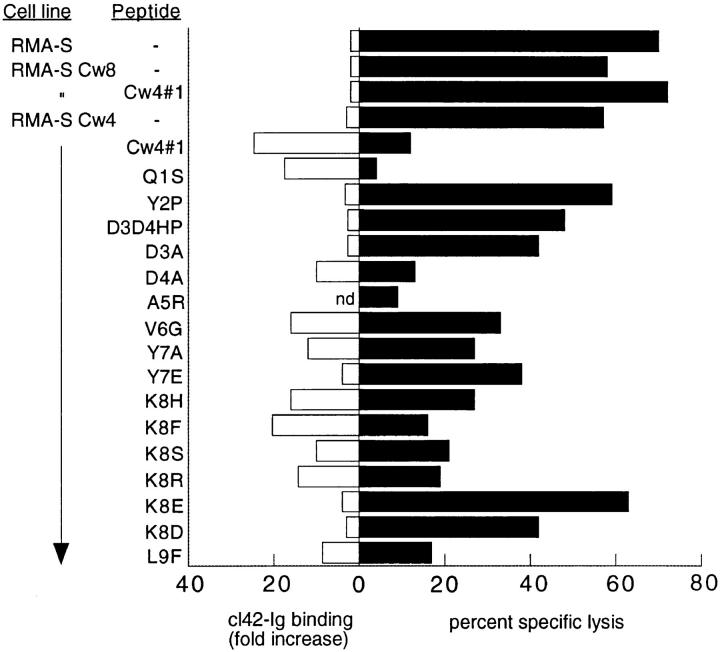
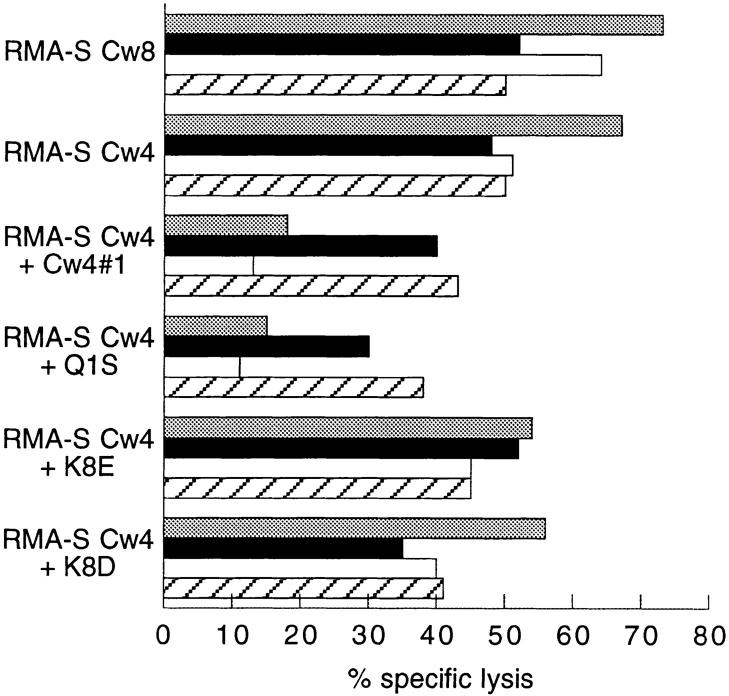
RMA-S-Cw4 cells loaded with peptide were tested in parallel for cl42-Ig binding and for inhibition of lysis by HLACw4–specific NK clones. There was a good correlation between cl42–Ig binding and the ability to induce protection from lysis by a representative peptide-sensitive HLA-Cw4– specific NK clone (Fig. 3). This peptide-dependent recognition of HLA-Cw4 by NK clones was reversible with the KIR cl42-reactive mAb HP3E4 (Fig. 4). In contrast with this reversion using anti-KIR mAb, anti-CD94 mAb DX22 had no effect, indicating that the CD94 complex in these cells did not contribute to the inhibitory effect. Thus, we show that HLA-Cw4–specific NK clones that are inhibited through the HP3E4-reactive KIR do exhibit peptide selectivity. This does not preclude the existence of other peptide-insensitive HLA-Cw4–specific NK clones that might express either singly or in combination with KIR other inhibitory receptors with potentially diverse and as yet unknown modes of interaction with class I. Indeed, such clones may have been used in previous studies in which HLA-C molecules on TAP-deficient cells provided protection from NK-mediated lysis (22).
Figure 3.
Recognition of peptide-stabilized surface HLA-C by an NK clone. RMA-S cells were incubated with peptides (100 μM) for 24 h at 25°C and tested for cl42–Ig binding (open bars) and for lysis of an HP3E4+ GL183− NK clone (closed bars). The cl42–Ig binding is expressed as fold increase of mean fluorescence intensity. The percent specific lysis is shown for an E/T ratio of 5:1. Similar results were obtained at other E/T ratios (data not shown). Two other experiments gave similar results.
Figure 4.
Effect of anti-KIR and anti-CD94 antibodies on recognition of peptide-loaded HLA-Cw4. A HP3E4+ CD94+ NK clone was assayed for the lysis of RMA-S-Cw4 loaded with the indicated peptides in the absence (shaded bars) or presence of anti-KIR (closed bars) (mAb HP3E4; 1:1,000 final dilution), anti-CD94 (open bars) (DX22; 2.5 μg/ml), or both antiKIR and anti-CD94 mAbs (hatched bars).
This study has identified a role for peptide in the specific interaction between HLA-Cw4 and the p58 inhibitory receptor cl42. The strength of this work is that recognition was determined by a direct binding assay with soluble KIR, thereby excluding any contribution by other NK receptors in the peptide-sensitive recognition of HLA-Cw4. Binding of a soluble form of KIR cl42 to HLA-Cw4 was not only dependent on the presence of a peptide, but it was also sensitive to the nature of the side chains at positions P7 and P8 of the nonamer peptide. This type of peptide-sensitive recognition of HLA class I by NK receptors was previously observed with HLA-B*2705–specific NK clones (18, 19). However, it is not known whether nonprotective B*2705bound peptides prevent the binding of p70 KIR. Both positively and negatively charged residues at P8 interfered with recognition of HLA-B*2705. In the case of HLA-Cw4, negatively charged residues at P8 were incompatible with binding of the p58 KIR cl42. These data establish that the sensitivity to peptide side chains P7 and P8 is a general feature shared by p58 and p70 KIR in their interaction with HLA class I.
Acknowledgments
We thank C. Winter for KIR–Ig fusion proteins; M. Weston for technical assistance; L. Lanier, M. LopezBotet, and S.Y. Yang for antibodies; P. Cresswell for transfected cells; R. Biassoni for plasmids; J. Coligan for peptides, and Hoffmann-La Roche for rIL-2.
References
- 1.Raulet DH, Held W. Natural killer cell receptors: the offs and ons of NK cell recognition. Cell. 1995;82:697–700. doi: 10.1016/0092-8674(95)90466-2. [DOI] [PubMed] [Google Scholar]
- 2.Gumperz JE, Parham P. The enigma of the natural killer cell. Nature (Lond) 1995;378:245–248. doi: 10.1038/378245a0. [DOI] [PubMed] [Google Scholar]
- 3.Correa I, Raulet DH. Binding of diverse peptides to MHC class I molecules inhibits target cell lysis by activated natural killer cells. Immunity. 1995;2:61–71. doi: 10.1016/1074-7613(95)90079-9. [DOI] [PubMed] [Google Scholar]
- 4.Orihuela M, Margulies DH, Yokoyama WM. The natural killer cell receptor Ly-49A recognizes a peptideinduced conformational determinant on its major histocompatibility complex class I ligand. Proc Natl Acad Sci USA. 1996;93:11792–11797. doi: 10.1073/pnas.93.21.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phillips JH, Chang CW, Mattson J, Gumperz JE, Parham P, Lanier LL. CD94 and a novel associated protein (94AP) form a NK cell receptor involved in the recognition of HLA-A, HLA-B, and HLA-C allotypes. Immunity. 1996;5:163–172. doi: 10.1016/s1074-7613(00)80492-6. [DOI] [PubMed] [Google Scholar]
- 6.Lazetic S, Chang C, Houchins JP, Lanier LL, Phillips JH. Human natural killer cell receptors involved in MHC class I recognition are disulfide-linked heterodimers of CD94 and NKG2 subunits. J Immunol. 1996;157:4741–4745. [PubMed] [Google Scholar]
- 7.Sivori S, Vitale M, Bottino C, Marcenaro E, Sanseverino L, Parolini S, Moretta L, Moretta A. CD94 functions as a natural killer cell inhibitory receptor for different HLA class I alleles: identification of the inhibitory form of CD94 by the use of novel monoclonal antibodies. Eur J Immunol. 1996;26:2487–2492. doi: 10.1002/eji.1830261032. [DOI] [PubMed] [Google Scholar]
- 8.Wagtmann N, Biassoni R, Cantoni C, Verdiani S, Malnati M, Vitale M, Bottino C, Moretta L, Moretta A, Long EO. Molecular clones of the p58 natural killer cell receptor reveal Ig-related molecules with diversity in both the extra- and intracellular domains. Immunity. 1995;2:439–449. doi: 10.1016/1074-7613(95)90025-x. [DOI] [PubMed] [Google Scholar]
- 9.Colonna M, Samaridis J. Cloning of immunoglobulin-superfamily members associated with HLA-C and HLA-B recognition by human natural killer cells. Science (Wash DC) 1995;268:405–408. doi: 10.1126/science.7716543. [DOI] [PubMed] [Google Scholar]
- 10.D'Andrea A, Chang C, Franz-Bacon K, McClanahan T, Phillips JH, Lanier LL. Molecular cloning of NKB1: a natural killer cell receptor for HLA-B allotypes. J Immunol. 1995;155:2306–2310. [PubMed] [Google Scholar]
- 11.Litwin V, Gumperz J, Parham P, Phillips JH, Lanier LL. NKB1: a natural killer cell receptor involved in the recognition of polymorphic HLA-B molecules. J Exp Med. 1994;180:537–543. doi: 10.1084/jem.180.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gumperz JE, Litwin V, Phillips JH, Lanier LL, Parham P. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J Exp Med. 1995;181:1133–1144. doi: 10.1084/jem.181.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagtmann N, Rajagopalan S, Winter CC, Peruzzi M, Long EO. Killer cell inhibitory receptors specific for HLA-C and HLA-B identified by direct binding and by functional transfer. Immunity. 1995;3:801–809. doi: 10.1016/1074-7613(95)90069-1. [DOI] [PubMed] [Google Scholar]
- 14.Dohring C, Scheidegger D, Samaridis J, Cella M, Colonna M. A human killer inhibitory receptor specific for HLA-A. J Immunol. 1996;156:3098–3101. [PubMed] [Google Scholar]
- 15.Pende D, Biassoni R, Cantoni C, Verdiani S, Falco M, Di Donato C, Accame L, Bottino C, Moretta A, Moretta L. The natural killer cell receptor specific for HLA-A allotypes: a novel member of the p58/p70 family of inhibitory receptors that is characterized by three immunoglobulin-like domains and is expressed as a 140-kD disulphide-linked dimer. J Exp Med. 1996;184:505–518. doi: 10.1084/jem.184.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moretta A, Vitale M, Bottino C, Orengo AM, Morelli L, Augugliaro R, Barbaresi M, Ciccone E, Moretta L. P58 molecules as putative receptors for major histocompatibility complex (MHC) class I molecules in human natural killer (NK) cells. Anti-p58 antibodies reconstitute lysis of MHC class I–protected cells in NK clones displaying different specificities. J Exp Med. 1993;178:597–604. doi: 10.1084/jem.178.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colonna M, Borsellino G, Falco M, Ferrara GB, Strominger JL. HLA-C is the inhibitory ligand that determines dominant resistance to lysis by NK1- and NK2-specific natural killer cells. Proc Natl Acad Sci USA. 1993;90:12000–12004. doi: 10.1073/pnas.90.24.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malnati MS, Peruzzi M, Parker KC, Biddison WE, Ciccone E, Moretta A, Long EO. Peptide specificity in the recognition of MHC class I by natural killer cell clones. Science (Wash DC) 1995;267:1016–1018. doi: 10.1126/science.7863326. [DOI] [PubMed] [Google Scholar]
- 19.Peruzzi M, Parker KC, Long EO, Malnati MS. Peptide sequence requirements for the recognition of HLA-B*2705 by specific natural killer cells. J Immunol. 1996;157:3350–3356. [PubMed] [Google Scholar]
- 20.Peruzzi M, Wagtmann N, Long EO. A p70 killer cell inhibitory receptor specific for several HLA-B allotypes discriminates among peptides bound to HLA-B*2705. J Exp Med. 1996;184:1585–1590. doi: 10.1084/jem.184.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falk CS, Steinle A, Schendel DJ. Expression of HLA-C molecules confers target cell resistance to some nonmajor histocompatibility complex–restricted T cells in a manner analogous to allospecific natural killer cells. J Exp Med. 1995;182:1005–1018. doi: 10.1084/jem.182.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mandelboim O, Reyburn HT, Valés-Gómez M, Pazmany L, Colonna M, Borsellino G, Strominger JL. Protection from lysis by natural killer cells of group 1 and 2 specificity is mediated by residue 80 in human histocompatibility leukocyte antigen C alleles and also occurs with empty major histocompatibility complex molecules. J Exp Med. 1996;184:913–922. doi: 10.1084/jem.184.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malnati MS, Lusso P, Ciccone E, Moretta A, Moretta L, Long EO. Recognition of virus-infected cells by natural killer cell clones is controlled by polymorphic target cell elements. J Exp Med. 1993;178:961–969. doi: 10.1084/jem.178.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson KS, Alexander J, Wei M, Cresswell P. Intracellular transport of class I MHC molecules in antigen processing mutant cell lines. J Immunol. 1993;151:3407–3419. [PubMed] [Google Scholar]
- 25.Long EO, Rosen-Bronson S, Karp DR, Malnati M, Sekaly RP, Jaraquemada D. Efficient cDNA expression vectors for stable and transient expression of HLADR in transfected fibroblast and lymphoid cells. Hum Immunol. 1991;31:229–235. doi: 10.1016/0198-8859(91)90092-n. [DOI] [PubMed] [Google Scholar]
- 26.Parker KC, Carreno BM, Sestak L, Utz U, Biddison WE, Coligan JE. Peptide binding to HLA-A2 and HLA-B27 isolated from Escherichia coli. Reconstitution of HLA-A2 and HLA-B27 heavy chain/beta 2-microglobulin complexes requires specific peptides. J Biol Chem. 1992;267:5451–5459. [PubMed] [Google Scholar]
- 27.Sidney J, del Guercio MF, Southwood S, Engelhard VH, Appella E, Rammensee H-G, Falk K, Rotzschke O, Takiguchi M, Kubo RT, et al. Several HLA alleles share overlapping peptide specificities. J Immunol. 1995;154:247–259. [PubMed] [Google Scholar]
- 28.Falk K, Rotzschke O, Grahovac B, Schendel D, Stevanovic S, Gnau V, Jung G, Strominger JL, Rammensee H-G. Allele-specific peptide ligand motifs of HLA-C molecules. Proc Natl Acad Sci USA. 1993;90:12005–12009. doi: 10.1073/pnas.90.24.12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan QR, Garboczi DN, Winter CC, Wagtmann N, Long EO, Wiley DC. Direct binding of a soluble natural killer cell inhibitory receptor to a soluble human leukocyte antigen-Cw4 class I major histocompatibility complex molecule. Proc Natl Acad Sci USA. 1996;93:7178–7183. doi: 10.1073/pnas.93.14.7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson RP, Trocha A, Buchanan TM, Walker BD. Recognition of a highly conserved region of human immunodeficiency virus type 1 gp120 by an HLA-Cw4– restricted cytotoxic T-lymphocyte clone. J Virol. 1993;67:438–445. doi: 10.1128/jvi.67.1.438-445.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biassoni R, Falco M, Cambiaggi A, Costa P, Verdiani S, Pende D, Conte R, Di Donato C, Parham P, Moretta L. Amino acid substitutions can influence the natural killer (NK)–mediated recognition of HLA-C molecules. Role of serine-77 and lysine-80 in the target cell protection from lysis mediated by “group 2” or “group 1” NK clones. J Exp Med. 1995;182:605–609. doi: 10.1084/jem.182.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]