Abstract
A cDNA encoding a novel human chemokine was isolated by random sequencing of cDNA clones from human monocyte-derived macrophages. This protein has been termed macrophagederived chemokine (MDC) because it appears to be synthesized specifically by cells of the macrophage lineage. MDC has the four-cysteine motif and other highly conserved residues characteristic of CC chemokines, but it shares <35% identity with any of the known chemokines. Recombinant MDC was expressed in Chinese hamster ovary cells and purified by heparin– Sepharose chromatography. NH2-terminal sequencing and mass spectrophotometry were used to verify the NH2 terminus and molecular mass of recombinant MDC (8,081 dalton). In microchamber migration assays, monocyte-derived dendritic cells and IL-2–activated natural killer cells migrated to MDC in a dose-dependent manner, with a maximal chemotactic response at 1 ng/ml. Freshly isolated monocytes also migrated toward MDC, but with a peak response at 100 ng/ml MDC. Northern analyses indicated MDC is highly expressed in macrophages and in monocyte-derived dendritic cells, but not in monocytes, natural killer cells, or several cell lines of epithelial, endothelial, or fibroblast origin. High expression was also detected in normal thymus and less expression in lung and spleen. Unlike most other CC chemokines, MDC is encoded on human chromosome 16. MDC is thus a unique member of the CC chemokine family that may play a fundamental role in the function of dendritic cells, natural killer cells, and monocytes.
Chemokines comprise a family of secreted proteins that attract and activate a variety of cell types, generally augmenting the immune response (reviewed in references 1–3). Chemokines have been classified into two subfamilies based on the relative positions of the first two of four conserved cysteine residues. In the CC subfamily, the first two cysteines are adjacent, whereas in the CXC subfamily, they are separated by one amino acid. The CC chemokines usually act on monocytes, T lymphocytes, and in some cases, eosinophils, basophils, or mast cells. In contrast, the CXC chemokines generally act upon neutrophils. Further, the chemokines macrophage inflammatory protein (MIP)1-1α (4), stromal-derived factor (5, 6), and Exodus (7) have been implicated in the regulation of hematopoiesis.
The repertoire of known human CC chemokines is expanding rapidly and now includes MIP-1α, MIP-1β, RANTES, I-309, monocyte chemotactic proteins 1, 2, and 3 (MCP-1, -2, -3; 8–13), and the recently described chemokines MCP-4 (14–16), eotaxin (17, 18), HCC-1 (19), thymus and activation regulated chemokine (TARC) (20), and Exodus (7). These proteins are 70–100 amino acids in length and have 25–70% identity with each other. Chemokines act through G protein–coupled receptors, which have a characteristic seven-transmembrane structure. Five CC chemokine receptors have been described: CCR-1 (21, 22) binds MIP1α, RANTES, and MCP-3 (23, 24); CCR-2 binds MCP-1 (25), MCP-3 (26), and MCP-4 (14–16); CCR-3 (18, 27) binds eotaxin, MCP-3, RANTES, and MCP-4 (15); CCR-4 (28) binds MIP-1α, RANTES, and MCP-1; and CCR-5 (29, 30) binds MIP-1α, MIP-1β, and RANTES. In addition, macrophage-tropic strains of HIV appear to require one of these receptors, primarily CCR-5, as a cofactor for infection (31–35).
The present study describes the cloning and characterization of a novel human CC chemokine, macrophage-derived chemokine (MDC). MDC is not closely related to the other chemokines and has not been previously identified in the publicly available databases. MDC is produced by macrophages and dendritic cells, and it is chemotactic for monocytes, monocyte-derived dendritic cells, and IL-2–activated natural killer cells.
Materials and Methods
Isolation of a cDNA-encoding MDC.
RNA Stat-60 (Tel-Test, Inc., Friendswood, TX) was used to isolate poly (A+) RNA from monocyte-derived macrophages from a normal human donor. cDNA generated from this RNA (Invitrogen Copy Kit; Invitrogen, San Diego, CA) was inserted into the mammalian expression vector pRc/CMV (Invitrogen) and used to transform Escherichia coli XL1-blue bacteria (Stratagene Corp., La Jolla, CA) (36). Plasmid DNA was isolated from randomly chosen individual transformants (Wizard miniprep purification system; Promega Corp., Madison, WI). Approximately 300–500 bp of each end of the plasmid inserts were sequenced on an automated sequencer (model 373; Applied Biosystems, Foster City, CA) and compared to the GenBank “nr” database using the BLAST (37) program (National Center for Biotechnology Information, Bethesda, MD, E-mail: Blast@ncbi.nlm.nih.gov). Among several hundred clones sequenced, one clone, designated pMP390, was identified as a unique sequence that contained significant homology to CC chemokines. To facilitate complete sequencing, the 2.9-kb insert of pMP390 was subcloned into the vector pBluescript SK− (Stratagene) and subjected to nested deletion (Erase-a-Base System; Promega Corp.).
Additional clones of MDC were isolated by probing the macrophage cDNA library with the insert of pMP390, which was purified by agarose gel electrophoresis and labeled (Random Primed DNA Labeling Kit; Boehringer Mannheim, Indianapolis, IN). Filters were hybridized according to standard protocols and stringently washed in 0.2× SSC and 0.2% SDS at 55°C, as described previously (38). Autoradiographs were exposed overnight at −80°C on Kodak XAR-5 film (Rochester, NY) with intensifying screens (Amersham, Arlington Heights, IL).
Production of Recombinant MDC.
PCR was used to amplify a fragment containing the entire coding region of the MDC cDNA clone (Fig. 1, bases 1–403), using the primers 5′-GACCAA GCTTGAGACATACAGGACAGAGCA and 5′-TGGATCTA GAAGTTGGCACAGGCTTCTGG. Restriction sites added to the primers are underlined. The PCR mix contained 0.2 μg of pMP390 plasmid DNA, 1.5 mM MgCl2, 50 mM KCl, 10 mM Tris, pH 8.4, 0.2 mM each dNTP, 10 μg/ml each primer, and 0.5 μl Taq polymerase (5 U/μl) (Boehringer Mannheim). The reactions were incubated for 4 min at 94°C, followed by 30 cycles of denaturation for 15 s at 94°C, annealing for 15 s at 60°C, and extension for 30 s at 72°C. The PCR fragment was cloned into the expression vector pDC1, a derivative of pRc/CMV in which the neomycin phosphotransferase gene had been replaced by the mouse dihydrofolate reductase gene from the vector pSV2-dhfr (vector #37146; American Type Culture Collection, Rockville, MD). The pDC1/MDC plasmid was linearized by restriction digestion within the vector sequence and electroporated into the Chinese hamster ovary (CHO) cell line DG44, which lacks the dihydrofolate reductase gene (39). Cells were electroporated in a 0.4-cm cuvette using a gene pulser (Bio Rad Labs., Hercules, CA) at 290 volts, 960 μF. Transfectants were selected by growth in α-medium (Catalogue No. 12000; GIBCO BRL, Gaithersburg, MD) plus 10% dialyzed FCS (Hyclone Labs., Logan, UT) in the absence of hypoxanthine and thymidine. Cells from several hundred transfected colonies were pooled and replated in α-medium containing 20 nM methotrexate (Sigma Chemical Co., St. Louis, MO). Colonies surviving this round of selection were isolated and expanded in α-medium containing 0.2–1.0% dialyzed FCS. MDC was isolated from the culture medium by passage over heparin–Sepharose CL-6B (Pharmacia, Uppsala, Sweden). The column was washed with 0.2 M NaCl in 20 mM Tris, pH 8, and eluted with 0.6 M NaCl in 20 mM Tris, pH 8. The eluted material was fractionated on an 18% acrylamide SDSPAGE gel (Novex, San Diego, CA) and electroblotted to polyvinylidene fluoride membrane (Millipore Corp., Bedford, MA). The band corresponding to MDC was sequenced on an automated sequencer (model 473A; Applied Biosystems).
Figure 1.

Sequence and deduced translation of the MDC cDNA. The leader sequence is in italics; the mature protein is in bold type. The mature NH2 terminus was confirmed by amino acid sequencing of recombinant MDC produced in CHO cells (GPYGANMEDSV . . .). Alu repeats (68) are present in the following positions: bases 1204–1482, 1530–1805, and 2335–2443. The cDNA sequence of MDC has been deposited in EMBL/GenBank/DDBJ under the accession number U83171.
The mature form of MDC protein (Fig. 1) was chemically synthesized by Gryphon Sciences (San Francisco, CA) using t-butyloxycarbonyl chemistries on a peptide synthesizer (430A; Applied Biosystems). Lyophilized protein was dissolved at 10 mg/ml in 4 mM HCl and immediately diluted to 0.1 mg/ml in PBS plus 0.1% BSA for storage at −80°C.
Production of MDC Antibodies.
A PCR fragment encoding a thrombin cleavage site and the mature form of MDC was inserted into the vector pGEX-3X (Pharmacia). XL-1 blue bacteria (Stratagene) were transformed with the resulting plasmid to generate a GST–MDC fusion protein. The protein was isolated from inclusion bodies, digested with thrombin (Sigma Chemical Co.), and fractionated by preparative SDS-PAGE (Tris glycine, 18% acrylamide). A gel slice containing the MDC fragment was excised and emulsified with adjuvant for immunization of rabbits for polyclonal sera or mice for monoclonal sera. mAbs were obtained from fusions of mouse spleen cells with NS-1 myeloma cells, according to standard protocols (40).
Cell Culture.
The human cell lines A549 (lung epithelial; CCL185; American Type Culture Collection), T84 (colon epithelial; CCL-248; American Type Culture Collection), and IMR-90 (lung fibroblast; CCL-186; American Type Culture Collection) were obtained. Cells were cultured in DME (GIBCO BRL) supplemented with 10% FCS (Hyclone Labs.), 25 mM Hepes, penicillin, and streptomycin. Immortalized human umbilical vein endothelial cells (I-HUVECs) were obtained (Dr. Jay Nelson, University of Oregon, Eugene, OR) and cultured in RPMI (GIBCO BRL) supplemented with 10% FCS (Hyclone Labs.), 400 μg/ml G418 (GIBCO BRL), 1 U/ml heparin (Sigma Chemical Co.), and 30 μg/ml endothelial cell growth factor (Collaborative Biomedical Products, Bedford, MA). A549 and IMR-90 cells were grown to 70–80% confluency and cultured in the presence or absence of 10 ng/ml TNF-α (R & D Syst. Inc., Minneapolis, MN) for 6 h. T84 cells were treated for 1 d with TNF-α (5 ng/ml), TGF-β (1 ng/ml; R & D Syst. Inc.), or interferon-γ (200 U/ml; PeproTech, Rocky Hill, NJ). For Northern blotting, monocytes were isolated from PBMC using histopaque gradients (Sigma Chemical Co.) and adherence to tissue culture plastic. Cells were cultured for 6 d to allow differentiation into macrophages (41).
PBMC for chemotaxis assays were obtained from buffy coats of healthy donors. Blood was washed once with PBS to remove plasma and platelets and centrifuged on Ficoll (Biochrom, Berlin, FRG) at 600 g at room temperature. PBMC were collected from the interface, washed twice with PBS, and resuspended in RPMI 1640 medium (Biochrom) with 1% FCS (Hyclone Labs.).
Dendritic cells were obtained from PBMC as previously reported (42), according to the procedure described by Sallusto et al. (43). In brief, purified monocytes were obtained from buffy coats by centrifugation through Ficoll and Percoll gradients. Cells were further purified by negative selection with a cocktail of antiCD19 and anti-CD2 magnetic beads. Cells were cultured in RPMI (Biochrom) containing 10% FCS (Hyclone Labs.), supplemented with 50 ng/ml GM-CSF and 20 ng/ml IL-13. They were normally used between days 6 and 8 of culture. These cells were >80% CD1a+, >90% MHC class II+, <10% CD14+, <2% CD3+, and <4% CD20+. For some experiments, cells were further depleted of CD14+ cells (<1% CD14+) by anti-CD14–coated Dynabeads (Unpath, Milan, Italy).
Natural killer cells were obtained from monocyte-depleted PBMC by centrifugation through discontinuous Percoll gradients (47, 49, and 52%), as previously described (44). Low density cells were further depleted of contaminating T lymphocytes by panning with anti-CD6 mAb (50 ng/ml). Contaminating monocytes were eliminated by incubation with anti-CD14–coated Dynabeads (Unpath). Purified preparations of natural killer cells (106/ml) were cultured with an irradiated lymphoblastoid cell line (105/ml) (44) in the presence of IL-2 (250 U/ml; Eurocetus, Milan, Italy). Proliferating cells had the phenotype of natural killer cells and were CD14− and CD3−.
Migration Assay.
Cell migration was evaluated using a chemotaxis microchamber technique, as previously described (42). 27μl aliquots of chemoattractant solution or control medium (RPMI 1640 with 1% FCS) were added to the lower wells of a chemotaxis chamber (NeuroProbe, Captain John, MD). A polycarbonate filter (5 μm pore size; NeuroProbe) was layered onto the wells and covered with a silicon gasket and top plate. For migration of natural killer cells, the filters were coated with 200 μg/ ml gelatin and 10 μg/ml fibronectin. 50 μl of cell suspension (0.7–1.5 × 106/ml) were seeded in the upper chamber. The chamber was incubated at 37°C in a humidified chamber in the presence of 5% CO2 for 1.5 to 2 h. After incubation, filters were removed and stained with Diff-Quik (Baxter s.p.a., Rome, Italy). Five high power oil-immersion fields (100×) were counted. Results are expressed as the mean number of migrated cells. Each experiment was performed in triplicate.
Northern Blotting.
The probe for Northern hybridizations was generated by PCR amplification of bases 102–461 of the MDC cDNA sequence using the primers 5′-TCTATCTAGAGGCCCCTACGGCGCCAACATGGAAG and 5′-AATGGATCCACAGCACGGAGGTGACCAAG. Restriction sites added to the primers are underlined. The fragment was gel purified and labeled with [32P]CTP and [32P]TTP (DuPont New England Nuclear, Boston, MA) by random priming (Boehringer Mannheim). RNA was isolated from cell lines and cultured macrophages using RNA Stat-60 (Tel-Test, Inc.). Total RNA (20 μg) was fractionated on 0.8% agarose formaldehyde gels, transferred to nitrocellulose, hybridized, and washed under stringent conditions as described (38). Autoradiographs were exposed at −80°C with intensifying screens for 3 h (glyceraldehyde phosphate dehydrogenase [GAPDH] and MDC) or 8 h (MCP-1). Multiple tissue Northern blots (Clontech, Palo Alto, CA) were probed and washed under stringent conditions according to the manufacturer's recommendations. Films were exposed at −80°C with intensifying screens for 16 h.
Chromosome Localization.
Hybrid mouse or hamster cell lines containing a single human chromosome (Coriell Institute, Camden, NJ) were screened by PCR for the presence of the MDC gene. Reaction conditions were as described above, using 0.5 μg of genomic DNA from each cell line as templates with the primers 5′-TATTGGATCCGTTCTAGCTCCCTGTTCTCC and 5′-CCAAGAATTCCTGCAGCCACTTTCTGGGCTC, which amplify bases 676–1,042 of the MDC cDNA sequence (Fig. 1). The identity of the PCR product from the positive cell line was verified by hybridization to an internal oligonucleotide complementary to bases 246–266 of the MDC cDNA.
Computer Analysis.
Protein comparisons (Fig. 2) were performed with the GeneWorks program (IntelliGenetics, Mountain View, CA).
Figure 2.
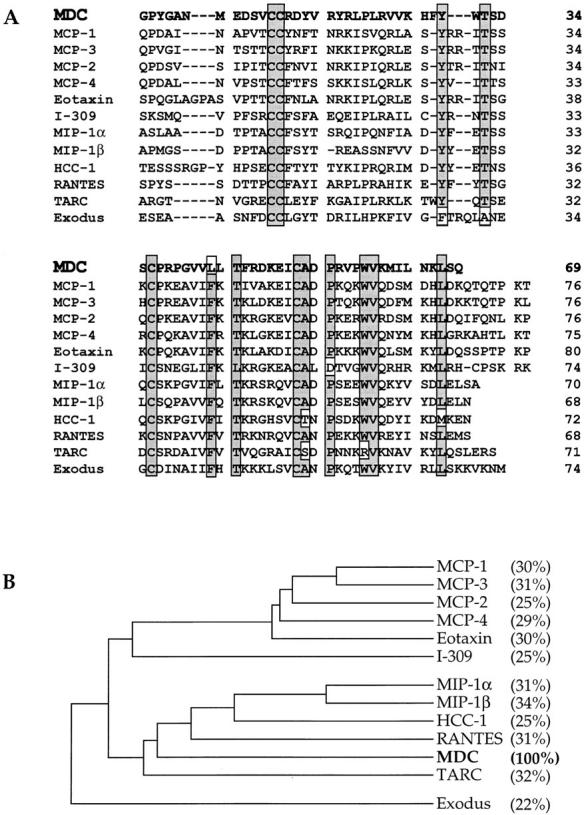
Comparison of the mature form of human MDC with the known human CC chemokines. (A) Direct comparison of amino acid sequences. Residues conserved in at least 10 of the 12 sequences are shaded; the exceptions are boxed and unshaded. Dashes are inserted to optimize alignment of the sequences. (B) Dendrogram analysis illustrating the similarity of the mature forms of the CC chemokines. Percent identity with MDC is indicated in parentheses.
Results
Random clones from a human macrophage cDNA library were partially sequenced and electronically compared to the GenBank Non-Redundant (nr) database of sequences (our manuscript in preparation). One clone contained a sequence that encoded a peptide with >30% identity to portions of RANTES, MIP-1α, and MIP-1β. This fragment was used to screen the same macrophage library for additional clones. The sequence of the full-length cDNA obtained is presented in Fig. 1. It contains an open reading frame of 93 amino acids that encodes a novel CC chemokine, designated MDC. The cDNA is considerably longer than that of other chemokines, and it includes three Alu repeats in the 3′ noncoding region.
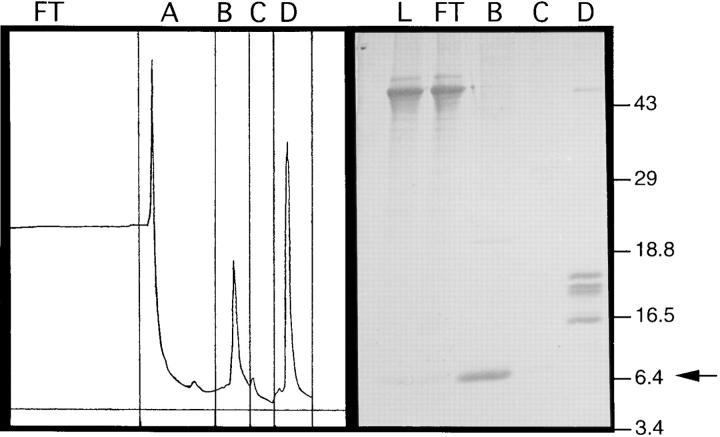
MDC has 28–34% amino acid identity with other CC chemokines. It contains the characteristic four-cysteine motif in addition to nine other residues that are very highly conserved in this family (Fig. 2). Comparison with other chemokines also suggests the first 24 amino acids of MDC constitute a leader sequence, which is consistent with von Heijne's rules governing signal cleavage (45). To confirm the predicted NH2 terminus of the mature peptide, the MDC cDNA was cloned into an expression vector for stable transfection of CHO cells. Protein was purified from culture supernatants by heparin–Sepharose chromatography and gel fractionation (Fig. 3). NH2-terminal sequencing of the band migrating at the expected position of MDC yielded the sequence GPYGANMEDSV, confirming the predicted NH2 terminus of the mature protein. Amino acid analysis and mass spectrophotometry of the mature protein were consistent with the predicted amino acid composition and molecular weight (8,081 dalton) (data not shown).
Figure 3.
Purification of recombinant MDC from CHO cells. A PCR fragment containing bases 1–403 of the MDC cDNA was subcloned into a mammalian expression vector driven by the CMV promoter and transfected into CHO cells. Culture supernatant from an individual transfected clone was harvested 4 d after cells reached confluency. MDC in the media was concentrated and purified by binding to a heparin-Sepharose column in 0.35 M NaCl. The left panel shows the column chromatogram. Fractions collected were flowthrough (FT), 0.35 M NaCl wash (A), 0.7 M NaCl elution (B), repeat of 0.7 M NaCl elution (C), and 1.5 M NaCl elution (D). The right panel shows SDS-PAGE analysis (18% Tris glycine) of the supernatant loaded onto the column (L) and fractions FT, B, C, and D. MDC eluted in fraction B (arrow). The gel was transferred to polyvinylidene difluoride membrane, and the MDC band was excised for NH2-terminal sequencing.
MDC Gene Expression in Human Cells.
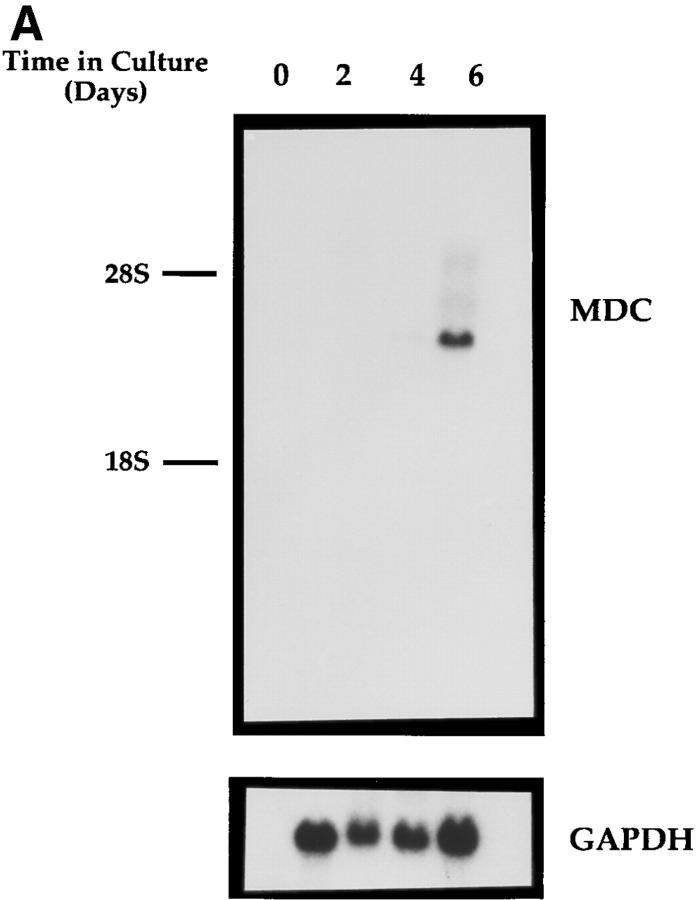
Because the MDC clone was isolated from a human macrophage cDNA library, its expression during differentiation of monocytes into macrophages was examined. Human monocytes from a single donor were cultured on a series of tissue culture plates, and cells from individual plates were harvested after 0, 2, 4, or 6 d. Under these conditions, monocytes differentiate into macrophages by day 6 (36, 41). A Northern blot of RNA from the cells harvested at each time point was probed with the MDC cDNA. No signal was detectable in RNA from freshly isolated monocytes, whereas a very strong signal was generated from cells that had differentiated into macrophages after 6 d of culture (Fig. 4 A).
Figure 4.
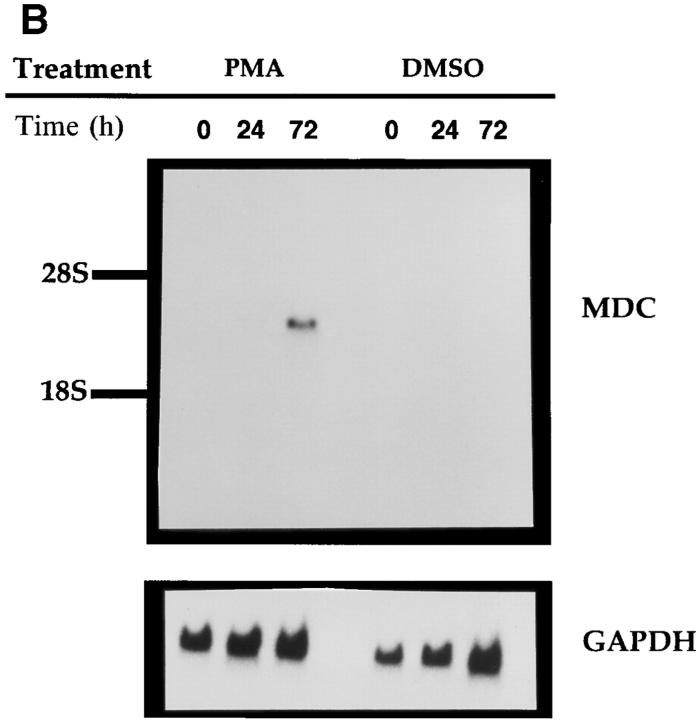
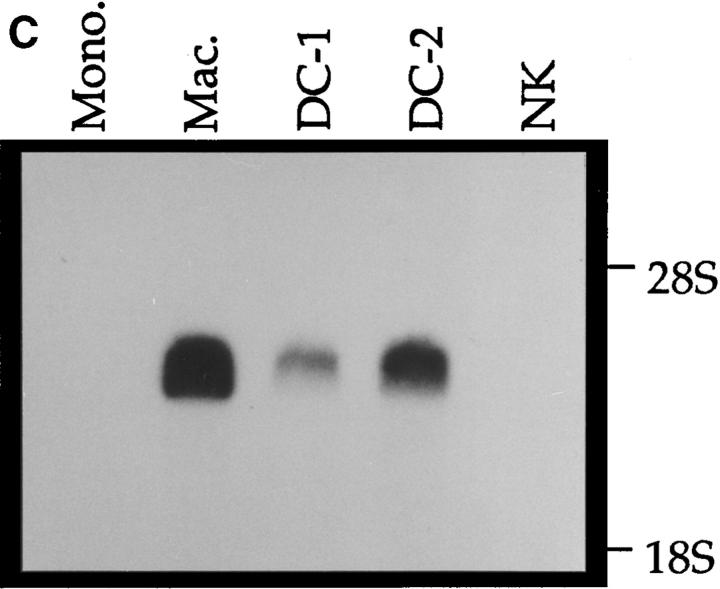
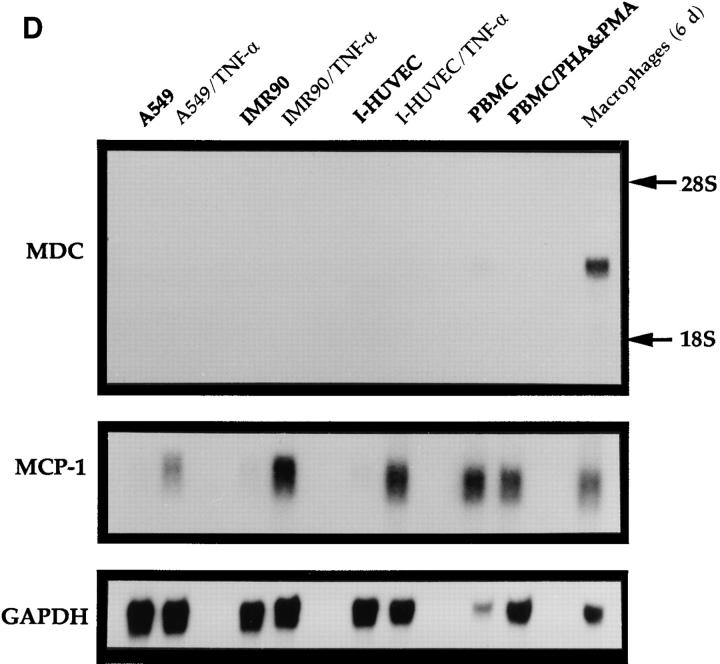
Northern analysis of MDC mRNA expression in human cells. Migration of MDC, 28S RNA, and 18S RNA is indicated. (A) Peripheral blood monocytes were allowed to differentiate into macrophages by incubation on plastic tissue culture dishes for 6 d (41). Total RNA was isolated from individual dishes on the indicated days, fractionated through a formaldehyde agarose gel, blotted, and probed with the radiolabeled MDC cDNA. The blot was subsequently probed for GAPDH message. (B) Expression of MDC in differentiated HL-60 cells. Cells were treated with PMA to induce differentiation to monocytic cells or with DMSO to induce granulocytic cells. MDC expression was analyzed by Northern blotting as in A. (C) Northern blot of MDC expression in human peripheral blood monocytes (Mono.), macrophages derived from these monocytes (Mac.), dendritic cells derived from PBMC of two different donors (DC-1 and DC-2), and natural killer cells derived from PBMC (NK). (D) Expression of MDC in cell cultures. Lung epithelial cell line A549, lung fibroblast line IMR90, and I-HUVEC without and with TNF-α stimulation; PBMC without and with stimulation by PHA and PMA; and monocyte derived macrophages after 6 d in culture. After probing with MDC, the filter was stripped and probed sequentially with MCP-1 and GAPDH.
MDC gene expression was examined further by treating the human cell line HL60 with either 1% DMSO or 50 ng/ml PMA. Treatment with DMSO induces differentiation of HL60 cells into a granulocytic cell type, whereas PMA induces their differentiation toward the macrophage lineage (46). After 3 d of PMA treatment, the macrophagelike cells clearly expressed MDC message, although the level of expression was less than that of monocyte-derived macrophages (Fig. 4 B). No MDC expression was seen after 1 d of PMA treatment or in untreated cells, nor was expression detectable in the granulocytic cells generated by treatment with DMSO for 1 or 3 d.
Northern blotting was also used to examine the expression of MDC in dendritic cells and natural killer cells derived from human PBMC (see Materials and Methods). High expression was observed in dendritic cells, which are of the mononuclear phagocyte lineage, whereas no MDC message was detectable in natural killer cells (Fig. 4 C). However, both of these cell types were capable of chemotaxis in response to MDC (see below).
Other human cell types analyzed for expression of MDC were unstimulated cultures of the lung epithelial line A549, the lung fibroblast line IMR90, I-HUVEC, and PBMC. In addition, to test the effect of proinflammatory cytokines on MDC expression, the A549, IMR90, and I-HUVECs were treated with TNF-α (10 ng/ml), and the PBMCs were treated with PHA (1 μg/ml) plus PMA (30 ng/ml). MDC mRNA was not detectable in the unstimulated cells by Northern analysis, and treatment with the cytokines did not induce MDC expression (Fig. 4 D). Induction of MCP-1 expression was readily apparent after these treatments.
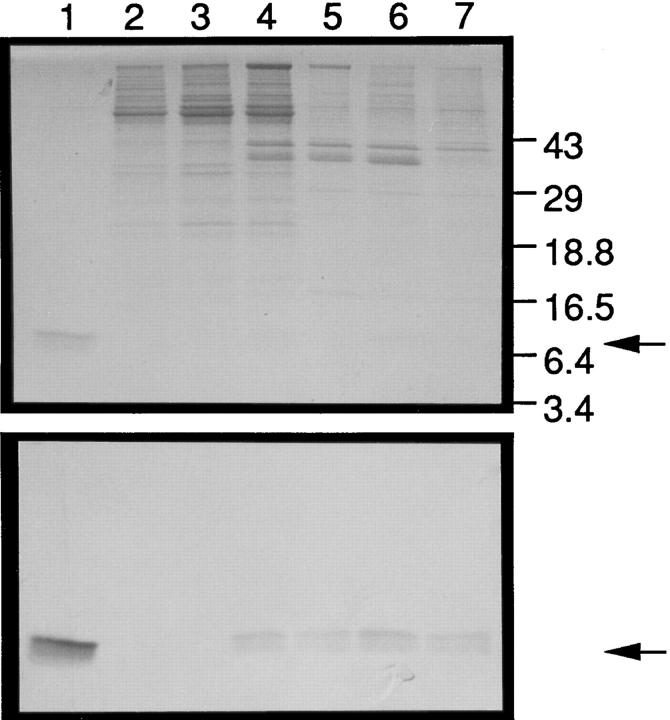
To correlate the expression of MDC protein with MDC mRNA, mAbs raised against recombinant MDC were used for Western analysis of culture supernatants from monocytederived macrophages and epithelial cell lines (Fig. 5). The results confirmed that the macrophages secreted MDC protein into the medium, whereas the epithelial cells did not. MDC protein expression was not affected by treatment of the macrophages with low density lipoprotein (LDL) or oxidized LDL.
Figure 5.
MDC protein expression in human monocyte–derived macrophages and unstimulated epithelial cell lines. Monocytes were differentiated into macrophages by incubation on tissue culture plastic for 6 d. LDL or oxidized LDL was then added for an additional 3 d. Culture supernatants from these macrophages or from epithelial lines were passed over heparin– Sepharose columns, and MDC was eluted with 0.6 M NaCl, fractionated by SDS-PAGE, and reacted with an mAb raised against recombinant MDC produced in bacteria. (Top) The Coomassie stained gel of the heparin– Sepharose elutates. (Bottom) The Western blot. Lanes 1, CHO-derived MDC; 2, A549 epithelial cell; 3, T84 epithelial cell; 4, macrophage, day 6; 5, macrophage, day 9; 6, macrophage plus LDL, day 9; 7, macrophage plus oxidized LDL, day 9. Arrows indicate the migration of MDC.
MDC Gene Expression in Human Tissues.
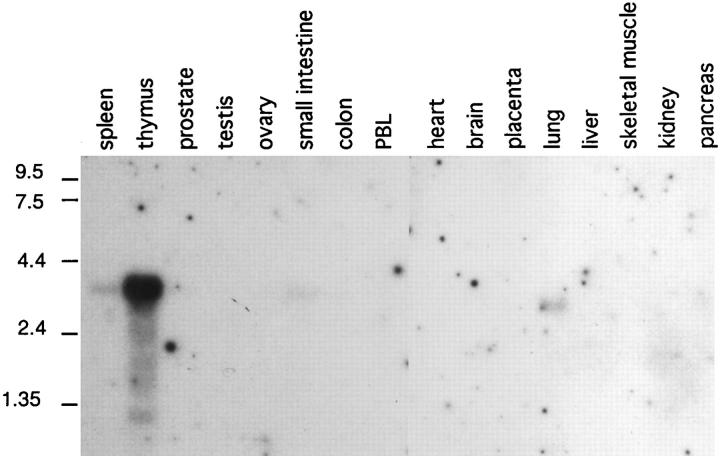
The expression pattern of MDC in normal human tissues was studied by Northern analysis. Greatest MDC gene expression was observed in the thymus, with much weaker expression in spleen and lung (Fig. 6). Very faint expression of MDC was seen in the small intestine, and no expression was detected in brain, colon, heart, kidney, liver, ovary, pancreas, placenta, prostate, skeletal muscle, testis, or peripheral blood leukocytes.
Figure 6.
Expression of MDC mRNA in normal human tissues. A multiple tissue Northern blot was probed with the MDC cDNA sequence and washed at high stringency (0.2× SSC, 50°C). The migration of RNA size markers is indicated in kb.
Biological Activity of MDC.
The mature form of MDC was chemically synthesized for use in biological assays. Synthetic chemokines have been shown to fold correctly and retain the biological activity of the natural species (47–49). To confirm that synthetic MDC was in fact correctly folded, formation of the disulfide bridges was confirmed by peptide mapping. The behavior of chemically synthesized MDC was identical to that of CHO-derived MDC in the following procedures: SDS-PAGE, heparin–Sepharose chromatography, and immunoprecipitation and Western blotting with mAbs or polyclonal antibodies raised against recombinant MDC (data not shown).
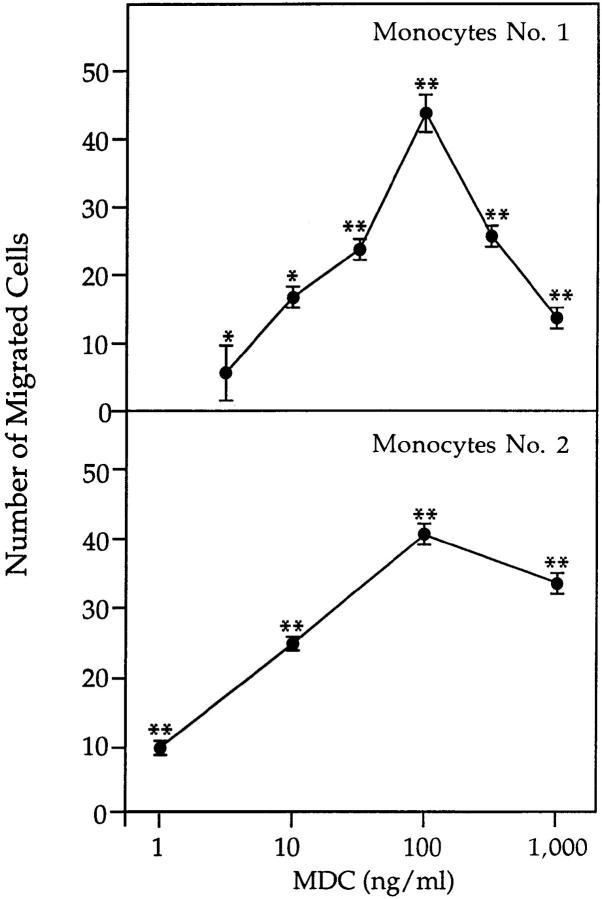
A microchamber migration assay was used to study the chemotactic activity of MDC upon dendritic cells and IL-2– activated natural killer cells, both derived from human PBMC. Both of these cell types migrated toward MDC with a bell-shaped dose-response curve. Migration became significant at 0.1 ng/ml (P <0.05) and reached a maximum at 1 ng/ml MDC (P <0.01) (Fig. 7). At the optimal concentration of 1 ng/ml, the number of dendritic cells that migrated to MDC was 87 ± 18% (n = 7) of that responding to 100 ng/ml MCP-3, a reference chemoattractant for dendritic cells (42).
Figure 7.
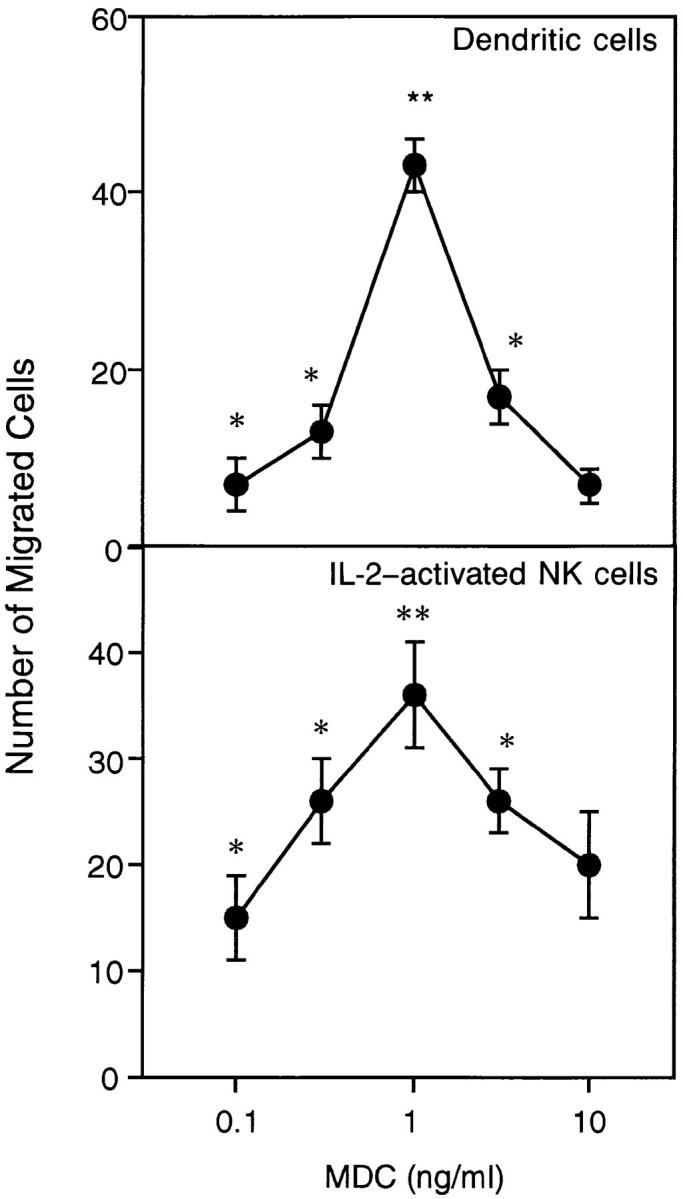
Chemotaxis of monocyte-derived dendritic cells and IL-2–activated natural killer cells induced by MDC. (Top) PBMC were isolated by density gradient centrifugation and depleted of CD19+ and CD2+ cells by antibody-coated magnetic beads. Dendritic cells were obtained by culturing the remaining cells for 6 to 8 d in RPMI containing containing 10% FCS, 50 ng/ml GM-CSF, and 20 ng/ ml IL-13. (Bottom) Natural killer cells were purified by discontinuous Percoll gradient centrifugation of monocyte-depleted PBMC, followed by negative selection of T cells by panning with an anti-CD6 mAb. Cells were cultured with an irradiated lymphoblastoid cell line in the presence of 250 U/ml IL-2. For chemotaxis assays, cells were added to the upper wells of a microchamber and medium (RPMI plus 1% FCS) with or without chemokine was added to the lower chamber. The figure shows representative experiments. Values represent the number of migrated cells (mean ± SE) after subtraction of basal migration (24 ± 3 for dendritic cells and 20 ± 6 for natural killer cells). *P <0.05), **P <0.01.
A similar assay was used to measure the chemotactic response of PBMC to MDC. Significant migration occurred at 1 ng/ml MDC (P <0.05), and the response peaked at 100 ng/ml (Fig. 8). Under these conditions, only monocytes had migrated through the filter. The peak number of cells was equivalent to that migrating to 100 ng/ml MCP-3 (data not shown).
Figure 8.
Chemotaxis of monocytes induced by MDC. Human PBMC were purified by density gradient centrifugation and resuspended in RPMI plus 1% FCS. Chemotaxis was assayed as described in Fig. 7; two independent experiments are illustrated. Under these conditions, only monocytes migrated through the filter. Basal migration of 15 ± 2 (top) and 16 ± 2 (bottom) was subtracted from the values shown. The response of these cells to 100 ng/ml MCP-3 was 42 ± 4 and to 10 nM fMLP was 54 ± 3.
Under similar assay conditions, MDC was not chemotactic for neutrophils at concentrations up to 1 μg/ml (data not shown).
Chromosome Localization.
PCR was used to screen for the MDC gene in genomic DNA isolated from a set of mouse–human or hamster–human chimeric cell lines, each of which retains a single human chromosome. The expected PCR product was produced only from the line containing human chromosome 16. The identity of the PCR band was confirmed by hybridization with an internal MDC oligonucleotide (data not shown).
Discussion
We have described the cloning and activity of a novel human CC chemokine, MDC. This sequence was not previously represented in the Expressed Sequence Tags (dbest) or Non-Redundant (nr) databases of GenBank. The amino acid sequence of MDC suggests that it is a member of the CC chemokine family, but it is not closely related to any of the known chemokines. The length of the 3′ noncoding region is unusually long and contains multiple Alu repeats. The size of the MDC transcript corresponds well to the size of the cDNA (Figs. 4 and 5), confirming that the entire cDNA is transcribed.
Our results indicate that MDC has a very specific pattern of expression. It was highly expressed by fully differentiated macrophages and monocyte-derived dendritic cells, but it was not expressed by freshly isolated monocytes, granulocytic cells, or natural killer cells (Fig. 4). The timing of MDC expression in macrophages cultured in vitro appears to be similar to that of the transferrin receptor, which is also strongly upregulated late in macrophage differentiation (6 d after plating; 50). The kinetics of induction of other macrophage-specific products is quite different. For example, platelet activating factor acetylhydrolase is highly upregulated by day 2 (36), and chitinase is strongly induced beginning at day 9 (51).
The activity and expression pattern of MDC have several implications in the function of dendritic cells. Immature dendritic cells are rapidly recruited to sites of inflammatory stimulation and are highly proficient in antigen uptake. After antigenic stimulation, they are again mobilized and migrate to draining lymph nodes. During this process of maturation, they lose their ability to process and present soluble antigen and become extemely potent stimulators of T lymphocytes (52–55). Consequently, dendritic cells have been implicated in organ transplant rejection, HIV infection, asthma, and induction of tolerance (56). Because MDC is highly expressed by dendritic cells and is also chemotactic for them, it may play an autocrine role in their accumulation at sites of inflammation. In addition, several other chemokines, including MCP-3, MIP-1α, and RANTES, are chemotactic for immature dendritic cells (42), but do not induce their maturation (Sozzani, S., and P. Allavena, unpublished data). Thus, chemokines may play a major role in the initial influx of immature dendritic cells. Administration of TNF-α also leads to accumulation of dendritic cells in vivo (57); however, this effect may be indirectly mediated by factors induced by TNF-α, including CC chemokines. Unlike the chemokines, TNF-α induces maturation of dendritic cells, decreasing their antigen uptake and increasing their ability to stimulate T cells (43).
The high expression of MDC in the thymus suggests it has an additional (or alternate) function in T cell development. One possible role is to attract or retain dendritic cells in the thymus to enhance stimulation of T cells. In addition, MDC may be chemotactic for T cells themselves and thereby aid in their aggregation with dendritic cells. Notably, the CC chemokine TARC, which is specifically chemotactic for T cells, has a tissue-specific expression pattern that is nearly identical to that of MDC, with very high expression in the thymus (20). TARC, which is ∼32% identical to MDC, is also the only other chemokine known to be encoded on chromosome 16 (58). The effect of MDC on T cells is currently being investigated.
Natural killer cells recognize a broad range of cytolytic targets and are believed to be involved in defense against viral infection, destruction of tumor cells, and regulation of hematopoiesis. The means by which they select a specific target cell is poorly understood, but their activity appears to involve an extensive set of cytokines that includes interleukins, interferons, and chemokines (59, 60). Granule exocytosis or chemotaxis of natural killer cells has been demonstrated in response to many CC chemokines, such as MCP-1, -2, and -3 (49, 61, 62), MIP-1α (63), MIP-1β (49), and RANTES (61). Further, these cells respond to the CXC chemokines IP-10 (63) and IL-8 (64), and migrate toward the C chemokine, lymphotactin (65). Our results indicate that MDC likewise induces directed migration of IL-2–activated natural killer cells, with an effective concentration in the low nanomolar range.
Monocytes also exhibited a dose-dependent chemotactic response to MDC. The magnitude of the response was similar to that of monocyte-derived dendritic cells and activated natural killer cells, but it occurred at a 100-fold higher concentration of MDC (Fig. 8). Thus, in vivo production of MDC may first attract dendritic cells or natural killer cells, and further accumulation of MDC may cause a subsequent influx of monocytes.
Induction of MDC expression does not follow the pattern typically observed for chemokines. These genes are generally not constitutively expressed, but they can be strongly induced by cytokine treatment (e.g., TNF-α induction of MCP-1; Fig. 4). In marked contrast, induction of MDC was not detected after proinflammatory stimulation of PBMC with PHA plus PMA or stimulation of lung epithelial cells (A549), lung fibroblast cells (IMR90), or I-HUVEC with TNF-α (Figs. 4 and 5). Similarly, treatment with LDL seemed to have little effect on production of MDC protein by macrophages, although a transient rise in MDC mRNA expression may not have been detected in this study. LDL treatment of macrophages appears to have diverse effects on production of other chemokines, either enhancing or quenching expression, depending on the activation state of the cells and modifications to the lipoprotein (66, 67).
The receptor responsible for binding MDC is not known, but preliminary results indicate it does not signal through the cloned chemokine receptors CCR1 or CCR2 (data not shown). Analysis of the receptors expressed by dendritic cells and natural killer cells may reveal the molecule(s) responsible for binding MDC. The chemotactic activity of MDC for these cells suggests that it may be clinically relevant in various physiological processes, including induction of immune responses and elimination of pathogenic microbes.
Acknowledgments
The authors wish to thank Christi Wood for DNA sequencing, Hai Le Trong for protein sequencing, Reyna Simon and Michael Siani for synthesis and chemical analysis of MDC, Mitch Fahning for production of mAbs, and Darcy Clark for supernatants from LDL-treated macrophages.
Footnotes
P. Allavena, S. Sozzani, and A. Mantovani were supported by Project AIDS, Istituto Superiore Sanitá, and Associazione Italiana per la Recerca sul Cancro.
1 Abbreviations used in this paper: CHO, Chinese hamster ovary; GAPDH, glyceraldehyde phosphate dehydrogenase; I-HUVEC, immortalized human umbilical vein endothelial cells; LDL, low density lipoprotein; MCP, monocyte chemotactic proteins; MDC, macrophage-derived chemokine; MIP, macrophage inflammatory protein; TARC, thymus and activation regulated chemokine.
References
- 1.Oppenheim JJ. Overview of chemokines. Adv Exp Med Biol. 1993;351:183–186. doi: 10.1007/978-1-4615-2952-1_19. [DOI] [PubMed] [Google Scholar]
- 2.Baggiolini M, Dewald B, Moser B. Interleukin-8 and related chemotactic cytokines-CXC and CC chemokines. Adv Immunol. 1994;55:97–179. [PubMed] [Google Scholar]
- 3.Schall TJ. Biology of the RANTES/SIS cytokine family. Cytokine. 1991;3:165–183. doi: 10.1016/1043-4666(91)90013-4. [DOI] [PubMed] [Google Scholar]
- 4.Eaves CJ, Cashman JD, Wolpe SD, Eaves AC. Unresponsiveness of primitive chronic myeloid leukemia cells to macrophage inflammatory protein 1 α, an inhibitor of primitive normal hematopoietic cells. Proc Natl Acad Sci USA. 1993;90:12015–12019. doi: 10.1073/pnas.90.24.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tashiro K, Tada H, Heilker R, Shirozu M, Nakano T, Honjo T. Signal sequence trap: a cloning strategy for secreted proteins and type I membrane proteins. Science (Wash DC) 1993;261:600–603. doi: 10.1126/science.8342023. [DOI] [PubMed] [Google Scholar]
- 6.Nagasawa T, Hirota S, Tachibana K, Takakura N, Nishikawa S, Kitamura Y, Yoshida N, Kikutani H, Kishimoto T. Defects of B-cell lymphopoiesis and bonemarrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature (Lond) 1996;382:635–638. doi: 10.1038/382635a0. [DOI] [PubMed] [Google Scholar]
- 7.Hromas, R., P. Gray, D. Chantry, R. Godiska, M. Krathwohl, K. Fife, G.I. Belle, J. Takeda, S. Aronica, M. Gordon et al. 1997. Cloning and characterization of Exodus, a novel beta chemokine. Blood. In press. [PubMed]
- 8.Nakao M, Nomiyama H, Shimada K. Structures of human genes coding for cytokine LD78 and their expression. Mol Cell Biol. 1990;10:3646–3658. doi: 10.1128/mcb.10.7.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown KD, Zurawski SM, Mosmann TR, Zurawski G. A family of small inducible proteins secreted by leukocytes are members of a new superfamily that includes leukocyte and fibroblast-derived inflammatory agents, growth factors, and indicators of various activation processes. J Immunol. 1989;142:679–687. [PubMed] [Google Scholar]
- 10.Schall TJ, Jongstra J, Dyer BJ, Jorgensen J, Clayberger C, Davis MM, Krensky AM. A human T cell–specific molecule is a member of a new gene family. J Immunol. 1988;141:1018–1025. [PubMed] [Google Scholar]
- 11.Miller MD, Hata S, de Waal R, Malefyt, Krangel MS. A novel polypeptide secreted by activated human T lymphocytes. J Immunol. 1989;143:2907–2916. [PubMed] [Google Scholar]
- 12.Matsushima K, Larsen CG, DuBois GC, Oppenheim JJ. Purification and characterization of a novel monocyte chemotactic and activating factor produced by a human myelomonocytic cell line. J Exp Med. 1989;169:1485–1490. doi: 10.1084/jem.169.4.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Damme J, Proost P, Lenaerts J-P, Opdenakker G. Structural and functional identification of two human, tumor-derived monocyte chemotactic proteins (MCP-2 and MCP-3) belonging to the chemokine family. J Exp Med. 1992;176:59–65. doi: 10.1084/jem.176.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uguccioni M, Loetscher P, Forssmann U, Dewald B, Li H, Hensche S, Lima, Li Y, Kreider B, Garotta G, Thelen M, et al. Monocyte chemotactic protein 4 (MCP4), a novel structural and functional analogue of MCP-3 and eotaxin. J Exp Med. 1996;183:2379–2384. doi: 10.1084/jem.183.5.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Zepeda EA, Combadiere C, Rothenber ME, Sarafi MN, Lavigne F, Hamid Q, Murphy PM, Luster AD. Human monocyte chemoattractant protein (MCP)-4 is a novel CC chemokine with activities on monocytes, eosinophils, and basophils induced in allergic and nonallergic inflammation that signals through the CC chemokine receptors (CCR)-2 and -3. J Immunol. 1996;157:5613–5626. [PubMed] [Google Scholar]
- 16.Godiska, R., D. Chantry, C.J. Raport, V.L. Schweickart, H. LeTrong, and P.W. Gray. 1997. Monocyte chemotactic protein-4: tissue-specific expression and signaling through CC chemokine receptor-2. J. Leukocyte Biol. In press. [DOI] [PubMed]
- 17.Kitaura M, Nakajima T, Imai T, Harada T, Combadiere C, Tiffany HL, Murphy PM, Yoshie O. Molecular cloning of human eotaxin, an eosinophil-selective CC chemokine, and identification of a specific eosinophil eotaxin receptor, CC chemokine receptor 3. J Biol Chem. 1996;271:7725–7730. doi: 10.1074/jbc.271.13.7725. [DOI] [PubMed] [Google Scholar]
- 18.Daugherty BL, Siciliano SJ, DeMartino JA, Malkowitz L, Sirotina A, Springer MS. Cloning, expression, and characterization of the human eosinophil eotaxin receptor. J Exp Med. 1996;183:2349–2354. doi: 10.1084/jem.183.5.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schulz-Knappe P, Mägert H-J, Dewald B, Meyer M, Cetin Y, Kubbies M, Tomeczkowski J, Kirchhoff K, Raida M, Adermann K, et al. HCC-1, a novel chemokine from human plasma. J Exp Med. 1996;183:295–299. doi: 10.1084/jem.183.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imai T, Yoshida T, Baba M, Nishimura M, Kakizaki M, Yoshie O. Molecular cloning of a novel T cell– directed CC chemokine expressed in thymus by signal sequence trap using Epstein-Barr virus vector. J Biol Chem. 1996;271:21514–21521. doi: 10.1074/jbc.271.35.21514. [DOI] [PubMed] [Google Scholar]
- 21.Gao JL, Kuhns DB, Tiffany HL, McDermott D, Li X, Francke U, Murphy PM. Structure and functional expression of the human macrophage inflammatory protein 1 α/RANTES receptor. J Exp Med. 1993;177:1421–1427. doi: 10.1084/jem.177.5.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neote K, DiGregorio D, Mak JY, Horuk R, Schall TJ. Molecular cloning, functional expression, and signaling characteristics of a C-C chemokine receptor. Cell. 1993;72:415–425. doi: 10.1016/0092-8674(93)90118-a. [DOI] [PubMed] [Google Scholar]
- 23.Combadiere C, Ahuja SK, Van Damme J, Tiffany HL, Gao JL, Murphy PM. Monocyte chemoattractant protein–3 is a functional ligand for CC chemokine receptors 1 and 2B. J Biol Chem. 1995;270:29671–29675. doi: 10.1074/jbc.270.50.29671. [DOI] [PubMed] [Google Scholar]
- 24.Ben-Baruch A, Xu L, Young PR, Bengali K, Oppenheim JJ, Wang JM. Monocyte chemotactic protein–3 (MCP3) interacts with multiple leukocyte receptors. C-C CKR1, a receptor for macrophage inflammatory protein-1 α/RANTES, is also a functional receptor for MCP3. J Biol Chem. 1995;270:22123–22128. doi: 10.1074/jbc.270.38.22123. [DOI] [PubMed] [Google Scholar]
- 25.Proost P, Wuyts A, Van Damme J. Human monocyte chemotactic proteins–2 and –3: structural and functional comparison with MCP-1. J Leukocyte Biol. 1996;59:67–74. doi: 10.1002/jlb.59.1.67. [DOI] [PubMed] [Google Scholar]
- 26.Franci C, Wong LM, Van Damme J, Proost P, Charo IF. Monocyte chemoattractant protein–3, but not monocyte chemoattractant protein–2, is a functional ligand of the human monocyte chemoattractant protein–1 receptor. J Immunol. 1995;1545:6511–6517. [PubMed] [Google Scholar]
- 27.Ponath PD, Qin S, Post TW, Wang J, Wu L, Gerard NP, Newman W, Gerard C, Mackay CR. Molecular cloning and characterization of a human eotaxin receptor expressed selectively on eosinophils. J Exp Med. 1996;183:2437–2448. doi: 10.1084/jem.183.6.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Power CA, Meyer A, Nemeth K, Bacon KB, Hoogewerf AJ, Proudfoot AE, Wells N. Molecular cloning and functional expression of a novel CC chemokine receptor cDNA from a human basophilic cell line. J Biol Chem. 1995;270:19495–19500. doi: 10.1074/jbc.270.33.19495. [DOI] [PubMed] [Google Scholar]
- 29.Raport CJ, Gosling J, Schweickart VL, Gray PW, Charo IF. Molecular cloning and functional characterization of a novel human CC chemokine receptor (CCR5) for RANTES, MIP-1β, and MIP–1α. J Biol Chem. 1996;271:17161–17166. doi: 10.1074/jbc.271.29.17161. [DOI] [PubMed] [Google Scholar]
- 30.Combadiere C, Ahuja SK, Tiffany HL, Murphy PM. Cloning and functional expression of CC CKR5, a human monocyte CC chemokine receptor selective for MIP-1α, MIP-1β, and RANTES. J Leukocyte Biol. 1996;60:147–152. doi: 10.1002/jlb.60.1.147. [DOI] [PubMed] [Google Scholar]
- 31.Deng H-K, Liu R, Ellmeier W, Choe S, Derya U, Burkhart M, DiMarzio P, Marmon S, Sutton RE, Hill CM, et al. Identification of a major co-receptor for primary isolates of HIV-1. Nature (Lond) 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 32.Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, Berger EA. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science (Wash DC) 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 33.Dragic T, Litwin V, Allaway GP, Martin SR, Haung Y, Nagashima KA, Cayanan C, Maddon PJ, Koup RA, Moore JP, et al. HIV-1 entry into CD4+cells is mediated by the chemokine receptor CC-CKR-5. Nature (Lond) 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 34.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, Wu L, Mackay CR, LaRosa G, Newman W, et al. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 35.Doranz BJ, Rucker J, Yi J, Smyth RJ, Samson M, Peiper SC, Parmentier M, Collman RG, Doms RW. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 36.Tjoelker LW, Wilder C, Eberhardt C, Stafforini DM, Dietsch G, Schimpf B, Hooper S, Le Trong H, Cousens LS, Zimmerman GA, et al. Anti-inflammatory properties of a platelet-activating factor acetylhydrolase. Nature (Lond) 1995;374:549–552. doi: 10.1038/374549a0. [DOI] [PubMed] [Google Scholar]
- 37.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–412. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 38.Godiska R, Chantry D, Dietsch GN, Gray PW. Chemokine expression in murine experimental allergic encephalomyelitis. J Neuroimmunol. 1995;5805:167–176. doi: 10.1016/0165-5728(95)00008-p. [DOI] [PubMed] [Google Scholar]
- 39.Urlaub G, Mitchell PJ, Kas E, Chasin LA, Funanage VL, Myoda TT, Hamlin J. Effect of gamma rays at the dihydrofolate reductase locus: deletions and inversions. Somatic Cell Mol Genet. 1986;12:555–566. doi: 10.1007/BF01671941. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., E.F. Fritsch, and T. Maniatis. 1987. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 41.Stafforini DM, Elstad MR, McIntyre TM, Zimmerman GA, Prescott SM. Human macrophages secrete platelet-activating factor acetylhydrolase. J Biol Chem. 1990;265:9682–9687. [PubMed] [Google Scholar]
- 42.Sozzani S, Sallusto F, Luini W, Zhou D, Piemontli L, Allavena P, VanDamme J, Valitutti S, Lanzavecchia A, Mantovani A. Migration of dendritic cells in response to formyl peptides, C5a and a distinct set of chemokines. J Immunol. 1995;155:3292–3295. [PubMed] [Google Scholar]
- 43.Sallusto F, Lanzavecchia A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor-α. J Exp Med. 1994;179:1109–1118. doi: 10.1084/jem.179.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Allavena P, Bianchi G, Giardina P, Polentarutti N, Zhou D, Introna M, Sozzani S, Mantovani A. Migratory response to human NK cells to monocyte-chemotactic proteins. Methods (Orlando) 1996;10:145–149. doi: 10.1006/meth.1996.0088. [DOI] [PubMed] [Google Scholar]
- 45.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perussia B, Lebman D, Ip SH, Rovera G, Trinchieri G. Terminal differentiation surface antigens of myelomonocytic cells are expressed in human promyelocytic leukemia cells (HL60) treated with chemical inducers. Blood. 1981;58:836–843. [PubMed] [Google Scholar]
- 47.Clark-Lewis I, Dewald B, Loetscher M, Moser B, Baggiolini M. Structural requirements for interleukin-8 function identified by design of analogs and CXC chemokine hybrids. J Biol Chem. 1994;269:16075–16081. [PubMed] [Google Scholar]
- 48.Gong JH, Clark-Lewis I. Antagonists of monocyte chemoattractant protein 1 identified by modification of functionally critical NH2-terminal residues. J Exp Med. 1995;181:631–640. doi: 10.1084/jem.181.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loetscher P, Seitz M, Clark-Lewis I, Baggiolini M, Moser B. Activation of NK cell by CC chemokines. J Immunol. 1996;156:322–327. [PubMed] [Google Scholar]
- 50.Hirata T, Bitterman PB, Mornex JF, Crystal RG. Expression of the transferrin receptor gene during the process of mononuclear phagocyte maturation. J Immunol. 1986;136:1339–1345. [PubMed] [Google Scholar]
- 51.Boot RG, Renkema HG, Strijland A, Jan van Zonneveld A, Aerts JMFG. Cloning of a cDNA encoding chitotriosidase, a human chitinase produced by macrophages. J Biol Chem. 1995;270:26252–26256. doi: 10.1074/jbc.270.44.26252. [DOI] [PubMed] [Google Scholar]
- 52.Steinman RM. The dendritic cell system and its role in immunogenicity. Annu Rev Immunol. 1991;9:271–296. doi: 10.1146/annurev.iy.09.040191.001415. [DOI] [PubMed] [Google Scholar]
- 53.Romani N, Schuler G. The immunologic properties of epidermal Langerhans cells as a part of the dendritic cell system. Springer Semin Immunopathol. 1992;13:265–279. doi: 10.1007/BF00200527. [DOI] [PubMed] [Google Scholar]
- 54.Austyn JM. Antigen uptake and presentation by dendritic leukocytes. Semin Immunol. 1992;4:227–236. [PubMed] [Google Scholar]
- 55.Austyn JM. New insights into the mobilization and phagocytic activity of dendritic cells. J Exp Med. 1996;183:1287–1292. doi: 10.1084/jem.183.4.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caux C, Liu Y-J, Banchereau J. Recent advances in the study of dendritic cells and follicular dendritic cells. Immunol Today. 1995;16:23–25. doi: 10.1016/0167-5699(95)80061-1. [DOI] [PubMed] [Google Scholar]
- 57.Cumberbatch M, Kimber I. Dermal tumour necrosis factor-alpha induces dendritic cell migration to draining lymph nodes, and possibly provides one stimulus for Langerhans' cell migration. Immunology. 1992;75:257–263. [PMC free article] [PubMed] [Google Scholar]
- 58.Nomiyama, H., T. Imai, J. Kusuda, R. Miura, D.F. Callen, and O. Yoshie. 1997. Assignment of the human CC chemokine gene TARC (SCYA17) to chromosome 16q13. Genomics. In press. [DOI] [PubMed]
- 59.Robertson MJ, Ritz J. Biology and clinical relevance of human natural killer cells. Blood. 1990;76:2421–2438. [PubMed] [Google Scholar]
- 60.Trinchieri G. Biology of natural killer cells. Adv Immunol. 1989;47:187–376. doi: 10.1016/S0065-2776(08)60664-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maghazachi AA, Al-Aoukaty A, Schall TJ. C-C chemokines induce the chemotaxis of NK and IL-2 activated NK cells. Role for G proteins. J Immunol. 1994;153:4969–4977. [PubMed] [Google Scholar]
- 62.Allavena P, Bianchi G, Zhou D, Van Damme J, Jîlek P, Sozzani S, Mantovani A. Induction of natural killer cell migration by monocyte chemotactic protein-1, -2 and -3. Eur J Immunol. 1994;24:3233–3236. doi: 10.1002/eji.1830241249. [DOI] [PubMed] [Google Scholar]
- 63.Taub DD, Sayers TJ, Carter CRD, Ortaldo JR. α and β chemokines induce NK cell migration and enhance NK cell mediated cytolysis. J Immunol. 1995;155:3877–3888. [PubMed] [Google Scholar]
- 64.Sebok K, Woodside D, Al-Aoukaty A, Ho AD, Gluck S, Maghazachi AA. IL-8 induces the locomotion of human IL-2–activated natural killer cells. J Immunol. 1993;150:1524–1534. [PubMed] [Google Scholar]
- 65.Bianchi G, Sozzani S, Zlotnik A, Mantovani A, Allavena P. Migratory response of human natural killer cells to lymphotactin. Eur J Immunol. 1996;26:3238–3241. doi: 10.1002/eji.1830261260. [DOI] [PubMed] [Google Scholar]
- 66.Wang N, Tabas I, Winchester R, Ravalli S, Rabbani LE, Tall A. Interleukin 8 induced by cholesterol loading of macrophages and expressed by macrophage foam cells in human atheroma. J Biol Chem. 1996;271:8837–8842. doi: 10.1074/jbc.271.15.8837. [DOI] [PubMed] [Google Scholar]
- 67.Hamilton TA, Major JA. Oxidized LDL potentiates LPS-induced transcription of the chemokine KC gene. J Leukocyte Biol. 1996;59:940–947. doi: 10.1002/jlb.59.6.940. [DOI] [PubMed] [Google Scholar]
- 68.Claverie J-M, Makalowski W. Alu alert. Nature (Lond) 1994;371:752. doi: 10.1038/371752a0. [DOI] [PubMed] [Google Scholar]