Abstract
The primary DNA lesion induced by malondialdehyde, a byproduct of lipid peroxidation and prostaglandin synthesis, is 3-(2′-deoxy-β-d-erythro-pentofuranosyl)-pyrimido[1,2-a]purin-10(3H)-one (M1G). When placed opposite cytosine (underlined) at neutral pH in either the d(GGTMTCCG)⋅d(CGGACACC) or d(ATCGCMCGGCATG)⋅ d(CATGCCGCGCGAT) duplexes, M1G spontaneously and quantitatively converts to the ring-opened derivative N2-(3-oxo-1-propenyl)-dG. Ring-opening is reversible on thermal denaturation. Ring-opening does not occur at neutral pH in single-stranded oligodeoxynucleotides or when T is placed opposite to M1G in a duplex. The presence of a complementary cytosine is not required to stabilize N2-(3-oxo-1-propenyl)-dG in duplex DNA at neutral pH. When N2-(3-oxo-1-propenyl)-dG is placed opposite to thymine in a duplex, it does not revert to M1G. A mechanism for the conversion of M1G to N2-(3-oxo-1-propenyl)-dG is proposed in which the exocyclic amino group of the complementary cytosine attacks the C8 position of the M1G exocyclic ring and facilitates ring opening via formation of a transient Schiff base. Addition of water to the Schiff base regenerates the catalytic cytosine and generates N2-(3-oxo-1-propenyl)-dG. These results document the ability of duplex DNA to catalyze the transformation of one adduct into another, which may have important consequences for mutagenesis and repair.
Malondialdehyde is a toxic and mutagenic metabolite produced by lipid peroxidation (1) and prostaglandin biosynthesis (2, 3). It is of interest in the etiology of human cancer because it is produced endogenously, and it is mutagenic in bacterial (4, 5) and mammalian cells (6) and is carcinogenic in rats (7). Malondialdehyde reacts with DNA (8–11), and, at physiological pH, the major adduct is an exocyclic pyrimidopurinone termed M1G (8, 9, 12–14). This 1,N2 exocyclic guanine adduct is unstable under basic conditions, rearranging to the ring-opened derivative N2-(3-oxo-1-propenyl)-dG (15) (Fig. 1). M1G has been identified in DNA from rodent (16) and human (17, 18) tissue samples, as have other exocyclic purine lesions (19–21), suggesting that these exocyclic lesions are formed in vivo. M1G represents the most abundant exocyclic DNA adduct present endogenously in human DNA, as quantitated by mass spectroscopic (11, 17, 22), postlabeling (23, 24), and immunochemical (25) techniques. Site-specific mutagenesis experiments indicate M1G is an efficient premutagenic lesion (26), suggesting that it is a mediator of human genetic disease.
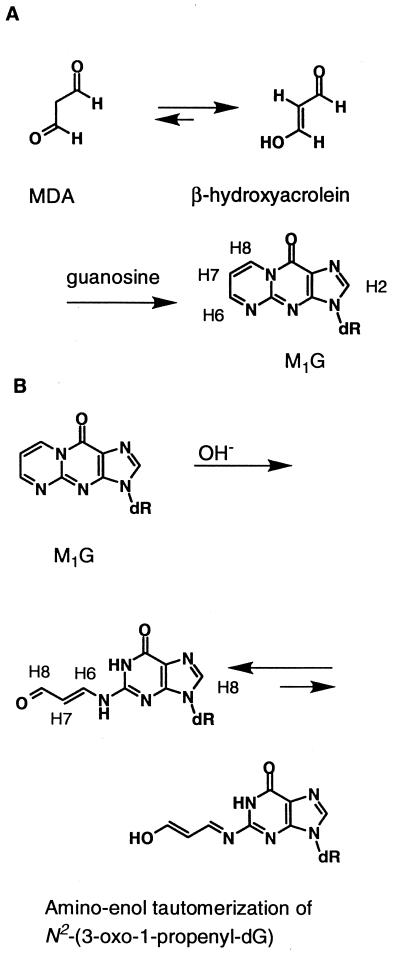
Figure 1.
(A) Formation of M1G from [1,2a] cyclization of deoxyguanosine by β-hydroxyacrolein. (B) Ring-opening to the N2-(3-oxo-1-propenyl)-dG derivative occurs at basic pH; the ring-opened species equilibrate between the favored amino and the enol tautomers. Note the numbering system for the M1G protons. In oligodeoxynucleotides, M1G is designated as M, and N2-(3-oxo-1-propenyl)-dG is designated as X.
The development of methodology to allow site-specific preparation of M1G-adducted oligodeoxynucleotides (15) allowed M1G to be introduced into the oligodeoxynucleotides d(GGTMTCCG) and d(ATCGCMCGGCATG). These oligodeoxynucleotides were derived from the hisD3052 mutation within the histidinol dehydrogenase gene in the corresponding frameshift-sensitive tester strain of Salmonella typhimurium (27, 28). In d(GGTMTCCG), M1G was centered in a palindromic sequence; in d(ATCGCMCGGCATG), M1G was inserted into an iterated CG repeat sequence. Both of these sequence contexts were of interest for site-specific mutagenesis studies and also for solution structural studies because both were suspected to be loci for frameshift mutations that caused reversion of the hisD3052 mutation. Studies using the structural analog 1,N2-propanodeoxyguanosine (PdG) revealed that PdG equilibrated between the anti- and syn-conformations of the glycosyl bond within each of these sequences (29, 30).
The present results reveal that, when combined with the appropriate complementary strands to form the d(GGTXTCCG)⋅d(CGGACACC) or d(ATCGCXCGGCATG)⋅ d(CATGCCG CGCGAT) duplexes, such that M1G is placed opposite cytosine, M1G undergoes quantitative transformation into the ring-opened N2-(3-oxo-1-propenyl)-dG derivative (Fig. 2). Ring-opening is reversible on thermal dissociation of the duplexes, such that M1G is regenerated. Ring-opening is not observed at pH 6.8 in the single-stranded oligodeoxynucleotides d(GGTMTCCG) and d(ATCGCMCGGCATG). When the N2-(3-oxo-1-propenyl)-dG derivative is placed complementary to thymine in duplex DNA, it does not revert to M1G, indicating that the presence of cytosine is not necessary to stabilize the ring-opened derivative. The conversion of M1G to N2-(3-oxo-1-propenyl)-dG is proposed to occur via formation of a transient Schiff base when the exocyclic amino group of the complementary cytosine attacks the C8 position of the M1G exocyclic ring. These data provide the first evidence for the catalyzed chemical transformation of one adduct into another by duplex DNA, which may have important consequences for mutagenesis and repair.
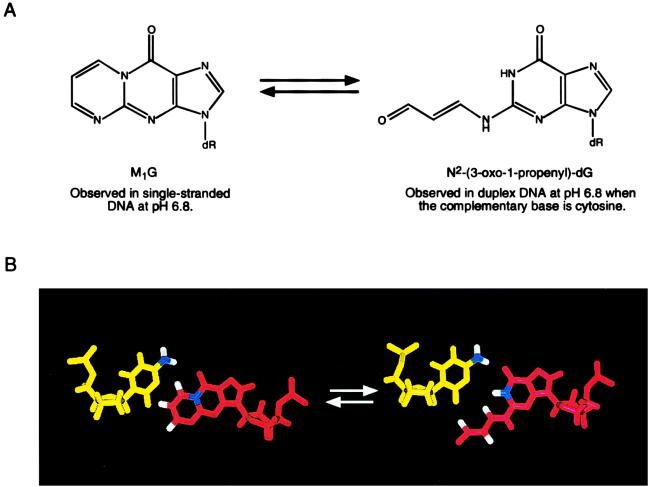
Figure 2.
(A) Analysis of d(GGTXTCCG)⋅d(CGGACACC) and d(ATCGCXCGGCATG)⋅d(CATGCCGCGCGAT) revealed that, at pH 6.8, M1G had undergone a quantitative chemical transformation into N2-(3-oxo-1-propenyl)-dG. On strand dissociation at neutral pH, M1G was regenerated. (B) Molecular modeling revealed that the exocyclic N4 amino group of the complementary cytosine was positioned in duplex DNA to catalyze opening of M1G, probably via attack at the C8 position of M1G.
MATERIALS AND METHODS
Materials.
The oligodeoxynucleotides d(GGTMTCCG) and d(ATCGCMCGGCATG) were synthesized by using the 2-(acetoxymethyl)-benzoyl protecting group, which was removed with anhydrous potassium carbonate/methanol (15). The M1G-modified oligodeoxynucleotides (15) were purified by reverse-phase and anion-exchange HPLC. The samples were maintained between pH 6 and pH 7 during purification. The adducted oligodeoxynucleotides were desalted by using gel filtration (BioGel P-2, Bio-Rad). Their purities were verified by 32P-end labeling followed by denaturing polyacrylamide gel electrophoresis. Unmodified oligodeoxynucleotides were purchased from Midland Certified Reagent (Midland, TX). They were purified by reverse-phase and anion-exchange HPLC. The sample purities were checked by using SureCheck oligodeoxynucleotide analysis kits (Amersham Pharmacia).
NMR Sample Preparation.
The NMR samples (0.5 ml) of d(GGTMTCCG) and d(ATCGCMCGGCATG) were prepared in 0.1 M NaCl, 10 mM NaH2PO4, and 50 μM Na2EDTA (pH 6.8). The complementary strands were prepared in 99.96% D2O. The solutions were lyophilized and exchanged with 99.96% D2O. Duplex samples were made by titrating the appropriate complementary strands into the NMR tube containing the modified strand of interest at 60°C until the integration of the M4 H2 resonance and the H8 resonance in the complementary strand reached a 1:1 ratio. Duplex samples were reconcentrated to 0.5 ml (0.7 mM strand concentration) in buffer of 0.1 M NaCl, 10 mM NaH2PO4, and 50 μM Na2EDTA (pH 6.8). Samples used for examining exchangeable protons were prepared in the above buffer containing 9:1 H2O:D2O. For monitoring pH, a microprobe (Ingold, Wilmington, MA) was inserted into NMR sample tube before and after experiments.
NMR.
Experiments were performed at the 1H frequency of 500.13 MHz at 10°C. 2,2-dimethyl-2-silapentane-5-sulfonate was used to reference the spectra. Phase-sensitive two-dimensional nuclear Overhauser effect spectra for resonance assignment were recorded by using time-proportional phase increment quadrature detection with a mixing time of 300 ms. For examination of exchangeable protons, experiments were carried out in 9:1 H2O/D2O at 5°C by using a field-gradient watergate pulse sequence (31) for water suppression. Phase-sensitive total correlation spectra were obtained by using a MLEV17 (32) spin lock pulse at 2 G for a mixing time of 100 ms with time-proportional phase increment quadrature detection. Typically, 1,024 real data in the t1 dimension and 2,048 real data in the t2 dimension were collected. The data were processed by using felix (Molecular Simulations, Inc. San Diego) on an Indigo2 workstation (Silicon Graphics, Mountain View, CA). The data in the t1 dimension were zero-filled to give a matrix of 2,048 × 2,048 real points. A skewed sinebell square apodization function with a 90° phase-shift was used in both dimensions.
To monitor the formation of N2-(3-oxo-1-propenyl)-dG in duplex DNA when cytosine was placed opposite M1G at neutral pH, an equimolar mixture of d(GGTMTCCG) and d(CGGACACC) was equilibrated at 60°C and pH 6.8. The sample was rapidly cooled to 5°C and then was maintained at that temperature. To monitor the formation of M1G in heat-denatured DNA at neutral pH, a duplex sample of d(GGTXTCCG)⋅d(CGGACACC) was equilibrated at 5°C and pH 6.8. The sample was rapidly heated to 60°C and then was maintained at that temperature. To monitor the formation of M1G when N2-(3-oxo-1-propenyl)-dG was placed opposite thymine in duplex DNA at neutral pH, an equimolar mixture of d(GGTMTCCG) and d(CGGATACC) was equilibrated at 60°C, pH 10. The sample was rapidly cooled to 5°C. Immediately thereafter, the pH was rapidly adjusted to pH 6.8. The sample was maintained at that temperature and pH. The presence of M1G or N2-(3-oxo-1-propenyl)-dG was monitored by integration of the H6 and H8 1H NMR resonances of M1G or N2-(3-oxo-1-propenyl)-dG. Alternatively, UV absorbance was monitored at 320 nm. For NMR experiments, the strand concentration of oligodeoxynucleotide was 0.7 mM, in buffer of 0.1 M NaCl, 10 mM NaH2PO4, and 50 μM Na2EDTA. For UV detection, the strand concentration was ≈10 μM in 1.0 M NaCl, 10 mM NaH2PO4, and 50 μM Na2EDTA.
Molecular Modeling.
Potential energy minimization calculations were performed by using xplor 2.4 (33) implemented with the charmm (34) force field. The empirical energy function (35) consisted of energy terms for bonds, bond angles, torsion angles, tetrahedral and planar geometry, hydrogen bonding, and nonbonded interactions, including van der Waals and electrostatic forces. The van der Waals energy term was approximated by using the Lennard-Jones potential energy function. The electrostatic term used the Coulomb function, based on a full set of partial charges (−1/residue). A distance dependent dielectric constant of 4 was applied for all calculations.
RESULTS
M1G Spontaneously Converts to N2-(3-oxo-1-propenyl)-dG at Neutral pH When Placed Opposite Cytosine in Duplex Oligodeoxynucleotides.
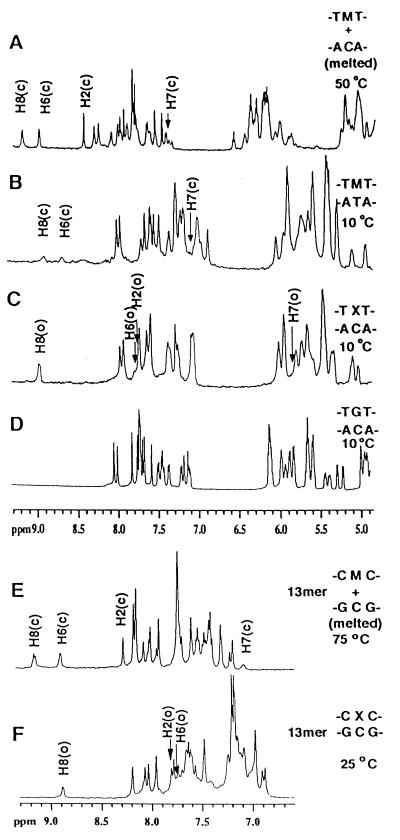
On addition of the complementary strand to the M1G-modified oligodeoxynucleotides at neutral pH, thus placing M1G opposite cytosine, spontaneous and quantitative conversion to N2-(3-oxo-1-propenyl)-dG occurred. This was observed both for the 5′-TMT-3′⋅5′-ACA-3′ and the 5′-CMC-3′⋅5′-GCG-3′ sequence contexts in d(GGTXTCCG)⋅d(CGGACACC) and d(ATCGCXCGGCATG)⋅d(CATGCCGCGCGAT). Below the melting temperature, the H6, H7, and H8 resonances derived from the malondialdehyde moiety were observed at ≈8.9, 5.8, and 7.8 ppm, respectively, consistent with conversion to N2-(3-oxo-1-propenyl)-dG. The imidazole proton H2 (corresponding to H8 in deoxyguanosine) shifted to 7.9 ppm. Small sequence-specific differences in the chemical shifts were observed in the two oligodeoxynucleotides. A coupling constant of JH7,H8 = 12 Hz indicated coupling from an aldehyde proton into a trans vinylic system. Likewise, JH6,H7 = 18 Hz was consistent with trans orientation about the vinylic bond. The long range coupling JH6,H8 was concluded to be <5Hz because it was not resolved (Fig. 3). The trans configuration also was suggested by the strong nuclear Overhauser effect between the propenyl H6 and H8 protons. The linewidths of the H6, H7, and H8 protons increased as compared with M1G, which was attributed to amino-enol tautomerism (Fig. 1). The NMR evidence argued that the amino tautomer predominated in both duplexes. This was indicated by observation of the aldehyde H8 resonance in the 9-ppm range (Fig. 3) and the characteristic aldehyde carbon resonance, which was observed in the 13C spectrum at 190 ppm.
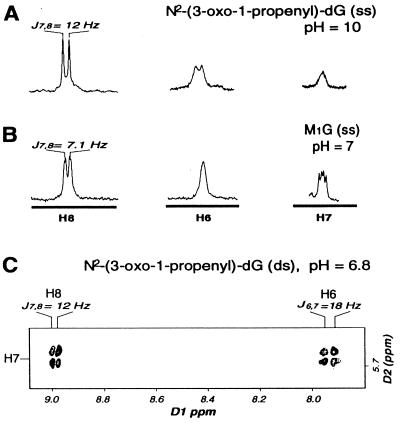
Figure 3.
Cytosine-mediated opening of M1G to N2-(3-oxo-1-propenyl)-dG is reflected by changes in scalar coupling constants. (A) 1H spectrum of N2-(3-oxo-1-propenyl)-dG resonances H6, H7, and H8 at pH 10. (B) 1H spectrum of M1G resonances H6, H7, and H8 at pH 7. (C) Expanded region of a 1H double quantum-filtered correlated spectroscopy spectrum of N2-(3-oxo-1-propenyl)-dG crosspeaks at pH 7.
For both d(GGTXTCCG)⋅d(CGGACACC) and d(ATCGCXCGGCATG)⋅d(CATGCCGCGCGAT), the opening of the exocyclic ring of M1G allowed observation of a resonance for the imino proton N1H of N2-(3-oxo-1-propenyl)-dG in double-stranded DNA (Fig. 4). This was not present in M1G because of the exocyclic 1,N2 ring of the modified guanine. This resonance was in both instances observed at ≈11.4 ppm, ≈1 ppm upfield of the chemical shift range normally observed for Watson-Crick hydrogen bonded G⋅C base pairs. The upfield shift of X4 N1H in both instances suggested that ring-opening of M1G to the N2-(3-oxo-1-propenyl)-dG derivative was not accompanied by base pair formation with the cytosine in the complementary strand.
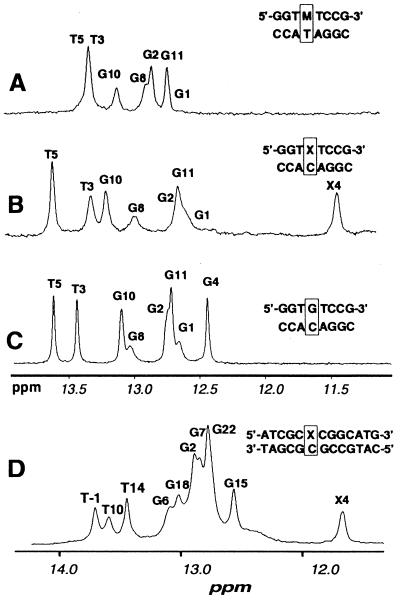
Figure 4.
1H NMR spectra of the far downfield imino proton region at 5°C and pH 6.8. (A) The unadducted duplex d(GGTGTCCG)⋅ d(CGGACACC). (B) The duplex d(GGTXTCCG)⋅d(CGGACACC). (C) The duplex d(GGTMTCCG)⋅d(CGGATACC). (D) The duplex d(ATCGCXCGGCATG)⋅d(CATGCCGCGCGAT).
The modified oligodeoxynucleotides remained in right-handed duplexes. Sequential 1H resonance assignments for the unmodified and modified strands were obtained from nuclear Overhauser effect spectroscopy, double quantum-filtered correlated spectroscopy, and total correlation spectra by using established protocols (36, 37). Complete sets of aromatic-to-anomeric proton sequential connectivities were observed for both modified and complementary strands. There were no interruptions to the sequential NOEs at or adjacent to the adducted site. The NMR data revealed that the expected cytosine H5 → H6 cross-peaks, thymine CH3 → H6 cross-peaks, and all expected H1′/H2′ and H1′/H2′′ cross-peaks were observed in their characteristic spectral regions. The magnitudes of the nuclear Overhauser effects between the H8 (imidazole) protons and the H1′ (anomeric) protons of the N2-(3-oxo-1-propenyl)-dG derivatives were consistent with the N2-(3-oxo-1-propenyl)-dG derivatives existing in the anti-conformation about the glycosyl bond χ.
Cytosine Accelerates the Rate of M1G Ring-Opening at Neutral pH.
At neutral pH, opening of M1G to N2-(3-oxo-1-propenyl)-dG occurred at an observable rate only when cytosine was placed opposite M1G in duplex DNA. For the 0.7 mM d(GGTMTCCG)⋅d(CGGACACC) sample containing 0.1 M NaCl, 10 mM Na2HPO4, and 50 μM Na2EDTA, ring-opening at 5°C was 90% complete in <5 min. For both d(GGTMTCCG)·d(CGGACACC) and d(ATCGCMCGGCATG)⋅d(CATGCCGCGCGAT), cytosine-promoted ring-opening was reversible when the duplexes were thermally denatured. On heating the duplexes above their melting temperatures, M1G was regenerated. For the 0.7 mM d(GGTXTCCG)⋅d(CGGACACC) sample containing 0.1 M NaCl, 10 mM Na2HPO4, and 50 μM Na2EDTA at pH 7, at 60°C N2-(3-oxo-1-propenyl)-dG was 90% converted to M1G in <5 min.
M1G in the single-stranded oligodeoxynucleotides, as well as M1G nucleotide, was stable indefinitely at neutral pH (15). The extent to which M1G and N2-(3-oxo-1-propenyl)-dG might be in rapid equilibrium, favoring M1G at neutral pH, was tested in D2O buffer by monitoring for deuterium exchange at M1G H8. No exchange was observed when the sample was monitored for a period of several weeks. To examine the role of the complementary cytosine in promoting ring opening of M1G, d(GGTMTCCG)⋅d(CGGATACC) was synthesized in which M1G was placed opposite T. M1G was stable at neutral pH in the M1G⋅T duplex and did not undergo ring-opening, evidenced by the observation of resonances for H6, H7, and H8, which were in agreement with the M1G in the single strand at pH 7 (Fig. 5). In that instance, the M1G H6, H7, and H8 resonances were assigned at ≈7.4, 9.0, and 9.3 ppm from total correlation spectroscopy experiments; the imidazole H2 (H8 in deoxyguanosine) resonances were at ≈8.4 ppm. Double quantum-filtered correlated spectra revealed JH6,H7 of ≈4 Hz and JH7,H8 of 7.1 Hz. Inspection of the imino region of the d(GGTMTCCG)⋅d(CGGATACC) mismatched duplex confirmed that M1G remained intact (Fig. 4). No resonance for a non-hydrogen-bonded guanine N1H proton was observed at or near 11.4 ppm. Integration of the imino region of the spectrum yielded a total of seven Watson-Crick base pairs. The imino resonance of the non-hydrogen-bonded thymine at the mismatch site was not observed, which was attributed to rapid exchange with solvent. This was consistent with the broadening of the M1G resonances and lower thermal stability of this duplex.
Figure 5.
Aromatic and anomeric regions of 1H NMR spectra at pH 6.8. (A) The unmodified duplex d(GGTGTCCG)⋅d(CGGACACC) at 10°C. (B) The duplex d(GGTXTCCG)⋅d(CGGACACC) at 10°C. (C) The duplex d(GGTMTCCG)⋅d(CGGATACC) at 10°C. The broadening of the M1G H6, H7, and H8 resonances was attributed to the low thermal stability of the M1G⋅T mismatch. An additional source could be conformational exchange of M1G about the glycosyl bond, as observed for its saturated analog PdG (38). (D) The duplex d(GGTXTCCG)⋅d(CGGACACC) at 50°C after thermal denaturation. (E) The duplex d(ATCGCXCGGCATG)⋅d(CATGCCGCGCGAT) at 10°C. (F) The duplex d(ATCGCXCGGCATG)⋅d(CATGCCGCGCGAT) after thermal denaturation.
In the absence of cytosine, M1G underwent measureable ring opening to N2-(3-oxo-1-propenyl)-dG only under basic conditions. Moreover, the rate of ring-opening at pH 10 was slower than in the presence of cytosine at neutral pH. For both d(GGTMTCCG) and d(ATCGCMCGGCATG) at pH 10, M1G underwent slow ring-opening. As investigated for d(GGTMTCCG) at 25°C, after 30 min, and at pH 10, the conversion of M1G to N2-(3-oxo-1-propenyl)-dG was 50% complete. About 90% of the M1G converted after 5 hr. A small quantity of M1G remained detectable in the spectrum after 24 hr at pH 10. Under these conditions, the H6, H7, and H8 resonances shifted to ≈8.0, 5.6, and 9.0 ppm. The imidazole H8 resonances were at ≈8 ppm. The scalar couplings JH6,H7 of 18 Hz and JH7,H8 of 12 Hz were observed. Ring opening was reversible in the single-stranded oligodeoxynucleotides on lowering pH. The reverse rate of conversion from N2-(3-oxo-1-propenyl)-dG to M1G was investigated in the single-stranded oligodeoxynucleotides at pH 6.5. In the case of d(GGTXTCCG), ≈50% of N2-(3-oxo-1-propenyl)-dG converted to M1G after 30 min. Almost all of the N2-(3-oxo-1-propenyl)-dG converted to M1G after 12 hr at pH 6.5, although a detectable amount of N2-(3-oxo-1-propenyl)-dG remained.
The Presence of Cytosine Opposite the Lesion Is Not Required to Stabilize N2-(3-oxo-1-propenyl)-dG in Duplex DNA.
N2-(3-oxo-1-propenyl)-dG was intentionally placed opposite thymine in duplex DNA at neutral pH to determine whether the ring-opened derivative would spontaneously recyclize to M1G in the absence of cytosine complementary to the lesion. For the 0.7 mM d(GGTXTCCG)⋅d(CGGATACC) sample containing 0.1 M NaCl, 10 mM Na2HPO4, and 50 μM Na2EDTA, no measureable recyclization occurred after 60 min at 5°C. Under these conditions, the N2-(3-oxo-1-propenyl)-dG derivative remained stable in duplex DNA. The stability of the N2-(3-oxo-1-propenyl)-dG lesion depended on maintenance of the DNA duplex. The Tm of the X⋅T mismatched duplex was below 20°C at neutral pH, and, as the temperature was raised further, significant denaturation caused the N2-(3-oxo-1-propenyl)-dG lesion to be rapidly recyclized to M1G.
DISCUSSION
Detection of the ring-opened N2-(3-oxo-1-propenyl)-dG derivative of M1G in d(GGTXTCCG)⋅d(CGGATACC) and d(ATCGCXCGGCATG)⋅d(CATGCCGCGCGAT) reveals the ability of duplex DNA to catalyze the chemical transformation of one adduct into another. The observation that this occurs when M1G is placed in either the 5′-TMT-3′ or 5′-CMC-3′ sequences opposite cytosine in the complementary strand, but not when M1G is positioned opposite thymine in the complementary strand, suggests that it represents a general phenomenon associated with the positioning of M1G opposite cytosine in the complementary strand of duplex DNA.
Cytosine-Catalyzed Ring-Opening of M1G.
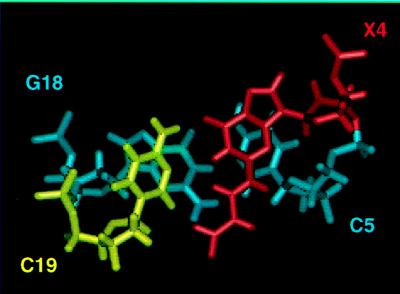
The observation that, in duplex DNA at neutral pH, M1G ring-opening occurred only when cytosine was placed opposite the adduct suggested the role of cytosine in catalyzing the ring-opening of M1G to the N2-(3-oxo-1-propenyl)-dG derivative. An alternative hypothesis, that the complementary cytosine acted by stabilizing N2-(3-oxo-1-propenyl)-dG by lowering its free energy, was considered. However, this was not consistent with the observation that N2-(3-oxo-1-propenyl)-dG was not formed in the absence of cytosine. Also, 1H NMR data in the 5′-CXC-3′⋅5′-GCG-3′ sequence showed that N2-(3-oxo-1-propenyl)-dG did not interact with cytosine but, rather, was oriented into the minor groove (Fig. 6). Watson–Crick base pairing was not observed between the N2-(3-oxo-1-propenyl)-dG derivative and the complementary cytosine. Finally, the observation that N2-(3-oxo-1-propenyl)-dG was stable when intentionally placed opposite thymine in duplex DNA suggested that cytosine was not necessary for stabilization of the ring-opened species.
Figure 6.
Structure obtained from molecular dynamics calculations by using NMR distance restraints for the (3-oxo-1-propenyl)-dG derivative of M1G in d(ATCGCXCGGCATG)⋅d(CATGCCGCGCGAT).
1H NMR data for the N2-(3-oxo-1-propenyl)-dG derivative contained in the 5′-CXC-3′⋅5′-GCG-3′ sequence provided a plausible explanation as to why, at neutral pH in duplex DNA, cytosine-promoted formation of N2-(3-oxo-1-propenyl)-dG was facile (Fig. 6). The ring-opened derivative exhibited the trans configuration of the propenal, as demonstrated by 1H J-coupling values. Consequently, both the amino proton and the 3-oxo-1-propenyl derivative were oriented into the minor groove, and the ring-opened species was easily accommodated by the DNA duplex with minimal structural perturbation. In contrast, modeling predicted that accommodation of the exocyclic ring of M1G would require substantial perturbation to duplex DNA, as observed for its fully saturated analog, 1,N2-propanodeoxyguanosine (PdG) (38).
Proposed Mechanism of M1G Ring-Opening.
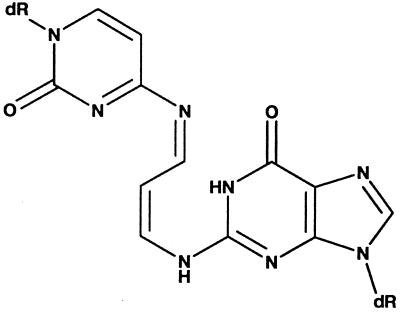
The spatial relationship between the exocyclic amino group of the cytosine complementary to M1G and the C8 position of M1G was examined by constructing a model duplex in which M1G in the anticonformation about the glycosyl bond χ was positioned opposite cytosine in the complementary strand, followed by potential energy minimization. The energy minimized structure suggested that, in the anti-conformation, the cytosine amino group was positioned to attack M1G at the major groove face of the exocyclic ring, i.e., from the N1 point of closure as opposed to the N2 side of the pyrimidopurinone moiety (Fig. 2B). This would facilitate ring opening at neutral pH, probably via formation of a transient Schiff base (Fig. 7). Addition of water to the Schiff base would regenerate the catalytic cytosine and would result in the formation of the N2-(3-oxo-1-propenyl)-dG derivative. This hypothesis derived support from UV and NMR spectroscopic experiments that revealed the existence of Schiff bases when M1G-dR was reacted with tris-hydroxymethylaminomethane (39) and with glycine (N.C.S.-B., H.M., G. Ramachandra Reddy, and L.J.M., unpublished work).
Figure 7.
Proposed Schiff base intermediate after attack by the exocyclic amino group of cytosine at C8 of M1G.
Biological Implications.
Structurally, M1G and its N2-(3-oxo-1-propenyl)-dG derivative are sufficiently different that their independent presence in DNA may result in significant differences in mutagenic potential and susceptibility to repair. The reversible transformation of M1G to the N2-(3-oxo-1-propenyl)-dG derivative may explain the observation that M1G located on viral genomes is more mutagenic when the base opposite the adduct is T than when it is C (40). M1G positioned opposite T in the M13 phage genomes was ≈5-fold more mutagenic than M1G positioned opposite C, but the mutation spectra were identical in the two vectors. When similar experiments were conducted with the stable M1G analog PdG, no difference in the frequency of mutations induced was detected between vectors containing C or T opposite the lesion. One interpretation is that M1G is considerably more mutagenic than N2-(3-oxo-1-propenyl)-dG, so that the frequency of mutations induced at the adduct site depends on the amount of M1G present at the replication fork. In vectors with M1G positioned opposite C, the adduct is predicted to be present as N2-(3-oxo-1-propenyl)-dG, and the mutation frequency would depend on the rate of its recyclization to M1G after helicase-catalyzed strand separation.
Summary.
The idea that DNA contains complementary binding pockets analogous to those of enzymes for their substrates has been advanced to explain the exquisite sequence specificity controlling the binding of many electrophiles to DNA (41). Our findings extend the enzyme–substrate analogy by showing that the DNA duplex actively catalyzes a chemical transformation in a site-specific manner. In this instance, the DNA duplex plays an active role in directing the metabolism of a genotoxic chemical, catalyzing the chemical transformation of one adduct into another.
Acknowledgments
This work was supported by National Institutes of Health Grants CA-55678 (to M.P.S.) and CA-47479 (to L.J.M.). J.P.W. received support from a training grant in Molecular Biophysics, Grant GM-08320, from the National Institutes of Health. Funding for the NMR spectrometer was supplied by National Institutes of Health Grant RR-05805 and Vanderbilt Center in Molecular Toxicology Grant ES-00267. This project was supported by Center Grant CA-68485, to the E. Bronson Ingram Cancer Center at Vanderbilt University.
ABBREVIATION
- PdG
propanodeoxyguanosine
References
- 1.Pryor W A, Stanley J P. J Org Chem. 1975;40:3615–3617. doi: 10.1021/jo00912a038. [DOI] [PubMed] [Google Scholar]
- 2.Hamberg M, Samuelsson B. J Biol Chem. 1967;242:5336–5343. [PubMed] [Google Scholar]
- 3.Diczfalusy U, Falardeau P, Hammarstrom S. FEBS Lett. 1977;84:271–274. doi: 10.1016/0014-5793(77)80704-7. [DOI] [PubMed] [Google Scholar]
- 4.Mukai F H, Goldstein B D. Science. 1976;191:868–869. doi: 10.1126/science.766187. [DOI] [PubMed] [Google Scholar]
- 5.Benamira M, Johnson K, Chaudhary A, Bruner K, Tibbetts C, Marnett L J. Carcinogenesis. 1995;16:93–99. doi: 10.1093/carcin/16.1.93. [DOI] [PubMed] [Google Scholar]
- 6.Yau T M. Mech Ageing Dev. 1979;11:137–144. doi: 10.1016/0047-6374(79)90031-9. [DOI] [PubMed] [Google Scholar]
- 7.Spalding J W. Natl Toxicol Progr Tech Rep Ser. 1988;331:5–13. [Google Scholar]
- 8.Marnett L J, Basu A K, O’Hara S M, Weller P E, Rahman A F M M, Oliver J P. J Am Chem Soc. 1986;108:1348–1350. [Google Scholar]
- 9.Basu A K, O’Hara S M, Valladier P, Stone K, Mols O, Marnett L J. Chem Res Toxicol. 1988;1:53–59. doi: 10.1021/tx00001a010. [DOI] [PubMed] [Google Scholar]
- 10.Chaudhary A K, Nokubo M, Oglesby T D, Marnett L J, Blair I A. J Mass Spectrom. 1995;30:1157–1166. [Google Scholar]
- 11.Chaudhary A K, Reddy R G, Blair I A, Marnett L J. Carcinogenesis. 1996;17:1167–1170. doi: 10.1093/carcin/17.5.1167. [DOI] [PubMed] [Google Scholar]
- 12.Seto H, Okuda T, Takesue T, Ikemura T. Bull Chem Soc Jpn. 1983;56:1799–1802. [Google Scholar]
- 13.Seto H, Seto T, Takesue T, Ikemura T. Chem Pharm Bull. 1986;34:5079–5085. [PubMed] [Google Scholar]
- 14.Reddy G R, Marnett L J. Chem Res Toxicol. 1996;9:12–15. doi: 10.1021/tx950105t. [DOI] [PubMed] [Google Scholar]
- 15.Reddy G R, Marnett L J. J Am Chem Soc. 1995;117:5007–5008. [Google Scholar]
- 16.Wang M Y, Liehr J G. Carcinogenesis. 1995;16:1941–1945. doi: 10.1093/carcin/16.8.1941. [DOI] [PubMed] [Google Scholar]
- 17.Chaudhary A K, Nokubo M, Reddy G R, Yeola S N, Morrow J D, Blair I A, Marnett L J. Science. 1994;265:1580–1582. doi: 10.1126/science.8079172. [DOI] [PubMed] [Google Scholar]
- 18.Wang M, Dhingra K, Hittleman W N, Liehr J G, de Andrade M, Li D. Cancer Epidemiol. 1996;5:705–710. [PubMed] [Google Scholar]
- 19.Nath R G, Chung F-L. Proc Natl Acad Sci USA. 1994;91:7491–7495. doi: 10.1073/pnas.91.16.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nath R G, Chung F-L. Proc Am Assoc Cancer Res. 1993;34:137. [Google Scholar]
- 21.O’Nair J, Barbin A, Guichard Y, Bartsch H. Carcinogenesis. 1995;16:613–617. doi: 10.1093/carcin/16.3.613. [DOI] [PubMed] [Google Scholar]
- 22.Rouzer C A, Chaudhary A K, Nokubo M, Ferguson D M, Reddy G R, Blair I A, Marnett L J. Chem Res Toxicol. 1997;10:181–188. doi: 10.1021/tx9601216. [DOI] [PubMed] [Google Scholar]
- 23.Vaca C E, Fang J L, Mutanen M, Valsta L. Carcinogenesis. 1995;16:1847–1851. doi: 10.1093/carcin/16.8.1847. [DOI] [PubMed] [Google Scholar]
- 24.Fang J L, Vaca C E, Valsta L M, Mutanen M. Carcinogenesis. 1996;17:1035–1040. doi: 10.1093/carcin/17.5.1035. [DOI] [PubMed] [Google Scholar]
- 25.Sevilla C L, Mahle N H, Eliezer N, Uzieblo A, O’Hara S M, Nokubo M, Miller R, Rouzer C A, Marnett L J. Chem Res Toxicol. 1997;2:172–180. doi: 10.1021/tx960120d. [DOI] [PubMed] [Google Scholar]
- 26.Fink S P, Reddy G R, Marnett L J. Proc Natl Acad Sci USA. 1997;16:8652–8657. doi: 10.1073/pnas.94.16.8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Hara S M, Marnett L J. Mutat Res. 1991;247:45–56. doi: 10.1016/0027-5107(91)90032-j. [DOI] [PubMed] [Google Scholar]
- 28.Benamira M, Marnett L J. Chem Res Toxicol. 1993;6:317–327. doi: 10.1021/tx00033a011. [DOI] [PubMed] [Google Scholar]
- 29.Moe J G, Reddy G R, Marnett L J, Stone M P. Chem Res Toxicol. 1994;7:319–328. doi: 10.1021/tx00039a008. [DOI] [PubMed] [Google Scholar]
- 30.Weisenseel J P, Moe J G, Reddy G R, Marnett L J, Stone M P. Biochemistry. 1995;34:50–64. doi: 10.1021/bi00001a007. [DOI] [PubMed] [Google Scholar]
- 31.Piotto M, Saudek V, Sklenar V. J Mol Biol. 1992;6:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 32.Bax A, Davis D G. J Magn Reson. 1985;65:355–360. [Google Scholar]
- 33.Brunger A T. xplor 3.1: A System for X-Ray Crystallography and NMR. New Haven, CT: Yale Univ. Press; 1992. [Google Scholar]
- 34.Brooks B R, Bruccoleri R E, Olafson B D, States D J, Swaminathan S, Karplus M. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 35.Nilsson L, Clore G M, Gronenborn A M, Brunger A T, Karplus M. J Mol Biol. 1986;188:455–475. doi: 10.1016/0022-2836(86)90168-3. [DOI] [PubMed] [Google Scholar]
- 36.Reid B R. Q Rev Biophys. 1987;20:2–28. doi: 10.1017/s0033583500004212. [DOI] [PubMed] [Google Scholar]
- 37.Patel D J, Shapiro L, Hare D. Q Rev Biophys. 1987;20:35–112. doi: 10.1017/s0033583500004224. [DOI] [PubMed] [Google Scholar]
- 38.Singh U S, Moe J G, Reddy G R, Weisenseel J P, Marnett L J, Stone M P. Chem Res Toxicol. 1993;6:825–836. doi: 10.1021/tx00036a012. [DOI] [PubMed] [Google Scholar]
- 39.Niedernhofer L J, Riley M, Schnetz-Boutand N, Sanduwaran G, Chaudhary A K, Reddy G R, Marnett L J. Chem Res Toxicol. 1997;10:556–561. doi: 10.1021/tx960191c. [DOI] [PubMed] [Google Scholar]
- 40.Niedernhofer L J. Ph.D. dissertation. Nashville, TN: Vanderbilt Univ.; 1996. [Google Scholar]
- 41.Warpehoski M A, Hurley L H. Chem Res Toxicol. 1988;1:315–333. doi: 10.1021/tx00006a001. [DOI] [PubMed] [Google Scholar]