Abstract
We isolated a new mouse gene that is highly expressed in thymocytes, testis, and brain. This gene, SRG3, showed a significant sequence homology to SWI3, a yeast transcriptional activator, and its human homolog BAF155. SRG3 encodes 1,100 amino acids and has 33–47% identity with SWI3 protein over three regions. The SRG3 protein contains an acidic NH2 terminus, a myb-like DNA binding domain, a leucine-zipper motif, and a proline- and glutamine-rich region at its COOH terminus. Rabbit antiserum raised against a COOH-terminal polypeptide of the SRG3 recognized a protein with an apparent molecular mass of 155 kD. The serum also detected a 170-kD protein that seems to be a mouse homologue of human BAF170. Immunoprecipitation of cell extract with the antiserum against the mouse SRG3 also brought down a 195-kD protein that could be recognized by an antiserum raised against human SWI2 protein. The results suggest that the SRG3 protein associates with a mouse SWI2. The SRG3 protein is expressed about three times higher in thymocytes than in peripheral lymphocytes. The expression of anti-sense RNA to SRG3 mRNA in a thymoma cell line, S49.1, reduced the expression level of the SRG3 protein, and decreased the apoptotic cell death induced by glucocorticoids. These results suggest that the SRG3 protein is involved in the glucocorticoid-induced apoptosis in the thymoma cell line. This implicates that the SRG3 may play an important regulatory role during T cell development in thymus.
Progenitor T cells arise in the bone marrow and migrate to thymus, where they continue to develop. During the T cell development, more than 95% of developing immature thymocytes die by apoptosis as a consequence of negative selection or lack of positive selection (1). This apoptotic death targets mainly the cortical double-positive (CD4+CD8+) thymocytes. In thymocytes, apoptosis can be triggered by several exogenous stimuli such as glucocorticoids (2–4), removal of growth factors (5, 6), exposure to γ-irradiation (7), and antigen binding involving the CD3/ TCR (8–10). The effect of glucocorticoids (GCs)1 is selective; the immature CD4+CD8+ thymocyte fraction is rapidly killed by GC treatment, whereas both the precursor population (TCR−CD4−CD8−) and mature thymocytes (CD4+ or CD8+) are relatively resistant (11). It was reported that GC is produced within the thymus (12), and that transgenic expression of anti-sense RNA to glucocorticoid receptor (GR) significantly affects the thymocyte development (13). These results suggest that endogenous GC produced in thymus may participate as an important regulatory molecule of normal thymic development (14, 15).
GCs, when complexed with an activated receptor, can induce or inhibit the expression of specific genes, which may be related to the induction of apoptosis. The transcriptional regulation of downstream genes by GCs requires not only GR itself but several additional transcription factors such as the SWI–SNF protein complex (16–20). For example, the rat GR, when expressed in yeast, requires SWI–SNF proteins for transcriptional activation of GR-responsive genes and the GR–SWI3 complexes were coimmunoprecipitated in yeast extract (19, 20). In addition, antibodies against SWI3 interfere with the ability of rat GR to activate transcription in Drosophila melanogaster nuclear extracts (19).
SWI3 is a subunit of the SWI–SNF complex that seems to facilitate transcriptional activation by antagonizing the repressive actions of chromatin (21–23). The other subunits of the SWI–SNF complex so far identified include the SWI1 (ADR6), SWI2 (SNF2), SNF5, SNF6, SNF11, and SWP73. The complex was initially identified in Saccharomyces cerevisiae (S. cerevisiae) as a positive regulator of HO, a gene involved in mating type switching (24, 25), and SUC2, a glucose-repressible gene that encodes the enzyme invertase (26, 27). These SWI gene products were subsequently found to be required for the transcriptional activation of many other genes (28–31). Such activities of the SWI–SNF proteins are closely interconnected and they seem to function as components of a complex that associates with genespecific activators (31–35). The functional significance of the SWI–SNF complex is reflected by the evolutionary conservation of these genes in higher eukaryotes. Several higher eukaryotic homologues of SWI–SNF genes such as Drosophila homeotic gene activator brm, hbrm (also known as hSNF2α) and BRG1 (also known as hSNF2β) have been identified (36–39). In addition, a human protein homologue of SNF5 (40, 41) and mouse BAF60 (42), which is homologous to SWP73, have been identified. Recently, distinct complexes containing the BRG1 or hbrm that have an in vitro activity similar to yeast SWI–SNF have been purified from human cell lines (42, 43). From these complexes, the human BAF155 and BAF170 proteins that are homologous to SWI3 protein were identified (42).
In this paper, we describe a newly isolated mouse gene, the SWI3-related gene (SRG3), expressed in thymus and encoding a protein that shows significant amino acid sequence homology to both yeast SWI3 and human BAF155 proteins. The SRG3 protein coimmunoprecipitates with a mouse SWI2-like protein, suggesting their forming a protein complex in vivo. In addition, our data show that the SRG3 is expressed at much higher level in thymus than in peripheral lymphocytes. Because the GC is proposed to be a regulatory molecule in thymocyte development in thymus, and the SWI-related proteins have an important role in GC-mediated gene regulation, the high level expression of SWI3-related gene (SRG3) in thymocytes may imply that SRG3 has a crucial role in thymocyte development as a mediator of GC-induced transcriptional activation and apoptotic cell death of thymocytes. As a first step of testing this hypothesis, we analyzed the effect of downregulation of SRG3 expression in a GC-sensitive thymoma cell line on GC-induced apoptosis.
Materials and Methods
Mice and Cells.
C57BL/6J mice were maintained in the Institute for Molecular Biology and Genetics (Seoul National University, Seoul, Korea). The yeast strain CY165 (MATα, swi3Δ :: trp1-Δ1, HO–lacZ, ura3-52, leu2-Δ1, his3-Δ200, ade2-101, lys-801) cells, and yCP50 plasmid containing the SWI3 gene were gifts from C. Peterson (University of Massachusetts, Worcester, MA). Yeast cells were grown in synthetic minimal medium (0.67% Bactoyeast nitrogen base without amino acids; GIBCO BRL, Gaithersburg, MD) supplemented with leucine, histidine, adenine, and lysine to a mid-log phase. The mouse thymoma cell line, S49.1, was purchased from American Type Culture Collection (ATCC, Rockville, MD) and grown in DMEM supplemented with 10% fetal bovine serum.
Isolation and Purification of Poly(A)+ RNA.
Total RNA was isolated by CsCl banding, as described by Chomczynski and Sacchi (44). Intact thymi and spleens were collected from 3-5-wk-old mice and used as sources for RNA. Poly(A)+ RNA was isolated by oligo(dT)–cellulose chromatography (45).
Preparation of Subtractive Probe.
∼10 μg of poly(A)+ RNA was heated at 65°C for 2 min and annealed with 1 mg of magnetic beads containing oligo (dT) (Dynabeads oligo [dT]25; Dynal, Inc., Great Neck, NY) for 30 min at room temperature (46). The annealed poly(A)+ RNA was separated in magnetic field, and used as templates for the synthesis of the first-strand cDNA. The second-stranded cDNA was synthesized by random priming using hexanucleotides and 200 μCi of [α-32P]dCTP (3,000 Ci/mmol). To prepare a subtractive probe, 200 μg of the first-stranded spleen cDNA conjugated with magnetic beads were mixed with the labeled probe. After incubating at 55°C for 1 h, the labeled probe DNAs hybridized to the first-stranded spleen cDNA were removed using magnetic field, and the remaining subtractive probe was used for the screening of thymic cDNA library. The cDNA library was obtained from M.M. Davis at Stanford University (Stanford, CA).
DNA Sequencing and Computer Analysis.
To determine the nucleotide sequences, the restriction fragments of the cloned gene were subcloned into pBluscript (SK−) vector (Stratagene Inc., La Jolla, CA). Nested deletions were generated by the Erasea-Base system (Promega Corp., Madison, WI). The nucleotide sequence was determined by dideoxy chain termination method (47) using Sequenase 2.0 kit (United States Biochemical Corp., Cleveland, OH). Homology searches of the nucleotide and deduced amino acid sequences with sequences were performed at the National Center for Biotechnology Information, using the BLAST network service (48).
Separation of T and B Cell.
Single cell suspensions were prepared from intact spleens and lymph nodes of C57BL/6J mice. After the red blood cells were removed, cells were resuspended in PBS (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4, pH 7.3). To isolate T cells, single cell suspensions were sequentially reacted with biotinylated H57-597 antibody, which is specific to αβ TCR, and streptavidin-conjugated microbeads. The B cells were reacted with microbead-conjugated goat anti– mouse IgM. The cell-magnetic bead conjugates were separated by a magnetic cell sorter (MACS; Miltenyi Biotec, GmbH, Bergisch Gladbach, FRG) (49). The purity of isolated populations were confirmed by FACS® (Becton Dickinson, Mountain View, CA) analysis and Northern blot assay using TCF-1, a T cell–specific gene, as a probe.
Overexpression and Purification of GST–fusion Protein.
For the construction of GST–fusion protein, COOH-terminal region of the SRG3 gene was inserted into pGEX4T-2 vector in frame. DH5α cells harboring recombinant plasmids with GST–3C fusion were grown overnight and diluted to 1:200 in 200 ml of LB medium. After incubation at 37°C for 2 h with vigorous shaking, the culture was treated with 1 mM IPTG and then incubated for 3 h to induce expression of the fusion protein. Cells were harvested and resuspended in a sample loading buffer (50 mM Tris–HCl, pH 6.8, 100 mM DTT, 4% SDS, 0.2% BPB, 20% glycerol), and boiled for 2 min. These lysates were analyzed by electrophoresis on polyacrylamide gel. The overexpressed protein was purified by glutathion–sepharose 4B affinity chromatography as described by Smith and Johnson (50). The polyclonal antiserum was prepared by immunizing New Zealand white rabbit with the purified fusion protein.
Immunoprecipitation and Immunoblot Analysis.
The immunoprecipitation of the SRG3 and SWI2-like protein was performed by a method described by Muchardt et al. (40) with some modifications. The single cell suspension of the mouse thymus was harvested in immunoprecipitation (IP) buffer (20 mM Hepes, pH 7.6, 10% glycerol, 25 mM MgCl2, 0.1 mM EDTA, 0.2% NP-40) containing 0.1 M potassium acetate and 2.25 μg/ml pepstatin, 10 μg/ml leupeptin, 1 μg/ml soybean inhibitor, 2 mM PMSF, and 0.1 mM DTT. The cells were sonicated and debris were pelleted by centrifugation. The extracts were precleared with protein A–sepharose suspension and anti-SRG3 or anti-hSWI2 rabbit antiserum were added. After overnight incubation at 4°C, the extracts were incubated with protein A–sepharose suspension. The beads were washed three times in IP buffer containing 0.6 M potassium acetate, and once with IP buffer without Hepes. The precipitate was eluted by boiling in SDS-PAGE loading buffer. For immunoblot analysis, the proteins separated on SDS-PAGE were electrotransferred to nitrocellulose paper, and incubated in blocking solution (3% non fat dry milk, 50 mM Tris–HCl, pH 7.5, 150 mM NaCl) with gentle agitation for 2 h. After the blot was incubated with the SRG3 antiserum or hSWI2 antiserum, the specific bands were detected by treating the blot with anti-rabbit IgG conjugated with alkaline phosphatase in blocking solution (100 mM Tris–HCl, pH 9.5, 100 mM NaCl, 10 mM MgCl2) containing 165 μg/ml BCIP and 330 μg/ml NBT.
Complementation of Yeast swi3− Mutant.
For complementation study, the SWI3–SRG3 hybrid gene encoding the NH2-terminal part of SWI3 and the COOH-terminal part of SRG3 was synthesized. The full-length SRG3 gene or the SWI3–SRG3 hybrid gene were inserted into the pRS316GU vector containing the URA3 promoter and the URA3 gene as auxotrophic marker. The resulting constructs were used to transform CY165 yeast cells that harbor swi3 − mutation and the HO–lacZ fusion gene construct. The Ura + cells were cultured in synthetic minimal media. Cells were collected when OD595 of the culture reached 0.5. β-gal assay was performed as described by Breeden and Nasmyth (51). The Cp15 construct that contains the SWI3 gene in yCP50 vector was also used to transform CY165 cells as a positive control.
Preparation of the Glycerol Density Gradient Sedimentation Fraction from Thymocytes Extract.
The thymocytes were prepared as a single cell suspension from thymi and homogenized in 700 μl extraction buffer (40 mM Hepes, pH 7.3, 200 mM NaCl, 0.5 mM DTT, 2 mM EDTA, and 2.25 μg/ml pepstatin, 10 μg/ml leupeptin, 1 μg/ml soybean inhibitor). After centrifugation at 12,000 rpm for 20 min at 4°C, the supernatant containing 7.2 mg of total protein in 400 μl was layered on the top of linear 18–40% 10 ml glycerol gradient cushion containing 40 mM Hepes, pH 7.3, 200 mM NaCl. After ultracentrifugation at 36,500 rpm in Beckman SW41 rotor for 20 h at 4°C, the samples were collected from the bottom of the tube by fractionation into 33 tubes. These fractionated samples were analyzed by immunoblotting with SRG3 or hSWI2 antiserum.
Construction of a Plasmid Expressing Anti-sense RNA of the SRG3 Gene.
A 2.8 kb XbaI fragment spanning 60 bases of 5′-untranslated region and 2772 bases of the SRG3 coding sequence was inserted into the pRc/CMV vector (Invitrogen, San Diego, CA) in the anti-sense orientation. The resulting plasmid was designated as pRcASRG3. The plasmid construct, pRcASRG3, or pRc/CMV was transfected into S49.1, a thymoma cell line (ATCC), by electroporation. The transfected cells were selected and maintained with 1 mg/ml Geneticin (GIBCO BRL, Gaithersburg, MD) in DMEM supplemented with 10% fetal bovine serum.
Induction and Measurement of Apoptosis.
The apoptotic cell death of thymoma cells were induced by treatment with 10 μM hydrocortisone (Sigma Chem. Co., St. Louis, MO) for 72 h at 37°C. After hydrocortisone treatment, the cells were harvested and lysed in lysis buffer (10 mM EDTA, 50 mM Tris–HCl, pH 8.0, 0.5% sodium lauryl sarcosine) containing the 100 μg/ml proteinase K at 55°C for 2 h. The DNA was extracted with phenol/chloroform and precipitated with ethanol. After RNA was removed by RNAse A treatment, the DNA was analyzed in agarose gel electrophoresis to visualize the fragmented DNA. The apoptotic cell death was also measured by flow cytometry as described by Nicoletti et al. (52). In brief, the harvested cells were fixed with 70% ethanol and stained with 50 μg/ml of propidium iodide (PI) and 10 μg/ml RNAse A for 30 min at 4°C. After washing with PBS buffer, stained cells were analyzed using the FACStar® (Becton Dickinson, Mountain View, CA) for the DNA content.
Results
Cloning and Characterization of the Mouse SWI3-related Gene (SRG3).
We made an attempt to isolate genes that are specifically expressed in thymus but not in spleen by subtractive hybridization. One of the clones isolated was found to be expressed preferentially in thymus and was found to have similar amino acid sequences to a part of SWI3 protein of S. cerevisiae. The isolated gene has an open reading frame of 3,300 bp encoding 1,100 amino acids (Fig. 1). The homology search in the GenBank sequence database of NCBI using the BLASTP program showed amino acid sequence similarity to the SWI3 protein of S. cerevisiae (Fig. 2). The new gene was named as SRG3 to emphasize its relatedness to SWI3 gene.
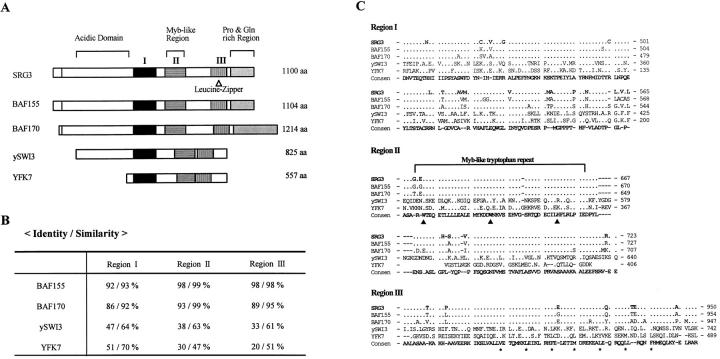
Figure 1.

Amino acid sequence of the SRG3 gene predicted from cDNA sequence. The predicted leucine-zipper motif is indicated by asterisks, and the myb-like tryptophan repeat is indicated by the closed triangles. The regions showing highest homology to the yeast SWI3 are underlined. These sequence data are available from EMBL/GenBank/DDBJ under accession number U85614.
Figure 2.
The comparison of amino acid sequences of the SRG3 with SWI3 and its human homologues, BAF155 and BAF170. The YFK7, another yeast homologue of SWI3, is also presented. The regions showing highest homology are shown by rectangles with distinctive fillings (A). The SRG3 and human homologues of SWI3 protein contain the proline- and glutamine-rich domains that lack in the yeast SWI3. Amino acid comparisons of the three regions (Region I, II, and III) are shown in B and C. The three homologous regions of SRG3 and SWI3 protein displayed 33–47% identity and 61–64% similarity. The amino acids that are identical to consensus sequences are indicated as dots (C).
The NH2-terminal part of the SRG3 is highly acidic; 23% of 231 amino acids (211–441) are either aspartate or glutamate. The COOH-terminal part has the leucine-zipper motif (53), and the proline- and glutamine-rich region. The proline and glutamine residues make up 44% over 150 amino acids (952–1,100). In addition, the myb-like tryptophan repeat domain or SANT (SWI3, ADA2, N-CoR, and TFIIIB B′′) domain (54), which may be involved in interaction with DNA, was also found in the middle part of this protein (Fig. 2 A). Therefore, this gene product seems to act as a transcriptional activator probably by interacting with other proteins or by interacting with DNA.
Recently, the human homologues of yeast SWI3, BAF155, and BAF170, were identified and designated as BRG1 associated factors (42). We found that the SRG3 protein has very high amino acid sequence homology to BAF155 protein (Fig. 2). They show 91% identity in their complete amino acid sequences, with the major deviations occurring at the NH2 termini. Therefore, we conclude that the SRG3 protein is the murine counterpart of human BAF155 protein. The SRG3 protein also matches well with the BAF170, another human SWI3 homologue; they are 60% identical and 70% similar at the amino acid level. In addition, the SRG3 protein has a considerable homology with the yeast YFK7 protein, which is also known as SWI3b (42) (Fig. 2).
The major similarity between the SWI3 and its mammalian homologues at the amino acid level is found over three regions (Fig. 2 B). The three regions of the SWI3 and SRG3 proteins shows 33–47% identity and 61–64% similarity. The NH2-terminal part of the SWI3 is also extremely acidic, yet similarity with the SRG3 is still very low. The leucine-zipper motifs in the region III of these proteins are different from SWI3 in that the third leucine of the SWI3 protein is replaced by phenylalanine in SRG3 and BAF155 (Fig. 2 C). In the COOH-terminal region, SWI3 lacks proline- and glutamine-rich regions found in its mammalian homologues. The data suggests that these proteins have similar protein structures and probably similar biochemical functions. However, there is divergence in structure and, probably, also in function between these proteins.
Identification of the SRG3 Gene Product.
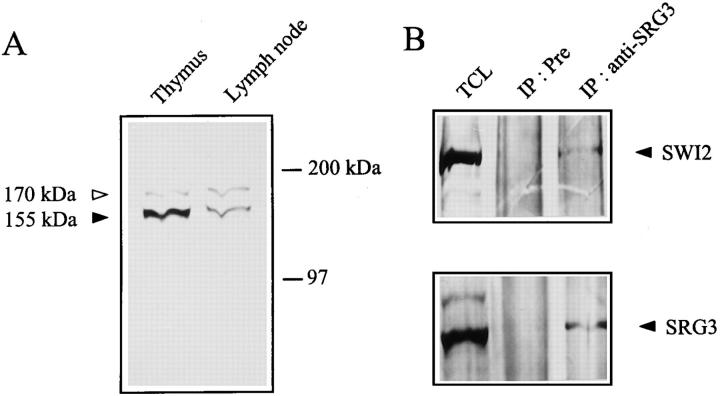
To identify the protein product of the SRG3 gene, polyclonal antibody was produced against the SRG3–GST fusion protein. The COOH-terminal part of the SRG3 gene was inserted inframe into the pGEX4T-2 plasmid containing the glutathione-S-transferase (GST) gene. The overexpressed GST– 3C fusion protein with ∼85 kD of molecular mass was used to immunize rabbits through subcutaneous injection. After primary and booster injections, polyclonal antiserum against GST–3C fusion protein was obtained. The antiserum was confirmed to recognize specifically the fusion protein (data not shown). To identify the SRG3 gene product, immunoblot analysis was performed with crude extracts prepared from thymus and lymph node. As shown in Fig. 3 A, two bands of ∼155 and 170 kD were observed; the 155-kD protein is likely to be SRG3. This is supported by the observations that the protein matches well to the size of the BAF155 in human (42), that the antiserum immunoprecipitates the 155-kD protein (Fig. 3 B), and that the intensity of the 155-kD protein band was specifically reduced when anti-sense RNA to the SRG3 is expressed in a cell line (see Fig. 6 A). The antiserum also recognized a 170-kD protein that seemed to be similar to the SRG3 protein in its structure. Considering the results of human SWI3 homologues (42), it is likely that the 170-kD protein may be the mouse counterpart of the human BAF170 protein. The SRG3 and BAF170 are quite similar to each other over regions I (86%), II (93%), and III (89%) (see Fig. 2 B), and anti-SRG3 antiserum seems to recognize a murine BAF170like protein as well as SRG3. Interestingly, SRG3 is expressed at a three times higher level in thymus than in lymph nodes; however, the 170-kD protein is expressed at similar levels in both tissues.
Figure 3.
Immunoblotting and immunoprecipitation of the SRG3 protein. The overexpressed COOH-terminal part of SRG3 gene in Escherichia coli system was used to immunize rabbits to produce the polyclonal antiserum. When thymus and lymph node extract were blotted with the SRG3 antiserum, bands at 155 and 170 kD were detected (A). When the extract was blotted with the hSWI2 antiserum, a band at 195 kD (B, top) was detected. After immunoprecipitating the extract with the SRG3 antiserum, the precipitates were blotted with the SRG3 antiserum (B, bottom) or the hSWI2 antiserum (B, top), displaying the 155- and 195-kD bands, respectively. Immunoprecipitation with preimmune serum and blotting with the SRG3 and hSWI2 antiserum dose not show any band (B). TCL, total cell lysate; IP:Pre, immunoprecipitation with the pre immune serum; IP: anti-SRG3, immunoprecipitation with the SRG3 antiserum.
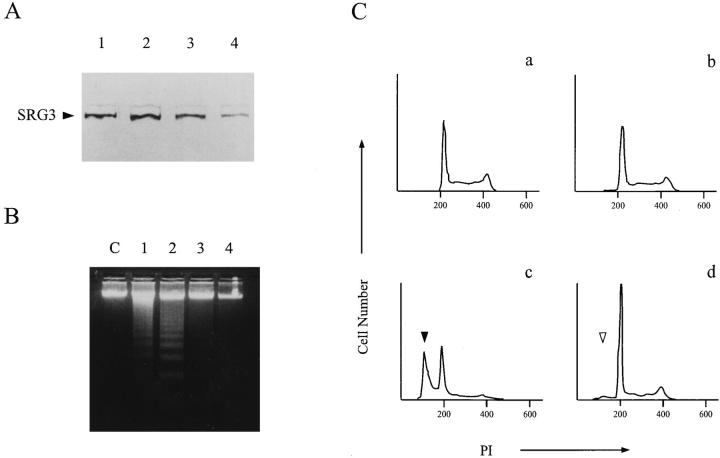
Figure 6.
Effect of SRG3 expression on GR mediated apoptosis. (A) Expression of the antisense RNA to SRG3 in thymoma cell line, S49.1, reduced the level of SRG3 protein. The pRcASRG3 plasmid expressing 2.9 kb of XbaI fragment (bases 1–2829) of SRG3 gene in anti-sense orientation under the control of CMV promoter was transfected into the S49.1 cells. The expression of the SRG3 protein in transfected cells was detected by immunoblotting using the SRG3 antiserum. lane 1, S49.1; lane 2, vector only; lane 3, pRcASRG3 transfectant, clone A; lane 4, pRcASRG3 transfectant, clone B. (B) Effects of SRG3 expression on apoptotic cell death induced by glucocorticoid treatment. The 10 mM of hydrocortisone was treated for 72 h (lanes 1–4) and the DNAs of each cell were electrophoresed on 2% agarose gel containing ethidium bromide. The DNA fragmentation was reduced as SRG3 expression level was reduced. Lane C, control (untreated S49.1); lane 1, S49.1; lane 2, vector only; lane 3, pRcASRG3 transfectant, clone A; lane 4, pRcASRG3 transfectant clone B. (C) FACS® analysis of the DNA contents of the cells transfected with vector only (a and c) and pRcASRG3 transfectant, clone B (b and d). The subdiploid peak (closed and open arrowheads) indicates apoptotic cells induced by glucocorticoid treatment (c and d).
We tested whether the SRG3 protein is associated with other proteins such as a mouse SWI2-like protein. After immunoprecipitation of cell extract with the antiserum followed by blotting with the same antiserum, the specific band of 155 kD was observed (Fig. 3 B, bottom). We have also employed the hSWI2 antiserum that recognized a protein with molecular mass ∼195 kD in thymocyte extract (Fig. 3 B). The 195-kD protein is similar to the size of hSWI2 (43) and seems to be a mouse homologue of hSWI2. When thymocyte extract was immunoprecipitated with the SRG3 antiserum and blotted with the hSWI2 antiserum, a specific band corresponding to the mouse SWI2like protein was detected (Fig. 3 B, top). These results suggest that the SRG3 associates with a SWI2-like protein and possibly with other SWI–SNF proteins.
The SRG3 Protein Does Not Complement the Yeast swi3− Mutant.
To test the possibility that the mouse SRG3 gene product complements the yeast swi3 − mutation, the S. cerevisiae host strain CY165 (swi3 −) containing the HO–lacZ gene (25) was transformed with the pRS316GU vector containing the SRG3 full coding sequence or SWI3–SRG3 hybrid gene at the downstream of the URA3 promoter. Cells transformed with the SWI3–SRG3 hybrid gene were confirmed to express the hybrid protein by Western blotting using the SRG3 and SWI3 antisera (data not shown). The growth rate of swi3 − mutant was very slow (33) and was not significantly changed after the transformation with either the SRG3 or the SWI3–SRG3 hybrid. In addition, expression of lacZ gene, controlled by the HO promoter which requires the SWI–SNF protein complex for transcriptional activation, was not induced after the transformations of SRG3 or SWI3–SRG3 hybrid. The level of expression of lacZ gene in the SRG3 transformant was only ∼22% of the mutant cells transformed with the yeast SWI3 (a positive control), whereas the LacZ expression level in mutant cells transformed only with a vector plasmid was ∼20% of the SWI3 transformed cells. The result was consistent in three independent experiments.
The SRG3 mRNA Is Expressed Highly in Thymocytes, Brain, and Testis.
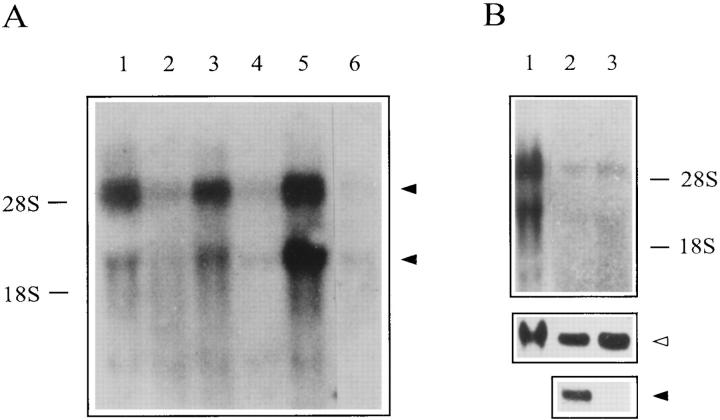
Northern blot analysis of SRG3 gene expression showed that the transcripts of this gene were 3.5 and 5 kb in size (Fig. 4 A). It seems that the 5.0-kb mRNA encodes the SRG3 protein because the 3.5-kb mRNA is not long enough to include the 3,300-base open reading frame, 5′- and 3′-untranslated regions, and poly(A) tail. At this point, it is not clear what the 3.5-kb mRNA species encodes for. However, when a 1.5-kb PstI fragment from the 3′-end of the SRG3 gene was used as a probe, only the 5-kb transcript was detected (data not shown), suggesting the possibility that these two transcripts are different at their 3′ termini. The SRG3 gene is expressed at higher levels in thymus, brain, and testis than in other tissues (Fig. 4 A). Northern blot analysis with RNAs isolated from separated splenic T and B cell populations showed that the two populations express similar level of SRG3. The separated population was highly pure, as judged by the Northern blot using TCF-1 gene, a T cell–specific gene (55, 56), as a probe (Fig. 4 B) and by FACS® analysis (data not shown). Interestingly, however, the level expressed in each peripheral lymphocyte population was only about 20% of that expressed in thymocytes (Fig. 4 B), as it was similarly shown by Western blot analysis (see Fig. 3 A).
Figure 4.
Northern blot analysis of SRG3 gene expression in different organs (A) and cell types (B). The same amount (15 μg) of total RNAs isolated from various tissues were analyzed by probing with a 1.8-kb HindIII fragment (bases 653–2361). (A) The lanes represent thymus (1), spleen (2), brain (3), lymph nodes (4), testis (5), and lung (6). Two transcripts of about 5 and 3.5 kb in size were expressed highly in thymus (lane 1), brain (lane 3), and testis (lane 5). (B) Both T and B cell expressed the SRG3. Lane 1, thymus; lane 2, T cells; lane 3, B cells. Normal T and B cells were separated from spleen and lymph nodes by magnetic activated cell sorter (miniMACS). The purity of the separated population was tested by probing the RNA blot with TCF-1 (closed arrowhead). Both B and T cells expressed about the same levels of SRG3, as judged by the control β-actin probe (open arrowhead).
The Function of the SRG3 Protein in Thymocytes.
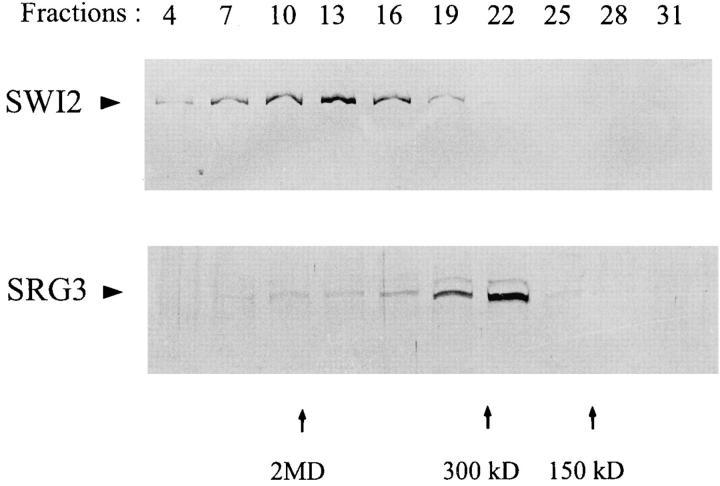
As shown in Fig. 3 A and Fig. 4, the level of SRG3 expression in thymus was much higher than the level in peripheral lymphocytes. Therefore, even though these proteins may form the SWI–SNF complex together with other proteins in mouse cells, it is also possible that the SRG3 protein exists independently of the SWI–SNF complex, especially in thymus. It has also been suggested that the SWI–SNF complexes are present as multiple forms in different tissues and cell lines (42). To see whether SRG3 protein exists mostly as a SWI– SNF complex, we fractionated the thymus extract according to the size of molecules by a glycerol density gradient. Each fraction was run on a gel and immunoblotted with the anti-SRG3 and the anti-hSWI2 antiserum. As shown in Fig. 5, the mouse SWI2 protein was mostly fractionated as a 2 MD complex as previously reported (42). However, the SRG3 protein was detected in a broad range of molecular masses, from 150 kD to 2 MD. Interestingly, a major portion of the protein was fractionated as 300 kD. The 170-kD protein was also fractionated similarly to SRG3 protein. These results suggest that a major portion of the SRG3 protein (and the 170 kD protein) exists independently of the SWI–SNF complex and that they may play a special role in developing thymocytes. This was similarly reported for BAF155 and BAF170 proteins; some BAF155 and BAF170 are not associated with human SWI–SNF complexes (42).
Figure 5.
Size fractionation of the SRG3 and SWI–SNF protein complexes. After total thymocytes extract was separated on glycerol density gradient sedimentation, the gradients were fractionated and immunoblotted with the SRG3 and hSWI2 antisera. The SRG3 protein was fractionated as separated complexes (300 kD) which are different from SWI2 complexes (2 MD). The blue–dextran (2 MD), thyroglobulin (669 kD), β-amylase (200 kD), and alcohol–dehydronase (150 kD) were used as standard molecular mass size markers.
One of the roles played by the SRG3 or the SWI–SNF complex in thymus may be mediating GR-induced transcriptional activation. One consequence of it is the induction of GC-mediated apoptosis in immature thymocytes. Thus, we have tested whether the SRG3 is required for the GC-mediated apoptosis in GC sensitive thymocyte cell line. A plasmid expressing the SRG3 gene in anti-sense orientation under the control of CMV promoter (pRcASRG3) was constructed. The pRcASRG3 construct was introduced into a GC-sensitive thymoma cell line, S49.1, through DNA transfection, and two clones, clone A and B, displaying reduced expression of SRG3 were selected. As shown in Fig. 6 A, clone A and B express ∼50 and 30% SRG3 protein of the vector transfectants, respectively. The DNA fragmentation induced by GC treatment was greatly reduced in these clones and this effect was more dramatic in clone B, which expressed lower level of SRG3 protein than clone A (Fig. 6 B). The reduction in apoptotic cell death of the pRcASRG3 transfectants was also confirmed by FACS® analysis of the DNA contents of the cells. After GC treatment, about 46% of the vector transfectants were subdiploid and apoptotic (Fig. 6 C, c); however, only about 4% of the treated clone B transfectants were apoptotic (Fig. 6 C, d). These results suggest that the SRG3 protein is involved in the GC-induced apoptosis of the thymoma cell line.
Discussion
The subunit proteins of the SWI–SNF complex were initially identified in S. cerevisiae as transcriptional activators of a set of genes. The SWI–SNF proteins seem to facilitate transcription by antagonizing the repressive actions of chromatin (21–23). The subunit proteins seem to function as components of a complex that associate with gene-specific activators. Mutations in any of the subunit genes resulted in very similar phenotypes and the phenotype of multiple defective swi − or snf − mutants was identical to that of a single swi − or snf − mutant (33). Besides, SWI–SNF proteins functioned interdependently in transcriptional activation (32). In addition, the SWI–SNF proteins were copurified and coimmunoprecipitated (31, 34) and were shown to be components of a large multisubunit complex (34, 35). Recently, human SWI–SNF complexes were shown to be present in multiple forms made up of 9–12 proteins and several of them were purified using the BRG1 antiserum (42, 43). There seems to exist several different forms of SWI–SNF complexes in a single cell and in different differentiated cell types (42). Based on this, it has been suggested that these complexes are involved in tissue-specific and developmental process–specific chromatin remodeling activity needed for the differentiation of a cell (42). The human homologues of yeast SWI3, the BAF155 and BAF170, were also identified and the two proteins seem to exist as core components in the same protein complex (42). SRG3 identified in the present study is homologous to human BAF155. The overall sequences are 94% similar to each other and the protein products are quite similar in size. The antiserum produced against SRG3 recognized an additional 170-kD protein, which seems to correspond to human BAF170, implying that there also exists two SWI3 homologues in mouse. In spite of this similarity, the expression patterns of BAF155 and SRG3 seem to be somewhat different. BAF155 is selectively expressed in muscle and heart but expressed at relatively low levels in liver and brain (42); however, SRG3 is expressed at much higher level in brain than in liver (Fig. 4 A). The selective expression patterns of BAF155 and BAF170 in different tissues were quite similar to each other. However, mouse SRG3 is expressed at higher level in thymus than in peripheral lymphoid tissues (Fig. 3); on the other hand, the 170-kD protein is expressed at similar levels in these tissues. Interestingly, some SRG3 proteins and the 170 kD proteins seem to exist independently of the SWI–SNF complex (Fig. 5). It is not yet conclusive whether some SRG3 and 170-kD proteins exist as heterodimers or homodimers. These results suggest that even though the human and mouse homologues of yeast SWI3 are structurally and functionally similar, they may play distinct roles in each species and in different tissues.
In yeast, the rat GR can activate transcription from a promoter bearing the GR-responsive element in the presence of glucocorticoid (GC) (57, 58). It has been reported that transcriptional activation by GR, which regulates the expression of a network of genes in a tissue-specific manner, is dependent on SWI1, SWI2, SWI3, SWP73 gene functions in yeast (19, 59). Furthermore, SWI3 was coimmunoprecipitated with GR in yeast extracts (19), suggesting that the two proteins interact directly upon activation of GR. Our study showed that SRG3 was highly expressed in developing thymocytes compared with mature peripheral T and B lymphocytes (Fig. 3). When the level of SRG3 protein was reduced to ∼50–30% of normal level of thymoma cells by expressing anti-sense RNA to the gene, apoptosis induced by GC on these cells was significantly reduced (Fig. 6). These results indicate that SRG3 is required for GC-induced apoptosis in the thymoma cells and suggests that SRG3 is an important factor for GC-mediated regulation of thymocyte development.
It has been hypothesized that GC might affect thymocyte development in a number of ways. Thymocytes respond to GC by apoptosis both in vitro and in vivo (2, 60). In vivo, the immature CD4+CD8+ thymocyte population is rapidly killed in the presence of GC, whereas both the CD4−CD8− precursor population expressing no TCR− and mature thymocytes (CD4+ or CD8+) cells expressing high levels of TCR are relatively resistant. Using anti-CD3 monoclonal antibodies as a model for negative selection, it has been found that pretreatment of mice with a GR antagonist, RU486, protects immature CD4+CD8+ thymocytes from apoptosis (61). Furthermore, it has been reported that radioresistent thymic epithelial cells constitutively produce GC (12). Thus, GC and GR may function as important regulators in normal thymic differentiation (14, 15). At this point, it is not clear whether GC-induced apoptosis of thymoma cells requires SRG3 as a component of the SWI– SNF complex or as a separate entity. In yeast, transcriptional activation by GR is blocked by disrupting any one of SWI1, SWI2, and SWI3 genes, suggesting the possibility that the SWI–SNF complex is required for the GC sensitivity of thymocytes. However, it is noteworthy that SRG3 protein is expressed at a much higher level in thymocytes than in peripheral T lymphocytes and that the major part of SRG3 protein may exist independently of the SWI–SNF complex in thymus. Furthermore, even though the antisense RNA expression reduced the level of SRG3 protein in the transfected cells, there still remained ∼50% (clone A) or 30% (clone B) of normal level of SRG3 protein (Fig. 6). Even in the case of the clone B transfectant, this is at least the similar level of protein found in peripheral T lymphocytes (Fig. 3). Therefore, it is likely that there still may be enough SRG3 protein left to form SWI–SNF complexes in the transfectants. These results suggest a possibility that SRG3 protein may function independently of the SWI– SNF complex in GC-mediated apoptosis. No matter how SRG3 functions in GC-mediated apoptosis in thymoma cells, either as a component of SWI–SNF complex or as an independent factor of the complex, our present results show that SRG3 protein is required for the process and possibly plays an important regulatory role during thymocyte development.
Acknowledgments
We thank Dr. M.M. Davis for the kind gift of mouse thymus cDNA library and Dr. C. Peterson for providing yeast strains used in this study.
This work was supported in part by the S.N.U.-Daewoo Research Fund (94-06-2068, 96-06-2078) and the Biotech 2000 project to R.H. Seong, and in part by grants from the Korea Science and Engineering Foundation, through the Research Center for Cell Differentiation, to S.D. Park and R.H. Seong.
Footnotes
1 Abbreviations used in this paper: GC, glucocorticoids; GR, glucocorticoid receptor; GST, glutathione-S-transferase; IP, immunoprecipitation; MACS, magnetic activated cell sorter; PI, propidium iodide.
References
- 1.von Boehmer H, Teh HS, Kisielow P. The thymus selects the useful, neglects the useless and destroys the harmful. Immunol Today. 1989;10:57–60. doi: 10.1016/0167-5699(89)90307-1. [DOI] [PubMed] [Google Scholar]
- 2.Wyllie AH. Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature (Lond) 1980;284:555–556. doi: 10.1038/284555a0. [DOI] [PubMed] [Google Scholar]
- 3.Cohen JJ, Duke RC. Glucocorticoid activation of a calcium-dependent endonuclease in thymocyte nuclei leads to cell death. J Immunol. 1984;132:38–42. [PubMed] [Google Scholar]
- 4.Compton MM, Cidlowski JA. Thymocyte apoptosis. A model of programmed cell death. TEM. 1992;3:17–23. doi: 10.1016/1043-2760(92)90087-h. [DOI] [PubMed] [Google Scholar]
- 5.Nieto M, Gonzalez A, Lopez A, Rivas, Diaz-Espada K, Gambon F. IL-2 protects against anti-CD3–induced cell death in human medullary thymocytes. J Immunol. 1990;145:1364–1368. [PubMed] [Google Scholar]
- 6.Williams G, Smith C, Spooncer E, Dexter T, Taylor D. Haemopoietic colony stimulating factors promote cell survival by suppressing apoptosis. Nature (Lond) 1990;343:76–79. doi: 10.1038/343076a0. [DOI] [PubMed] [Google Scholar]
- 7.Sellins KS, Cohen JJ. Gene induction by γ-irradiation leads to DNA fragmentation in lymphocytes. J Immunol. 1987;139:3199–3206. [PubMed] [Google Scholar]
- 8.McConkey DJ, Hartzell P, Amador-Pérez JF, Orrenius S, Jondal M. Calcium-dependent killing of immature thymocytes by stimulation via the CD3/T cell receptor complex. J Immunol. 1989;143:1801–1806. [PubMed] [Google Scholar]
- 9.Smith CA, Williams GT, Kingston R, Jenkinson EJ, Owen JJT. Antibodies to CD3/T-cell receptor complex induce death by apoptosis in immature T cells in thymic cultures. Nature (Lond) 1989;337:181–184. doi: 10.1038/337181a0. [DOI] [PubMed] [Google Scholar]
- 10.Shi Y, Bissonnette RP, Parfrey N, Szalay M, Kubo RT, Green DR. In vivo administration of monoclonal antibodies to the CD3 T cell receptor complex induces cell death (apoptosis) in immature thymocytes. J Immunol. 1991;146:3340–3346. [PubMed] [Google Scholar]
- 11.Jondal, M., Y. Xue, D.J. McConkey, and S. Okret. 1995. Thymocyte apoptosis by glucocorticoids and cAMP. In Apoptosis in immunology. G. Kroemer and C. Martinez-A., editors. Springer-Verlag, Berlin. 67–79. [DOI] [PubMed]
- 12.Vacchio MS, Papadopoulos V, Ashwell JD. Steroid production in the thymus: implications for thymocyte selection. J Exp Med. 1994;179:1835–1846. doi: 10.1084/jem.179.6.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.King LB, Vacchio MS, Dixon K, Hunziker R, Margulies DH, Ashwell JD. A targeted glucocorticoid receptor antisense transgene increases thymocyte apoptosis and alters thymocyte development. Immunity. 1995;3:647–656. doi: 10.1016/1074-7613(95)90135-3. [DOI] [PubMed] [Google Scholar]
- 14.Zacharchuk CM, Mercep M, Chakraborti P, Simons SS, Jr, Ashwell JD. Programmed T lymphocyte death: cell activation- and steroid-induced pathways are mutually antagonistic. J Immunol. 1990;145:4037–4045. [PubMed] [Google Scholar]
- 15.Iwata M, Hanaoka S, Sato K. Rescue of thymocytes and T cell hybridomas from glucocorticoid-induced apoptosis by stimulation via the T cell receptor/CD3 complex: a possible in vitromodel for positive selection of the T cell repertoire. Eur J Immunol. 1991;21:643–648. doi: 10.1002/eji.1830210316. [DOI] [PubMed] [Google Scholar]
- 16.Schüle R, Muller M, Kaltschmidt C, Renkawitz R. Many transcription factors interact synergistically with steroid receptors. Science (Wash DC) 1988;242:1418–1420. doi: 10.1126/science.3201230. [DOI] [PubMed] [Google Scholar]
- 17.Strähle U, Schmid W, Schütz G. Synergistic action of the glucocorticoid receptor with transcription factors. EMBO (Eur Mol Biol Organ) J. 1988;7:3389–3395. doi: 10.1002/j.1460-2075.1988.tb03212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imai E, Miner JN, Mitchell JA, Yamamoto KR, Granner DK. Glucocorticoid receptor–cAMP response element-binding protein interaction and the response of the phosphoenolpyruvate carboxykinase gene to glucocorticoids. J Biol Chem. 1993;268:5353–5356. [PubMed] [Google Scholar]
- 19.Yoshinaga SK, Peterson CL, Herskowitz I, Yamamoto KR. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science (Wash DC) 1992;258:1598–1604. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]
- 20.Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brmgenes potentiates transcriptional activation by the glucocorticoid receptor. EMBO (Eur Mol Biol Organ) J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kruger W, Herskowitz I. A negative regulator of HOtranscription, SIN1 (SPT2), is a nonspecific DNA-binding protein related to HMG1. Mol Cell Biol. 1991;11:4135–4146. doi: 10.1128/mcb.11.8.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirschhorn JN, Brown SA, Clark CD, Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992;6:2228–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- 23.Peterson CL, Tamkun JW. The SWI-SNF complex: a chromatin remodeling machine? . TIBS. 1995;20:143–146. doi: 10.1016/s0968-0004(00)88990-2. [DOI] [PubMed] [Google Scholar]
- 24.Stern M, Jensen R, Herskowitz I. Five SWI genes are required for expression of the HOgene in yeast. J Mol Biol. 1984;178:853–868. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- 25.Sternberg PW, Stern MJ, Clark I, Herskowitz I. Activation of the yeast HOgene by release from multiple negative controls. Cell. 1987;48:567–577. doi: 10.1016/0092-8674(87)90235-2. [DOI] [PubMed] [Google Scholar]
- 26.Carlson M, Osmond BC, Botstein D. Mutants of yeast defective in sucrose utilization. Genetics. 1981;98:25–40. doi: 10.1093/genetics/98.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neigeborn L, Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. . Genetics. 1984;108:845–858. doi: 10.1093/genetics/108.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Estruch F, Carlson M. SNF6 encodes a nuclear protein that is required for expression of many genes in Saccharomyces cerevisiae. . Mol Cell Biol. 1990;10:2544–2553. doi: 10.1128/mcb.10.6.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laurent BC, Treitel MA, Carlson M. The SNF5 protein of Saccharomyces cerevisiaeis a glutamine- and proline-rich transcriptional activator that affects expression of a broad spectrum of genes. Mol Cell Biol. 1990;10:5616–5625. doi: 10.1128/mcb.10.11.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Happel AM, Swanson MS, Winston F. The SNF2, SNF5 and SNF6 genes are required for Ty transcription in Saccharomyces cerevisiae. . Genetics. 1991;128:69–77. doi: 10.1093/genetics/128.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Côté J, Quinn J, Workman L, Peterson CL. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science (Wash DC) 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 32.Laurent BC, Treitel MA, Carlson M. Functional interdependence of the yeast SNF2, SNF5, and SNF6proteins in transcriptional activation. Proc Natl Acad Sci USA. 1991;88:2687–2691. doi: 10.1073/pnas.88.7.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peterson CL, Herskowitz I. Characterization of the yeast SWI1, SWI2, SWI3genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- 34.Cairns BR, Kim Y-J, Sayre MH, Laurent BC, Kornberg RD. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc Natl Acad Sci USA. 1994;91:1950–1954. doi: 10.1073/pnas.91.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peterson CL, Dingwall A, Scott MP. Five SWI/SNFgene products are components of a large multisubunit complex required for transcriptional enhancement. Proc Natl Acad Sci USA. 1994;91:2905–2908. doi: 10.1073/pnas.91.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okabe I, Bailey LC, Attree O, Srinivasan S, Perkel JM, Laurent BC, Carlson M, Nelson DL, Nussbaum RL. Cloning of human and bovine homologs of SNF2/ SWI2: a global activator of transcription in yeast S. cerevisiae. . Nucleic Acids Res. 1992;20:4649–4655. doi: 10.1093/nar/20.17.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamkun JW, Deuring R, Scott MP, Kissinger M, Pattatucci AM, Kaufman TC, Kennison JA. brahma: A regulator of Drosophilahomeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 38.Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature (Lond) 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 39.Chiba H, Muramatsu M, Nomoto A, Kato H. Two human homologues of Saccharomyces cerevisiae SNF2/ SWI2 and Drosophilabrahma are transcriptional coactivators cooperating with the estrogen receptor and the retinoic acid receptor. Nucleic Acids Res. 1994;22:1815–1820. doi: 10.1093/nar/22.10.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muchardt C, Sardet C, Bourachot B, Onufryk C, Yaniv M. A human protein homology to Saccharomyces cerevisiaeSNF5 interacts with the potential helicase hbrm. Nucleic Acids Res. 1995;23:1127–1132. doi: 10.1093/nar/23.7.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kalpana GV, Marmon S, Wang W, Crabtree GR, Goff SP. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science (Wash DC) 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- 42.Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 43.Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR. Nucleosome disruption and enhancement of activator binding by a human SWI/SNF complex. Nature (Lond) 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 44.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol– chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 45.Aviv H, Leader P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid–cellulose. Proc Natl Acad Sci USA. 1972;69:1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodriguez IR, Chader GJ. A novel method for the isolation of tissue-specific genes. Nucleic Acids Res. 1992;20:3528. doi: 10.1093/nar/20.13.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tools. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 49.Miltenyi S, Müller W, Weichel W, Radbruch A. High gradient magnetic cell separation with MACS. Cytometry. 1990;11:231–238. doi: 10.1002/cyto.990110203. [DOI] [PubMed] [Google Scholar]
- 50.Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia colias fusion with glutathione-S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 51.Breeden L, Nasmyth K. Cell cycle control of the yeast HOgene: cis- and trans-acting regulators. Cell. 1987;48:389–397. doi: 10.1016/0092-8674(87)90190-5. [DOI] [PubMed] [Google Scholar]
- 52.Nicoletti I, Migliorati G, Pagliacci MC, Grignani F, Riccardi C. A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. J Immunol Methods. 1991;139:271–279. doi: 10.1016/0022-1759(91)90198-o. [DOI] [PubMed] [Google Scholar]
- 53.Landschulz WH, Johnson PF, Mcknight SL. The leucine zipper, a hypothetical structure common to a new class of DNA binding proteins. Science (Wash DC) 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 54.Aasland R. The SANT domain: a putative DNAbinding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. TIBS. 1996;21:87–88. [PubMed] [Google Scholar]
- 55.van de Wetering M, Oosterwegel M, Dooijes D, Clevers H. Identification and cloning of TCF-1, a T lymphocyte–specific transcription factor containing a sequencespecific HMG box. EMBO (Eur Mol Biol Organ) J. 1991;10:123–132. doi: 10.1002/j.1460-2075.1991.tb07928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verbeek S, Izon D, Hofhuls F, Robanus-Maandag E, Riele H, van de Wetering M, Oosterwegel M, Wilson A, MacDonald HR, Clevers H. An HMG-box–containing T-cell factor required for thymocyte differentiation. Nature (Lond) 1995;374:70–74. doi: 10.1038/374070a0. [DOI] [PubMed] [Google Scholar]
- 57.Schena M, Yamamoto KR. Mammalian glucocorticoid receptor derivatives enhance transcription in yeast. Science (Wash DC) 1988;241:965–967. doi: 10.1126/science.3043665. [DOI] [PubMed] [Google Scholar]
- 58.Schena M, Freedman LP, Yamamoto KR. Mutations in the glucocorticoid receptor zinc finger region that distinguish interdigitated DNA binding and transcriptional enhancement activities. Genes Dev. 1989;3:1590–1601. doi: 10.1101/gad.3.10.1590. [DOI] [PubMed] [Google Scholar]
- 59.Cairns BR, Levinson RS, Yamamoto KR, Kornberg RD. Essential role of Swp73p in the function of yeast Swi/Snf complex. Genes Dev. 1996;10:2131–2144. doi: 10.1101/gad.10.17.2131. [DOI] [PubMed] [Google Scholar]
- 60.Gonzalo JA, Gonzàlez-Garcia A, Martìnez C -A, and G. Kroemer. Glucocorticoid mediated control of the activation and clonal deletion of peripheral T cells in vivo. . J Exp Med. 1993;177:1239–1246. doi: 10.1084/jem.177.5.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jondal M, Okret S, McConkey D. Killing of immature CD4+CD8+ thymocytes in vivoby anti-CD3 or 5′-(N-ethyl)-carboxamide adenosine is blocked by glucocorticoid receptor antagonist RU-486. Eur J Immunol. 1993;23:1246–1250. doi: 10.1002/eji.1830230608. [DOI] [PubMed] [Google Scholar]