Abstract
The chemokine receptors CXCR4, CCR2B, CCR3, and CCR5 have recently been shown to serve along with CD4 as coreceptors for HIV-1. The tropisms of HIV-1 strains for subgroups of CD4+ cells can be explained, at least partly, by the selective use of G protein–coupled receptors (GPCRs). We have identified a novel human gene, STRL33, located on chromosome 3 that encodes a GPCR with sequence similarity to chemokine receptors and to chemokine receptor–like orphan receptors. STRL33 is expressed in lymphoid tissues and activated T cells, and is induced in activated peripheral blood lymphocytes. When transfected into nonhuman NIH 3T3 cells expressing human CD4, the STRL33 cDNA rendered these cells competent to fuse with cells expressing HIV-1 envelope glycoproteins (Envs). Of greatest interest, STRL33, in contrast with CXCR4 or CCR5, was able to function as a cofactor for fusion mediated by Envs from both T cell line–tropic and macrophage-tropic HIV-1 strains. STRL33-transfected Jurkat cell lines also supported enhanced productive infection with HIV-1 compared with control Jurkat cells. Despite the sequence similarities between STRL33 and chemokine receptors, STRL33-transfected cell lines did not respond to any in a panel of chemokines. Based on the pattern of tissue expression of the STRL33 mRNA, and given the ability of STRL33 to function with Envs of differing tropisms, STRL33 may play a role in the establishment and/or progression of HIV-1 infection.
The seven transmembrane domain G protein–coupled receptors (GPCRs)1 constitute a large family of proteins that signal in response to agonists as diverse as polypeptide hormones, neurotransmitters, odorants, light, and cytokines (1, 2). Within this superfamily is the group of receptors for the chemokines, cytokines that are produced by many tissues and cell types and whose best-described activities are as chemotactic factors for leukocytes, including lymphocytes (3). To date, 10 human receptors that signal in response to chemokines have been described, designated by the most recent nomenclature CXCR1–4 for the receptors for chemokines in the CXC subfamily and CCR1, 2A, 2B, and 3–5 for the receptors for chemokines in the CC subfamily. In addition, a number of orphan GPCRs have been identified whose sequences suggest that they may be receptors for as yet unidentified chemokines (4).
Recent work has revealed novel relationships between chemokines–chemokine receptors and infection with HIV-1. Understanding these relationships has shed light on mechanisms of HIV-1 entry into cells and on the tropisms of HIV-1 strains for specific populations of CD4+ cells. All HIV-1 strains can infect primary T cells. However, as a result of differences among their envelope glycoproteins (Envs), some HIV-1 strains are able to infect macrophages (M-tropic) but not immortalized T cell lines; some, among them laboratory-adapted strains, are able to infect immortalized T cell lines (TCL-tropic) much more efficiently than they infect macrophages; and some strains are readily able to infect both types of cells. Analysis of clinical HIV-1 isolates suggests that M-tropic strains are critical for establishing and maintaining infection (reviewed in references 5, 6).
The CC chemokines MIP-1α, MIP-1β, and RANTES were found to suppress infection of lymphocytes by M-tropic HIV-1 strains (7). This report was followed by the functional cDNA cloning of a cofactor (designated fusin) that, along with CD4, mediates fusion and entry of TCL-tropic HIV-1 strains (8). Fusin, previously identified as an orphan GPCR, has recently been shown to be a receptor for the CXC chemokine SDF-1 and has been renamed CXCR4 (9, 10).
The discovery of the activity of CXCR4 was followed by the demonstration that chemokine receptor CCR5 can function as a cofactor with CD4 for cell fusion and/or viral entry and infection by M-tropic strains of HIV-1 (11–16). CCR5 has been demonstrated to play a critical role in the establishment of HIV-1 infection, because homozygosity for an allele encoding a fusion-defective CCR5, present in ∼1% of the Caucasian population, is strongly associated with resistance to HIV-1 infection (17–20). Nonetheless, HIV-1 infection has been reported in an individual homozygous for the defective CCR5 gene (21), indicating that under some circumstances CCR5 is not essential for infection. Identifying the range of coreceptors that can be used by HIV-1 is important, particularly if therapies targeting specific coreceptors place the virus under selective pressure.
As part of studies on the roles of chemokines in lymphocyte biology, we designed experiments to identify novel chemokine receptors in human T cells. In this report, we describe the molecular cloning of a cDNA for STRL33, a gene encoding a GPCR related to known chemokine receptors that is expressed in lymphoid tissues and activated T cells, and is induced in activated PBL. We demonstrate that STRL33, in marked contrast with CXCR4 and CCR5, can function with CD4 as a cofactor for cell fusion mediated by Envs of both M-tropic and TCL-tropic strains of HIV-1 and we show that transfection with STRL33 significantly enhances the ability of Jurkat cells to support productive infection with HIV-1. These data suggest that STRL33 may play a role in the establishment and/or the course of HIV-1 infection.
Materials and Methods
Cell Culture.
Jurkat, SUP-T1, U937, human embryonic kidney (HEK) 293, HeLa, and NIH 3T3 cells were obtained from American Type Culture Collection (ATCC, Rockville, MD). CEM clone 12D7 was obtained from Dr. G. Poli (National Institute of Allergy and Infectious Diseases, National Institutes of Health). Tumor-infiltrating lymphocytes (TIL) R4, R8, F9, and B10, prepared from human melanomas, were obtained from Dr. J.R. Yannelli (National Cancer Institute). EBV414 is an EBVtransformed B lymphoblastoid cell line obtained from Dr. R. Siliciano (Johns Hopkins University). Jurkat, SUP-T1, CEM, U937, and EBV414 cells were grown in RPMI-1640 with 10% fetal bovine serum (FBS). Elutriated PBL and monocytes were obtained from a normal donor by the Department of Transfusion Medicine (National Institutes of Health), and PBL were purified further by banding on Ficoll–Paque. For activation, 5 × 106 PBL/ml were cultured with 10 μg/ml PHA-P and 1 μM ionomycin for 3 d before harvesting for RNA. 293 cells were grown in MEM plus 10% horse serum. HeLa and NIH 3T3 cells were grown in DMEM with 10% FBS. TIL were grown in either RPMI-1640 with 10% FBS or in AIM-V (GIBCO BRL, Gaithersburg, MD), in each case supplemented with 500 U/ml IL-2, and the cells were stimulated periodically with 250 ng/ml PHA plus irradiated allogeneic PBMC.
Cloning of STRL33 cDNAs.
Total RNA was prepared from the F9 TIL using TRIzol reagent (GIBCO BRL, Gaithersburg, MD), poly(A)+ RNA was selected using oligo(dT) cellulose (Collaborative Biomedical Products, Bedford, MA), and first-strand cDNA was synthesized using oligo(dT) primers and the SuperScript Preamplification System (GIBCO BRL) according to the instructions of the manufacturer. For amplification, primer pools were designed based on transmembrane domain (TMD) II and TMD VII amino acid sequences from the human sequences for IL-8RA, IL-8RB, CCR1, and CCR2 and the murine homologues of IL-8RB and CCR1 and were 5′-GA(T/C)(C/T)TI(C/ T/G)TITT(T/C)(G/T/C)(C/T)I(T/C/A)TIACI(T/C)TICC, and 5′-CCIA (T/C)(A/G)AAI (G/A) (C/T) (A/G) TAIA (T/A/G) IA ( G / A/T/C)IGG(A/G)TT, respectively. Amplifications were done with cDNA synthesized from 0.015 μg of poly(A)+ RNA, with 1.5 μM of each primer pool in a 20 μl reaction volume with Taq polymerase and reagents from Perkin Elmer (Norwalk, CT), according to the instructions of the manufacturer. PCR was done using 30 cycles of denaturation at 94°C for 0.5 min, annealing at 45°C for 2 min, and chain extension at 72°C for 1.5 min.
1 μl from the first PCR was used in a second PCR done identically to the first and the products of the second reaction were separated on a 1.5% agarose gel from which fragments of the predicted size of ∼670 bp were purified and inserted by blunt end ligation into the vector pNOTA/T7 (5′ 3′ Prime, Inc., Boulder, CO). 88 ampicillin-resistant bacterial transformants were picked and, to eliminate known sequences, hybridizations were done with radiolabeled oligonucleotide probes for receptors CCR1, CCR2, CCR3, CXCR4, BLR1, EBI1 and STRL22. Among the inserts in the nonhybridizing colonies was a novel sequence designated STRL33.
Using poly(A)+ RNA from F9 TIL a λ ZAP Express (Stratagene, La Jolla, CA) cDNA library was prepared according to the instructions of the manufacturer. 1.4 × 106 recombinant phages from the nonamplified library were screened using a radiolabeled STRL33 probe. 10 positive phages were plaque-purified and the pBK-CMV (Stratagene, La Jolla, CA) plasmids containing STRL33 inserts were recovered by in vivo excision according to the instructions of the manufacturer. Manual and/or automated dideoxy sequencing was done for the entire cDNA clone STRL33.1, the 5′ nontranslated region of clone STRL33.2, the 5′ nontranslated region and open reading frame (ORF) of clone STRL33.3, and portions of other cDNA clones, some of which were obtained using RT–PCR.
Northern Blot Analysis.
Total RNA was prepared as above. DNAs used for probes were the following: IL8RA, IL8RB, CCR3, EBI1, and BLR1 genomic fragments and CCR2B cDNA obtained from Dr. P. Murphy (National Institute of Allergy and Infectious Diseases); STRL33, CCR1, CCR4, CCR5, CXCR4, and CMKBRL1 cDNAs that we isolated either from our λ library or by RT–PCR from TIL mRNA; and an STRL22 genomic fragment isolated as described (22). Hybridizations to leukocyte RNA were performed as described with washes in 0.1× SSC, 0.1% SDS at 50°C (23). Hybridizations with an oligonucleotide probe to 18S rRNA were as described (24). The blot of poly(A)+ RNA from human tissues was obtained from Clontech (Palo Alto, CA). Hybridizations were done according to the instructions of the manufacturer with washes as described above. Autoradiography/fluorography was done using an intensifying screen.
Production and Analysis of STRL33-transfected Cell Lines.
An EcoRI– EarI fragment containing the complete STRL33 ORF was isolated from the pBK-CMV/STRL33.1 plasmid and inserted into pCEP4 (Invitrogen, Carlsbad, CA) and pCIneo (Promega Corp., Madison, WI). The pCIneo/STRL33 DNA was transfected into HEK 293 cells by calcium phosphate precipitation (25) and the pCEP4/STRL33 DNA and pCEP4 without a cDNA insert were transfected into Jurkat cells by electroporation (26). Selection was in 200 μg/ml hygromycin B (Sigma, St. Louis, MO) and 1 mg/ml G418 (GIBCO BRL, Gaithersburg, MD) for pCEP4-transfected cells and pCIneo-transfected cells, respectively. Individual colonies of resistant 293 cells were cloned and expanded and Jurkat lines were derived by limiting dilution after the electroporation. Lines expressing the highest levels of STRL33 mRNA were used to test responses to chemokines using the fluorometric calcium flux assay as described (27), or for infection with HIV-1 as described below. Recombinant HuMig was obtained by infecting High Five cells of Trichoplusia ni (Invitrogen, Carlsbad, CA), as will be described elsewhere, and was purified by column chromatography as described (27). IP-10, MCP-1, MCP-2, MCP-3, RANTES, MIP-1α, MIP-1β, platelet factor 4, IL-8, and lymphotactin were purchased from Pepro Tech (Rocky Hill, NJ). MCP-4 was a gift from Dr. A. Luster (Harvard University). I309 and SDF-1 were gifts from R&D Systems (Minneapolis, MN).
Assays for Activity of STRL33 as a Fusion Cofactor.
Assays were done using a vaccinia-based Escherichia coli lacZ reporter gene assay for fusion between two cell populations, one expressing an HIV-1 Env and the other expressing CD4 (28). Vaccinia-mediated expression of GPCRs was achieved by transfection of 7 × 106 NIH 3T3 cells using DOTAP lipofectin (Boehringer Mannheim, Indianapolis, IN) with 10 μg of plasmids containing DNA encoding the GPCRs linked to the bacteriophage T7 promoter. The plasmid vectors were pCIneo (Promega Corp., Madison, WI) for STRL33 and pCDNA3 for CXCR4 (8) and CCR5 (11); as a negative control, pCIneo lacking an insert was used. After 4–5 h, the transfected cells were infected at 10 PFU/cell with recombinant vaccinia viruses vCB-3 encoding human CD4 (29) and vTF7-3 encoding T7 RNA polymerase (30). A separate population of HeLa cells was coinfected with vaccinia virus vCB-21R–LacZ containing lacZ encoding β-galactosidase (β-Gal), under control of a T7 promoter (31) and one of the following Env-encoding vaccinia viruses: vCB-41 encoding the LAV (NL4.3) Env (32), vCB-39 encoding the ADA Env (32), vCB-28 encoding the JR-FL Env (32), vCB-32 encoding the SF-162 Env (32), vCB-43 encoding the Ba-L Env (32), vSC60 encoding the IIIB Env (Chakrabarti, S., and B. Moss, personal communication), and vCB-16 encoding the nonfusogenic Unc Env (32). For experiments using the 89.6 Env, before infection with vCB-21R–LacZ, HeLa cells were transfected with 10 μg of the pSC59 plasmid (Chakrabarti, S., and B. Moss, personal communication) containing DNA encoding the 89.6 Env (33), a gift of R. Collman (University of Pennsylvania School of Medicine). Infected cells were incubated overnight at 31°C. Duplicate samples of 105 NIH 3T3 target cells and 105 Env-expressing cells were mixed; after 2.5 h cells were lysed and β-Gal activity was measured as described (28).
Infectivity Assay for HIV-1.
Virus stocks of HIV-1ELI1 were prepared by transfection of 293 cells with molecular clone pELI1 (34), and the amount of virus was determined by RT activity (35). 2 × 106 cells of the Jurkat cell lines were infected with the amount of virus corresponding to 105 cpm of RT activity as described (35). Virus production was measured by the appearance of RT activity in the medium.
Results and Discussion
To identify novel chemokine receptors expressed in T cells, we used RT–PCR with poly(A)+ RNA prepared from TIL line F9 and pools of degenerate primers based on conserved sequences in the TMDs of known chemokine receptors. We isolated a sequence encoding a novel GPCR, designated STRL33, for seven TMD receptors from lymphocytes clone 33. Southern blot analysis of human genomic DNA digested with BamHI, HindIII, and PstI revealed a single STRL33 gene, and using STRL33-specific primers and DNA prepared from a panel of human–hamster hybrid cell lines, STRL33 was localized to chromosome 3 (data not shown). This raises the possibility that STRL33 is in the cluster of genes for chemokine receptors CCR1, 2, 3, and 5 at 3p21, although the CCR proteins show >50% amino acid identity among themselves, demonstrating significantly closer relationships than what is seen in comparisons with STRL33 (see below).
Screening of a nonamplified λ cDNA library prepared from F9 TIL revealed an abundance for STRL33 mRNA of ∼0.01%. 10 cDNA clones were isolated and by restriction enzyme digestion, three size classes were identified. Representative cDNAs from each class, STRL33.1, STRL33.2, and STRL33.3, respectively, were evaluated in more detail by restriction analysis and partial sequencing, revealing that size differences among the clones were due to 5′ nontranslated regions that differed not only in length but in their sequences. The complete sequence of STRL33.1, the sequence of the 5′ nontranslated region of STRL33.2, and the sequence of the 5′ nontranslated region and ORF of STRL33.3 have been submitted to GenBank with accession numbers U73529, U73530, and U73531, respectively. STRL33.1 contained 1,897 nucleotides, excluding the poly(A) tail, with an ORF encoding a predicted protein of 342 amino acids (Fig. 1). The predicted initiator codon was in a favorable context for initiation (36) and could be assigned unambiguously because, with the reading frame fixed by comparison with other GPCRs, it was the first ATG after an in-frame stop codon in cDNA STRL33.3, and it was also the first in-frame ATG in cDNAs STRL33.1 and STRL33.2. As noted above, ORF sequence was also determined from cDNAs other than STRL33.1, and the sequence of the entire STRL33.1 ORF was confirmed in independent clones. However, differences were noted between the ORFs of STRL33.1 and STRL33.3 at two positions: a second position change in the 25th codon of STRL33.3 (GAC → GCC), replacing D25 with A, and a silent third position change in the 103rd codon of STRL33.3 . The silent change in the 103rd codon of STRL33.3 was present in a second independent clone, whereas the 25th codon change has not been independently verified. We presume that at least the silent change represents a true polymorphism in the F9 TIL. The 5′ nontranslated regions of STRL33.1, STRL33.2, and STRL33.3 were 30, 135, and 1,462 nucleotides, respectively, and sequence comparisons revealed that differences among these regions were due, at least in part, to alternative splicing (data not shown). Not surprisingly, the long additional 5′ nontranslated sequence of STRL33.3 contained many ATG sequences (but no significant ORFs), suggesting that the STRL33.3 mRNA may not be translated efficiently, and although we could detect a STRL33.3-specific mRNA by Northern blot analysis (see below), the STRL33.3 cDNA may be derived from an incompletely processed mRNA. Extensive processing, yielding mRNAs with alternative 5′ exons, is well documented among the chemoattractant receptors (2), and incomplete processing of mRNAs in lymphocytes has been suggested as a possible mechanism for the regulation of translation (37).
Figure 1.
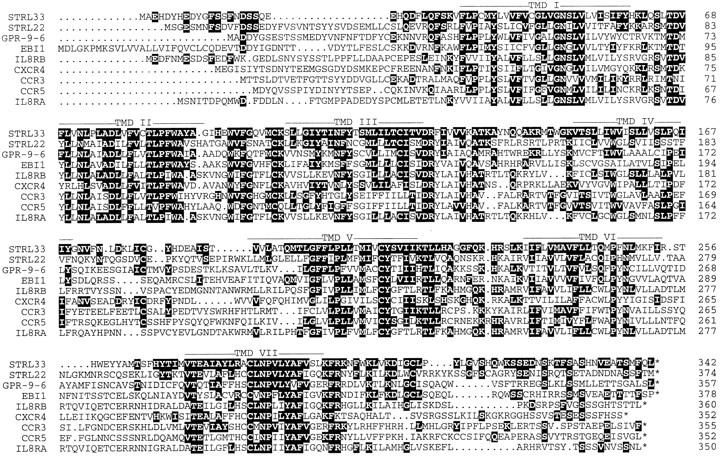
Alignment of the STRL33 predicted amino acid sequence with the selected GPCRs STRL22, GPR-9-6, EBI1, IL-8RB, CXCR4, CCR3, CCR5, and IL-8RA. Numbers at the right indicate the positions of the residues at the end of each line of sequence. Solid backgrounds highlight matches between STRL33 and the other receptors. Dots indicate gaps introduced for optimal alignments. Putative TMDs I–VII are indicated by bars. The alignments were generated using the PileUp program of the Wisconsin Sequence Analysis Package of Genetics Computer Group (Madison, WI).
Comparison of STRL33 with sequences in the GenBank database as of March 30, 1997, using BLAST (38) revealed no identical sequences but greatest similarity to orphan GPCRs and related chemokine receptors. Matches were found between sequences in the 3′ nontranslated region of STRL33 and several sequences in the EST database for which no significant homologies had been identified. Alignments between STRL33 and selected related sequences is shown in Fig. 1. Percent identities between STRL33 and orphan receptors STRL22 (22), GPR-9-6 (GenBank accession number U45982), and EBI1 (39) are 37, 32, and 32%, respectively, and between STRL33 and chemokine receptors IL-8RB (CXCR2), CXCR4, CCR3, CCR5, and IL-8RA (CXCR1) are 30, 30, 30, 29, and 28%, respectively. STRL22 is an orphan receptor gene that, like STRL33, was isolated from the F9 TIL (22).
Fig. 1 shows that similarities among the receptors are greatest in the TMDs, as is typical of GPCRs. Like other GPCRs, STRL33 includes a site for N-linked glycosylation in the NH2-terminal domain (N16), cysteines in extracellular loops one and two (C102 and C180), and multiple serines in the COOH terminal domain (1). While there is no signature sequence motif for the chemokine receptors, the STRL33 sequence does contain some features characteristic of chemokine receptors, including an acidic NH2terminal domain with paired acidic residues (E8 and D9, E21 and E22) (2), a short basic third intracellular loop, an alanine in place of the proline that is conserved in nonchemokine receptor GPCRs in the second intracellular loop (A134), a paired cysteine and tyrosine in TMD V (C210 and Y211), and a cysteine in TMD VII (C282). In contrast, some residues typical for chemokine receptors are absent from STRL33, including cysteine residues in the NH2-terminal domain and in the third extracellular loop. Multiple sequence alignment (PileUp, Genetics Computer Group, Madison, WI) places STRL33 in a group of orphan receptors, including STRL22, GPR-9-6, and EBI1, separate from the groupings of the CXCRs on one hand and the CCRs on the other (data not shown).
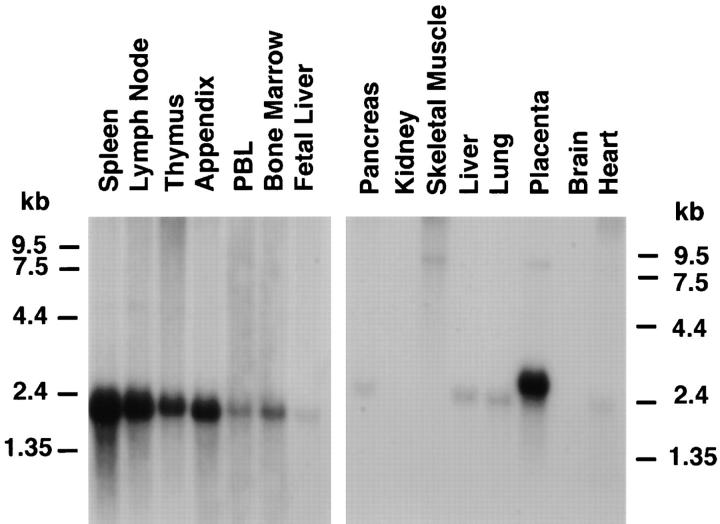
We analyzed RNA expression by Northern blot of total RNA for STRL33 and other related receptors in leukocyte populations and lines and in human tissues. As shown in Fig. 2 A, the STRL33 cDNA probe hybridized to a broad band at ∼2 kb that was prominent in both CD4+ (R4, F9, and B10) and CD8+ (R8) TIL with low level signal in PBL but not in other cells tested, including immortalized CD4+ T cell lines. Using a probe from the 5′ nontranslated sequences specific for the STRL33.3 mRNA, we also detected a band at ∼3.6 kb in the F9 and B10 TIL after long exposure, not shown in Fig. 2 A. Fig. 2 B demonstrates that in vitro activation of PBL induces expression of STRL33 to levels comparable to that seen in the TIL.
Figure 2.
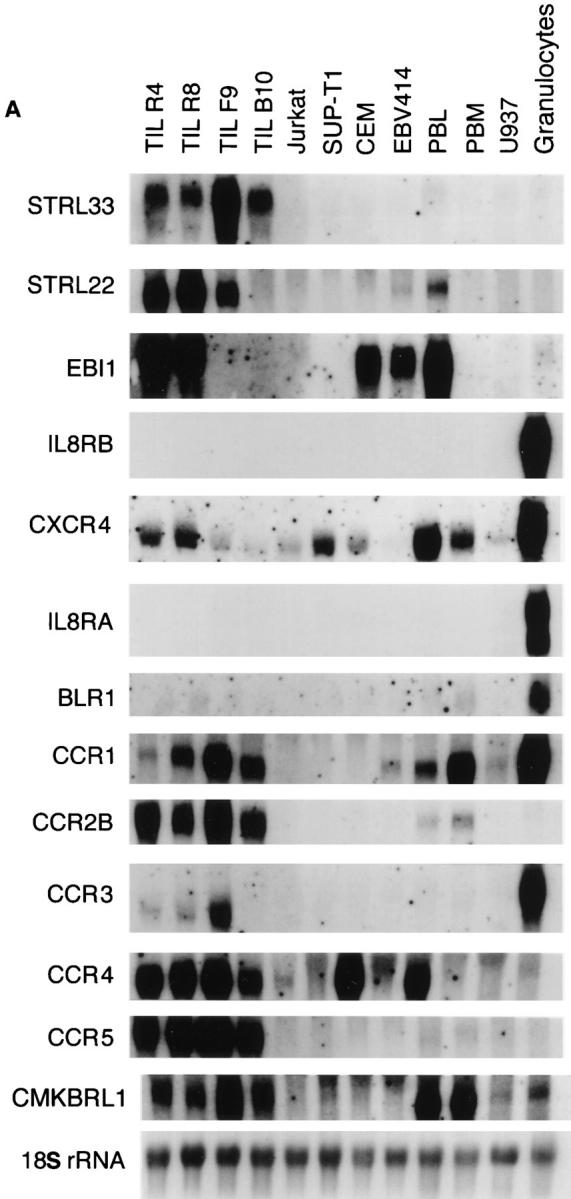
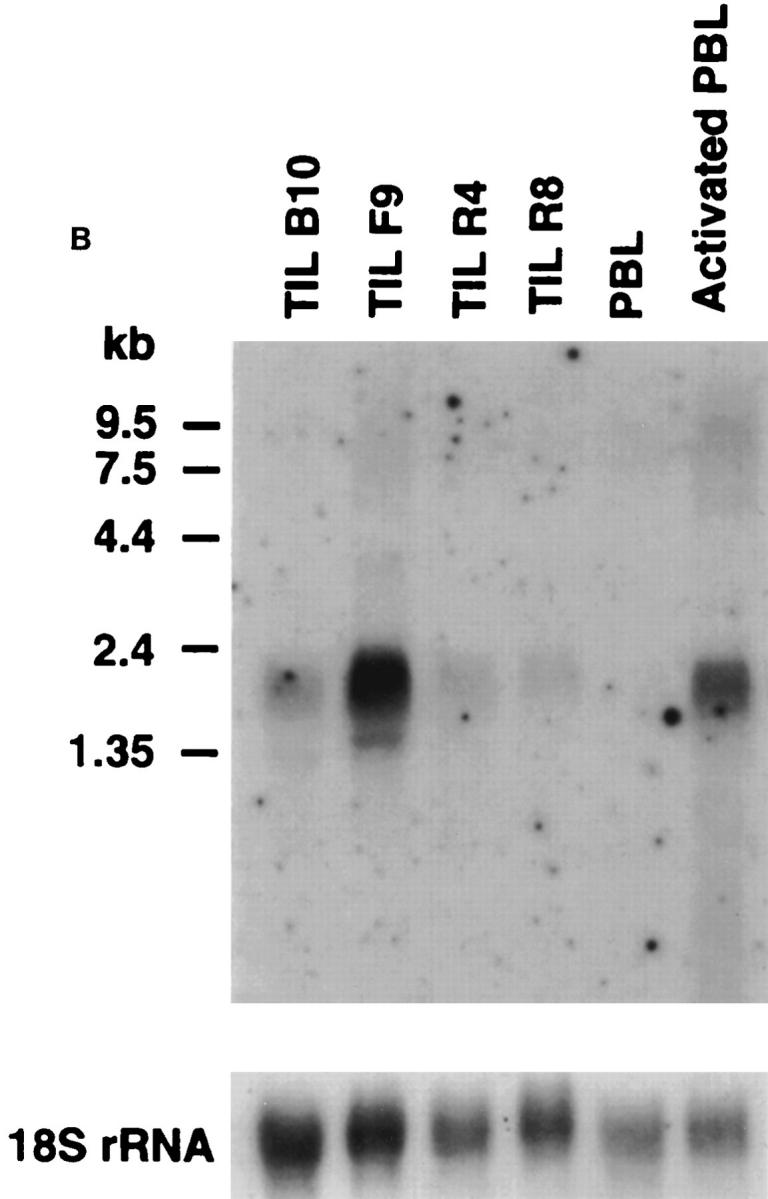
Expression of STRL33 and other GPCR genes. (A) The expression of STRL33, genes for known chemokine receptors, and genes for selected orphan GPCRs in leukocytes. 15 μg of total RNA were electrophoresed on 1.2% agarose–formaldehyde gels, transferred to nitrocellulose membranes, and hybridized with the probes indicated on the left. A total of six membranes were used for hybridizations, and adequate removal of signal was documented before repeat probings. Film exposure times ranged from overnight for the IL-8RA and IL-8RB blots to 13 d for the CXCR4 blot. Probings were done using an oligonucleotide complementary to 18S rRNA in order to demonstrate amounts of RNA loaded per lane and a representative blot is shown. (B) The expression of STRL33 in activated PBL. 25 μg of total RNA from TIL, and freshly isolated and activated PBL were analyzed as in A and hybridized with a 32P-labeled STRL33 ORF probe and a probe for 18S rRNA.
Fig. 2 A shows significant differences in expression of various chemokine receptor and orphan receptor genes among the leukocytes. Of particular interest is the demonstration of heterogeneity of receptor gene expression among T cell preparations. In general, receptor gene expression is higher among the TIL than in T cell lines or freshly isolated PBL, although even among the TIL there are significant differences. For example, the CD4+ F9 TIL show a significant signal for CCR3. This is noteworthy, because although CCR3 can serve in vitro as a coreceptor for HIV-1 (13), speculation on a role for CCR3 in systemic HIV-1 infection has been constrained by the assumption that among leukocytes CCR3 expression is limited to eosinophils.
Fig. 3 shows the expression of the STRL33 gene in selected human tissues. There is an mRNA species of ∼2.1 kb prominently expressed in lymphoid tissue; a species of ∼2.5 kb in placenta; low abundance species of 2.1-2.4 kb expressed in pancreas, liver, lung, and heart; and low abundance larger species in a variety of tissues. The conspicuous expression of the STRL33 gene in T cells, activated PBL, and lymphoid tissues is consistent with the presumption that STRL33 is a chemokine receptor.
Figure 3.
The expression of STRL33 in human tissues. Blots were prepared by the supplier (Clontech, Palo Alto, CA) from 1.2% agarose–formaldehyde gels containing ∼2 μg poly(A)+ RNA per lane. Hybridizations were done using a 32P-labeled STRL33 ORF probe and blots were washed according to the instructions of the manufacturer. The blot prepared from lymphoid tissue (left) was exposed for 2 d, and the blot from other selected tissues (right) was exposed for 8 d.
HEK 293 cells were transfected with an expression vector containing the STRL33.1 ORF, and cell lines were derived as described in Materials and Methods. Cell lines expressing the highest levels of STRL33 RNA were tested in a fluorometric calcium flux assay for responses to chemokines. The STRL33-transfected cells were tested with platelet factor 4, IL-8, IP-10, HuMig, SDF-1, MIP-1α, MIP-1β, RANTES, MCP-1, MCP-2, MCP-3, MCP-4, I309, and lymphotactin, and no responses were found.
The demonstrations that receptors for both CXC and CC chemokines can function as cofactors for HIV-1 entry into cells led us to test STRL33 in an assay designed to detect fusion between two cell populations: NIH 3T3 cells expressing T7 RNA polymerase, human CD4, and either STRL33 or CXCR4 or CCR5 , and HeLa cells expressing Envs from HIV-1 isolates with differing tropisms. Fusion between the two cell populations resulted in expression of β-Gal. As a negative control, we used the Unc Env, a mutant protein that cannot mediate fusion due to a deletion of the gp120/gp41 cleavage site.
The results of the fusion assays are shown in Fig. 4. In Fig. 4 A, NIH 3T3 cells expressing CD4 plus CXCR4 fused well with cells expressing Envs from TCL-tropic LAV (NL4.3) and IIIB, whereas with Envs from the M-tropic strains only weak (ADA) or negligible (SF162, Ba-L, and JR-FL) fusion was observed. Cells expressing CD4 plus CCR5 showed the opposite specificity: they fused well with cells expressing the Envs from the M-tropic strains ADA, SF162, Ba-L, and JR-FL and not the Envs from TCL-tropic LAV (NL4.3) and IIIB. These results are consistent with previous reports (8, 11–15).
Figure 4.
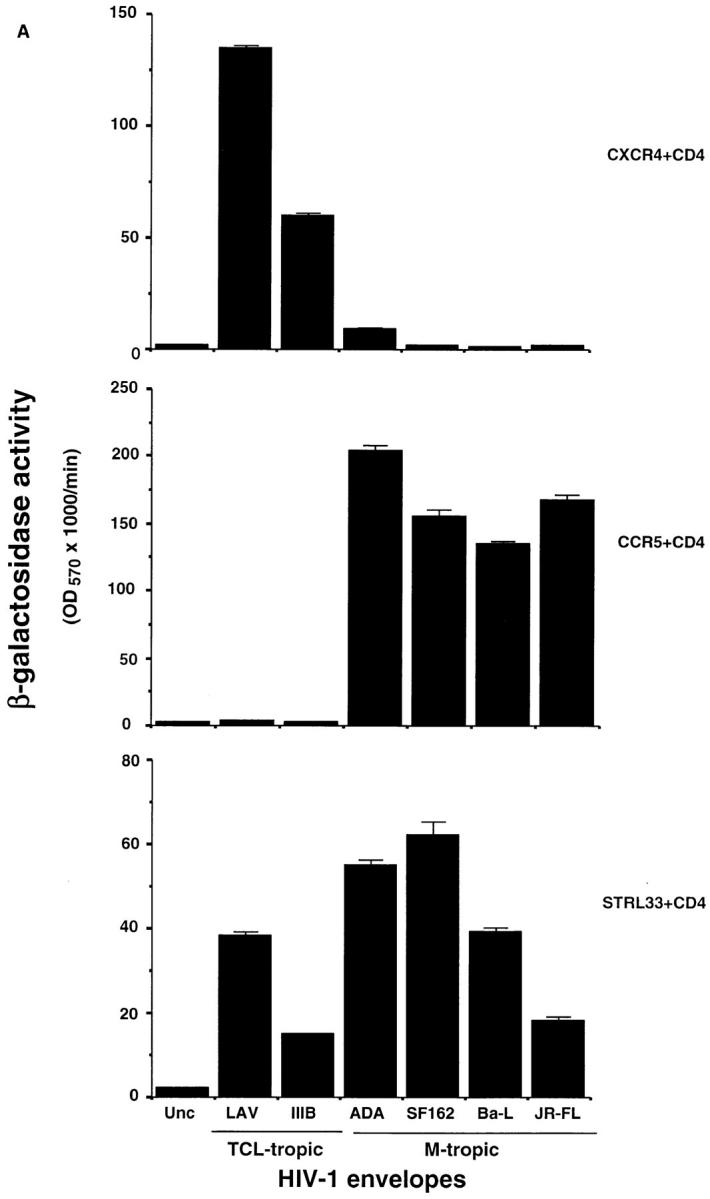
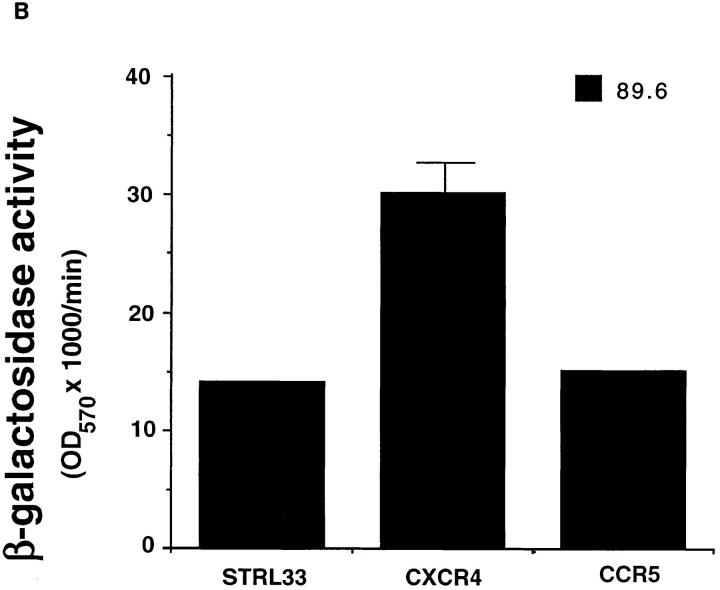
Activity of STRL33 as a fusion cofactor. (A) NIH 3T3 cells were transfected with DNAs encoding CXCR4 or CCR5 or STRL33, and infected with vaccinia recombinants encoding CD4 and T7 RNA polymerase. HeLa cells were infected with a vaccinia recombinant containing lacZ under control of a T7 promoter and infected separately with vaccinia recombinants encoding the indicated Envs. Unc is a mutant Env that cannot be cleaved to gp120 and gp41 and cannot mediate fusion. Cell fusion was quantified by measuring β-Gal activity. NIH 3T3 cells transfected with the STRL33 cDNA but not infected with virus vCB-3 encoding CD4 did not fuse with cells expressing any of the Envs (data not shown). Results of one experiment are shown. STRL33 also mediated fusion with cells expressing both TCL-tropic and M-tropic Envs in four additional experiments. (B) Cell fusion experiment done as in A, except that here the DNA encoding the Env, 89.6, was introduced into HeLa cells by transfection of plasmid DNA, resulting in lower levels of fusion overall as compared with A. The value obtained for Unc has been subtracted from each of the values obtained using the 89.6 Env. Results are shown from one experiment. Similar activity for STRL33 was found in another experiment using the 89.6 Env.
In contrast with the restricted specificities of CXCR4 and CCR5, STRL33 functioned with CD4 as a fusion cofactor for cells expressing Envs from both TCL-tropic and M-tropic strains. Additionally, as shown in Fig. 4 B, STRL33, like CXCR4 and CCR5, mediated fusion with the dualtropic Env 89.6. Negligible β-Gal activity, equivalent to levels seen using the Unc Env, was detected in fusion assays using CD4-expressing NIH 3T3 cells transfected with a control vector lacking the STRL33 cDNA insert, or in assays using NIH 3T3 cells transfected with the STRL33 cDNA but not expressing CD4 (data not shown).
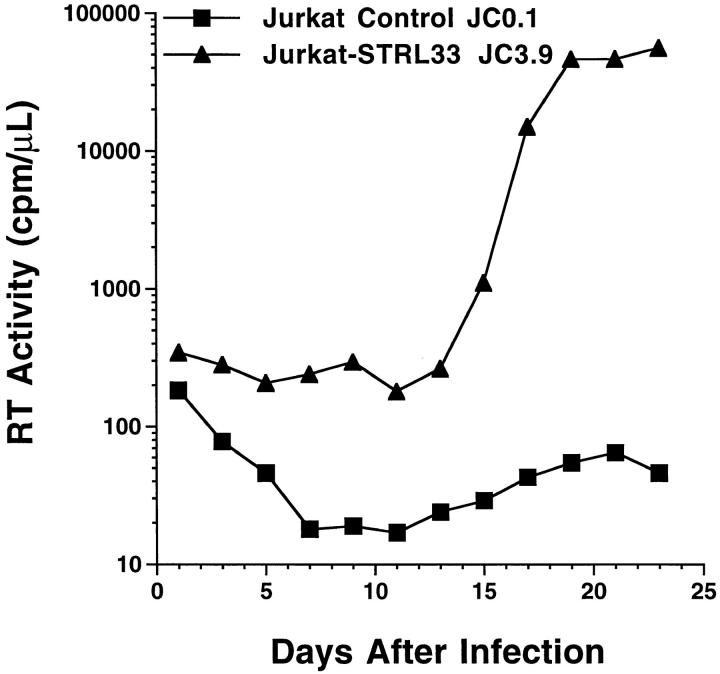
We tested the ability of STRL33 to support productive infection with HIV-1. We transfected Jurkat cells, which express CD4 and CXCR4, but not other known HIV-1 coreceptors, with vector containing the STRL33.1 cDNA, or with vector control, and cell lines were derived under selection by limiting dilution. We infected the STRL33transfected and control cells with HIV-1ELI1, a molecularly cloned isolate with properties of a primary virus (34, 40, 41). HIV-1ELI1 has been shown to grow poorly in Jurkat cells (34) and CXCR4 does not function efficiently as a coreceptor for the HIV-1ELI1 Env (13). As shown in Fig. 5, the STRL33-transfected cells supported productive infection with HIV-1ELI1 under conditions where control transfected cells yielded no detectable virus. Although in experiments using higher innocula of virus, HIV-1ELI1, as reported (34), could infect cultures of control Jurkat cells, the activity of STRL33 was also clear in these experiments, because HIV1ELI1 production by the control cultures lagged significantly behind HIV-1ELI1 production in cultures of STRL33-transfected cells (data not shown). Enhanced infection with HIV-1ELI1 in STRL33-transfected Jurkat cells was demonstrated in four experiments using a total of three independent STRL33-transfected cell lines.
Figure 5.
HIV-1 infection of STRL33-transfected Jurkat cells. 2 × 106 cells were infected with HIV-1ELI1 corresponding to 105 cpm of RT activity. Samples were taken every 2 d for determining RT activity from cultures of the control-transfected Jurkat cell line JC0.1 (▪) and from cultures of the STRL33.1-transfected Jurkat cell line JC3.9 (▴).
The identification of STRL33 adds to the recent discoveries on the roles of chemokine receptors in the pathobiology of HIV-1 infection. STRL33 is a novel GPCR that can function with CD4 to mediate fusion with cells bearing HIV-1 Envs from laboratory-adapted TCL-tropic, M-tropic, and dual-tropic strains. In this regard, STRL33 can mediate fusion with a wider range of Envs than can the major cofactors CXCR4 and CCR5. Recent literature has supported the strong correlation between assays for Env/coreceptor–mediated fusion and infection (8, 11–16), and we have demonstrated that STRL33 can significantly enhance the ability of HIV-1 to infect cells. While the role of STRL33 in the biology of viral infection is unknown, the STRL33 gene is expressed in cells and tissues that are among the natural targets for HIV-1. Further work will determine whether STRL33 represents an important host component in the transmission of HIV-1 and the progression of AIDS.
Footnotes
Thanks are due to J. Yannelli, P. Murphy, S. Ahuja, C. Combadière, Y. Chang, R. Siliciano, and M. Tsang for reagents and/or helpful discussions.
1 Abbreviations used in this paper: β-Gal, β-galactosidase; Envs, envelope glycoproteins; FBS, fetal bovine serum; GPCRs, G protein–coupled receptors; HEK, human embryonic kidney; M-tropic, macrophage tropic; ORF, open reading frame; TCL-tropic, T cell line tropic; TIL, tumorinfiltrating lymphocytes; TMD, transmembrane domain.
References
- 1.Savarese TM, Fraser CM. In vitromutagenesis and the search for structure–function relationships among G protein–coupled receptors. Biochem J. 1992;283:1–19. doi: 10.1042/bj2830001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murphy P. The molecular biology of leukocyte chemoattractant receptors. Annu Rev Immunol. 1994;12:593–633. doi: 10.1146/annurev.iy.12.040194.003113. [DOI] [PubMed] [Google Scholar]
- 3.Schall TJ, Bacon K. Chemokines, leukocyte trafficking and inflammation. Curr Opin Immunol. 1994;6:865–873. doi: 10.1016/0952-7915(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 4.Raport CJ, Schweickart VL, Chantry D, Eddy RLJ, Shows T, Godiska R, Gray PW. New members of the chemokine receptor gene family. J Leuk Biol. 1996;59:18–23. doi: 10.1002/jlb.59.1.18. [DOI] [PubMed] [Google Scholar]
- 5.Miedema F, Meyaard C, Koot M, Klein MR, Roos MT, Groenink M, Fouchier RA, Van't Wout A. Changing virus–host interactions in the course of HIV-1 infection. Immunol Rev. 1994;140:35–72. doi: 10.1111/j.1600-065x.1994.tb00864.x. [DOI] [PubMed] [Google Scholar]
- 6.Goudsmit J. The role of viral diversity in HIV pathogenesis. J Acquired Immune Defic Syndr Hum Retrovirol. 1995;10:S15–S19. [PubMed] [Google Scholar]
- 7.Cocchi F, DeVico AL, Garzino-Demo A, Arya SK, Gallo RC, Lusso P. Identification of RANTES, MIP-1α, and MIP-1β as the major HIV-suppressive factors produced by CD8+ T cells. Science (Wash DC) 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 8.Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G-protein–coupled receptor. Science (Wash DC) 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 9.Bleul CC, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer TA. The lymphocyte chemoattractant SDF-1 is a ligand for LESTER/fusin and blocks HIV-1 entry. Nature (Lond) 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 10.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J-L, Arenzana-Seisdedos F, Schwartz O, Heard J-M, Clark-Lewis I, Legler DF, et al. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line–adapted HIV-1. Nature (Lond) 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 11.Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, Berger EA. CC CKR5: A RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science (Wash DC) 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 12.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton RE, Hill CM, et al. Identification of a major co-receptor for primary isolates of HIV-1. Nature (Lond) 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 13.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, Wu L, Mackay CR, LaRosa G, Newman W, et al. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 14.Doranz BJ, Rucker J, Yi Y, Smyth RJ, Samson M, Peiper SC, Parmentier M, Collman RG, Doms RW. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 15.Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA, Cayanan C, Maddon PJ, Koup RA, Moore JP, Paxton WA. HIV-1 entry into CD4+cells is mediated by the chemokine receptor CC-CKR-5. Nature (Lond) 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 16.Simmons G, Wilkinson D, Reeves JD, Dittmar MT, Beddews S, Weber J, Carnegie G, Desselberger U, Gray PW, Weiss RA, Calpham PR. Primary, syncytium-inducing human immunodeficiency virus type I isolates are dual-tropic and most can use either Lestr or CCR-5 as co-receptors for virus entry. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber C-M, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature (Lond) 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 18.Liu R, Paxton WA, Choe S, Ceradini D, Martin SR, Horuts R, MacDonald ME, Stahlmann H, Koup RA, Candau NR. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 19.Dean M, Carrington M, Winkler C, Huttley GA, Smith MW, Allikmets R, Goedert JJ, Buchbinder SP, Vittinghoff E, Gomperts E, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Science (Wash DC) 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 20.Zimmerman PA, Buckler-White A, Alkhatib G, Spalding T, Kubofcik J, Combadiere C, Weissman D, Cohen O, Rubbert A, Lam G, et al. Inherited resistance to HIV-1 conferred by an inactivating mutation in CC chemokine receptor 5: studies in populations with contrasting clinical phenotypes, defined racial background and quantified risk. Mol Med. 1996;3:23–36. [PMC free article] [PubMed] [Google Scholar]
- 21.Biti R, French R, Young J, Bennetts B, Stewart GS. HIV-1 infection in an individual homozygous for the CCR5 deletion allele. Nature Med. 1997;3:252–253. doi: 10.1038/nm0397-252. [DOI] [PubMed] [Google Scholar]
- 22.Liao F, Lee H-H, Farber JM. Cloning of STRL22, a new human gene encoding a G protein–coupled receptor related to chemokine receptors and located on chromosome 6q27. Genomics. 1997;40:175–180. doi: 10.1006/geno.1996.4544. [DOI] [PubMed] [Google Scholar]
- 23.Vanguri P, Farber J. Identification of CRG-2: an interferon-inducible mRNA predicted to encode a murine monokine. J Biol Chem. 1990;265:15049–15057. [PubMed] [Google Scholar]
- 24.Amichay D, Gazzinelli RT, Karupiah G, Moench TR, Sher A, Farber JM. The gene for chemokines MuMig amd Crg-2 are induced in protozoan and viral infections in response to IFN-γ with patterns of tissue expression that suggest nonredundant roles in vivo. . J Immunol. 1996;157:4511–4520. [PubMed] [Google Scholar]
- 25.Kingston RE. Transfection of DNA into eukaryotic cells. Curr Prot Mol Biol. 1996;9:11–19. [Google Scholar]
- 26.Memon SA, Petrak D, Moreno MB, Zacharchuk CM. A simple assay for examining the effect of transiently expressed genes on programmed cell death. J Immunol Meth. 1995;180:15–24. doi: 10.1016/0022-1759(94)00294-7. [DOI] [PubMed] [Google Scholar]
- 27.Liao F, Rabin RL, Yannelli JR, Koniaris LG, Vanguri P, Farber JM. Human Mig chemokine: biochemical and functional characterization. J Exp Med. 1995;182:1301–1314. doi: 10.1084/jem.182.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nussbaum O, Broder CC, Berger EA. Fusogenic mechanisms of enveloped-virus glycoproteins analyzed by a novel recombinant vaccinia virus–based assay quantitating cell fusion–dependent reporter gene activation. J Virol. 1994;68:5411–5422. doi: 10.1128/jvi.68.9.5411-5422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Broder CC, Dimitrov DS, Blumenthal R, Berger EA. The block to HIV-1 envelope glycoproteinmediated membrane fusion in animal cells expressing human CD4 can be overcome by a human cell component(s) Virology. 1993;193:483–491. doi: 10.1006/viro.1993.1151. [DOI] [PubMed] [Google Scholar]
- 30.Fuerst TR, Niles EG, Studier FW, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesized bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alkhatib G, Broder CC, Berger EA. Cell type– specific cofactors determine human immunodeficiency virus type 1 tropism for T-cell lines versus primary macrophages. J Virol. 1996;70:5478–5494. doi: 10.1128/jvi.70.8.5487-5494.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Broder CC, Berger EA. Fusogenic selectivity of the envelope glycoprotein is a major determinant of human immunodeficiency virus type 1 tropism for CD4+ T-cell lines vs. primary macrophages. Proc Natl Acad Sci USA. 1995;92:9004–9008. doi: 10.1073/pnas.92.19.9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Collman R, Balliet JW, Gregory SA, Friedman H, Kolson DL, Nathanson N, Srinivasan A. An infectious molecular clone of an unusual macrophage-tropic and highly cytopathic strain of human immunodeficiency virus type 1. J Virol. 1992;66:7517–7521. doi: 10.1128/jvi.66.12.7517-7521.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peden K, Emerman M, Montagnier L. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV1MAL, and HIV-1EL1 . Virology. 1991;185:661–672. doi: 10.1016/0042-6822(91)90537-l. [DOI] [PubMed] [Google Scholar]
- 35.Peden, K.W.C., and M.A. Martin. 1995. Virological and molecular genetic techniques for studies of established HIV isolates. In HIV: A Practical Approach. Vol. 1. Virology and Immunology. J. Karn, editor. Oxford University Press, Oxford, UK. 21–46.
- 36.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acid Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozak M. Interpreting cDNA sequences: some insights from studies on translation. Mammalian Genome. 1996;7:563–574. doi: 10.1007/s003359900171. [DOI] [PubMed] [Google Scholar]
- 38.Altschul SF, Gish W, Miller W, Mayers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 39.Birkenbach M, Josefsen K, Yalamanchili R, Lenoir G, Kieff E. Epstein–Barr virus–induced genes: first lymphocyte-specific G protein–coupled peptide receptors. J Virol. 1993;67:2209–2220. doi: 10.1128/jvi.67.4.2209-2220.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alizon M, Wain-Hobson S, Montagnier L, Sonigo P. Genetic variability of the AIDS virus: nucleotide sequence analysis of two isolates from African patients. Cell. 1986;46:63–74. doi: 10.1016/0092-8674(86)90860-3. [DOI] [PubMed] [Google Scholar]
- 41.Willey RL, Martin M, Peden KWC. Increase in soluble CD4 binding to and CD4-induced dissociation of gp120 from virions correlates with infectivity of human immunodeficiency virus type 1. J Virol. 1994;68:1029–1039. doi: 10.1128/jvi.68.2.1029-1039.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]