Abstract
IL-7R-deficient mice have severely impaired expansion of early lymphocytes and lack γδ T cells. To elucidate the role of IL-7R on γδ T cell development, we analyzed the rearrangements of TCR-α, β, γ, and δ genes in the thymus of the IL-7R-deficient mice. Southern blot analysis with a Jγ1 probe revealed that more than 70% of Jγ1 and Jγ2 alleles are recombined to form distinct Vγ1.2–Jγ2 and Vγ2–Jγ1 fragments in control mice. On the contrary, no such recombination was detected in the mutant mice. The rearrangements in the TCR-α, β, and δ loci were comparably observed in control and mutant mice. PCR analysis indicated that the V–J recombination of all the Vγ genes is severely hampered in the mutant mice. The mRNA of RAG-1, RAG-2, Ku-80, and terminal deoxynucleotidyl transferase (TdT) genes was equally detected between control and mutant thymi, suggesting that the expression of common recombination machinery is not affected. These data demonstrated that the V–J recombination of the TCR γ genes is specifically blocked in the IL-7R-deficient mice and suggested the presence of highly specific regulation for TCR γ gene rearrangement.
IL-7 is a growth factor for early B and T cell precursors. It was first characterized by its ability to support the growth of pre-B cells. Subsequently, it has been shown to support survival and growth of early thymocytes and promote rearrangement of TCR β and γ chains in fetal thymus and fetal liver cultures (1, 2). In vivo administration of neutralizing antibodies to IL-7 and IL-7R resulted in the inhibition of both B and T lymphopoiesis (3, 4). Finally, IL-7- and IL-7Rdeficient mice have severely impaired expansion of early lymphocytes (5, 6).
γδ T cells have unique features in contrast with αβ T cells (7, 8). γδ T cells expressing specific Vγ chain appear as several successive waves in the developing thymus and each of them shows specific tissue distribution in the adult mouse. However, little is known about the mechanism of γδ T cell development. In fetal thymic organ culture, addition of IL-7 promotes expansion of mature γδ T cells but prevents generation of mature αβ T cells (9). The epithelial cells in the skin and the gut produce IL-7 (10, 11), and dendritic epidermal T cells proliferate in response to IL-7 (10). Additionally, IL-7 induced rearrangement of Vγ2 and Vγ4, but not Vγ3 or Vγ5 genes, and sustained expression of RAG-1 and RAG-2 genes (1, 2). Collectively, these results suggested that IL-7 may be involved in development and maintenance of γδ T cells in the thymus and the periphery.
Although T and B lymphopoiesis is severely hampered, decreased but certain numbers of αβ T cells and B cells exist in the periphery of the IL-7R-deficient mice, and they normally respond to mitogenic stimuli such as Con A and LPS (12). In contrast, γδ T cells are completely absent in the IL-7R-deficient mice as well as in IL-2R-γ- and Jak3-deficient mice: no γδ T cells were detected in fetal and adult thymus, spleen, skin, small intestine, and liver of IL-7Rdeficient mice (12–14). Two possibilities can be considered to explain the lack of γδ T cells in the IL-7R-deficient mice. The one is that γδ T cell precursors may completely depend on IL-7 for their survival and/or proliferation. The other is that IL-7 may be a key factor for the induction of the TCR γ gene rearrangements in T cell precursors.
To test the latter hypothesis, we analyzed the recombination status of TCR loci in the αβ T cells remaining in IL-7R-deficient mice. The V–J recombination was almost completely blocked in the TCR γ locus in the mutant thymus, whereas the TCR α, β, and δ loci were rearranged at comparable levels with control thymus. These results clearly demonstrated that the signal from IL-7R plays an indispensable role on the induction of TCR γ gene rearrangement. Thus, the mouse TCR γ locus will provide a unique system to analyze the mechanism of cytokine-induced gene rearrangements.
Materials and Methods
Mice.
IL-7R-deficient mice were established by replacing the exon 2 with a PGK–neo cassette as described (12). Animals heterozygous (+/−) and homozygous (−/−) for the IL-7R mutation were on the (129/Ola × C57BL/6)F3 hybrid background. The age of fetuses was determined by scoring for the appearance of a vaginal plug and taking as day 0 the morning on which the mating plug was observed. All mice were maintained under the specific pathogen-free conditions in the Animal Center for Biomedical Research, Faculty of Medicine (The University of Tokyo).
Southern Blot Analysis.
Thymocyte genomic DNA was digested with HindIII or EcoRI restriction enzyme and electrophoresed through 0.7% agarose gel. The DNA was transferred to polyvinylidene difluoride filters (Immobilon; Millipore, Bedford, MA) and hybridized with 32P-labeled probes. The following fragments were used as probes: Jγ1, a 1.1 kb StyI–HindIII fragment containing the Jγ1 segment and its 3′ flanking region of genomic DNA from KN6 (15); Jα1, a 3.5 kb EcoRI–HindIII fragment of TA28.1 (16); Jδ1, a 2.5 kb SacI fragment of pCDS17 (17); Jβ2, a 2.3 kb EcoRI fragment of mouse genomic Jβ region (18). To confirm equal loading of genomic DNA, the membranes were hybridized with a 1.3 kb KpnI fragment of mouse RAG-2 cDNA (19). Southern blots were analyzed and radioactivity was quantitated using a Bio-image Analyzer (Fujix BAS2000; Fuji Film, Tokyo, Japan). The percentage of rearranged alleles was calculated by normalizing with the radioactivity of the RAG-2 probe.
PCR Analysis.
Thymocyte DNA was prepared from fetuses at day 17 of gestation and mice at 4 wk old. PCR was carried out in a 25 μl reaction mixture containing 0.5 ng template DNA (0.5 μg for TCR β genes), 50 pmol each primer, 200 mM each dNTP, and 2.5 U Taq DNA polymerase. For TCR γ genes, samples were amplified for 30 cycles of 45 s at 94°C, 2 min at 50°C, and 1 min at 72°C. For TCR δ and β genes, PCR was performed as described previously (20–23). The PCR products were electrophoresed in 3% agarose gel, blotted onto nylon membranes, and hybridized with 32P-labeled oligonucleotide probes. PCR primers are as follows: Vγ1.1 and Vγ1.2, 5′-CTTCCATATTTCTCCAACACAGC-3′; Jγ2, 5′-ACTATGAGCTTTGTTCCTTCTG-3′; Jγ4, 5′-ACTACGAGCTTTGTCCCTTTGG-3′; 5′ RAG-2, 5′-CACATCCACAAGCAGGAAGTACAC-3′; 3′ RAG-2, 5′-GGTTCAGGGACATCTCCTACTAAG-3′. Vγ2, Vγ3, Vγ4, Vγ5, Jγ1, Vδ1, Vδ4, Vδ5, Jδ1, Vβ8.2, Dβ2, and Jβ2 primers were described previously (20–23). Oligonucleotide probes used are as follows: Jγ2, 5′-CAAATACCTTGTGAAAGCCCGAGC-3′; Jγ4, 5′-CAAATATCTTGACCCATGATGTGC-3′. Jγ1, Jδ1, and Jβ2 oligonucleotide probes were described previously (20, 22). Radioactivity was analyzed using the Bio-image Analyzer.
RT-PCR Analysis.
Total RNA was isolated using the AGPC method as described (24). RNA samples were treated with RQ-1 RNase-free DNase (Promega Corp., Madison, WI) to remove contaminating genomic DNA. Oligo (dT)-primed cDNA was prepared by Molony murine leukemia virus RNase H− reverse transcriptase (GIBCO BRL, Gaithersburg, MD) at 37°C for 1 h. PCR was carried out for 25 cycles of 1 min at 94°C, 1 min at 50°C for hypoxanthine phosphoribosyl transferase (HPRT) or at 55°C for others, and 1 min at 72°C. The PCR products were electrophoresed in 3% agarose gel, blotted onto nylon membranes, and hybridized with 32P-labeled probes. The following DNA fragments were used as probes: RAG-1, a 1.4-kb EcoRI fragment of mouse RAG-1 cDNA (19); terminal deoxynucleotidyl transferase (TdT), a 1.3-kb EcoRI–EcoRV fragment of mouse TdT cDNA, M16-1b (25); Ku-80, a 540-bp PCR fragment of Ku-80 cDNA; HPRT, a 350-bp PCR fragment of HPRT cDNA. The RAG-2 probe was described above. The following PCR primers are used: 5′ RAG-1, 5′-GCAATGAGGAAGTGAGTCTGGA-3′; 3′ RAG-1, 5′-CTGAGGAAGGTATTGACACGGA-3′; 5′ Ku-80, 5′-AGAGGACACTATTCAAGGGTAC-3′; 3′ Ku-80, 5′-AGACACTGGTACAATCGCTGAA-3′; 5′ TdT, 5′-ACTGCGACATCTTAGAGTCA-3′; 3′ TdT, 5′-CTTCCCCTTAGTCCTGTCAT-3′; 5′ HPRT, 5′-CTCGAAGTGTTGGATACAGG-3′; 3′ HPRT, 5′-TGGCCTATAGGCTCATAGTG-3′. Radioactivity was analyzed using the Bio-image Analyzer.
Results
V–J Recombination of TCR γ Genes Is Blocked in IL-7Rdeficient Mice.
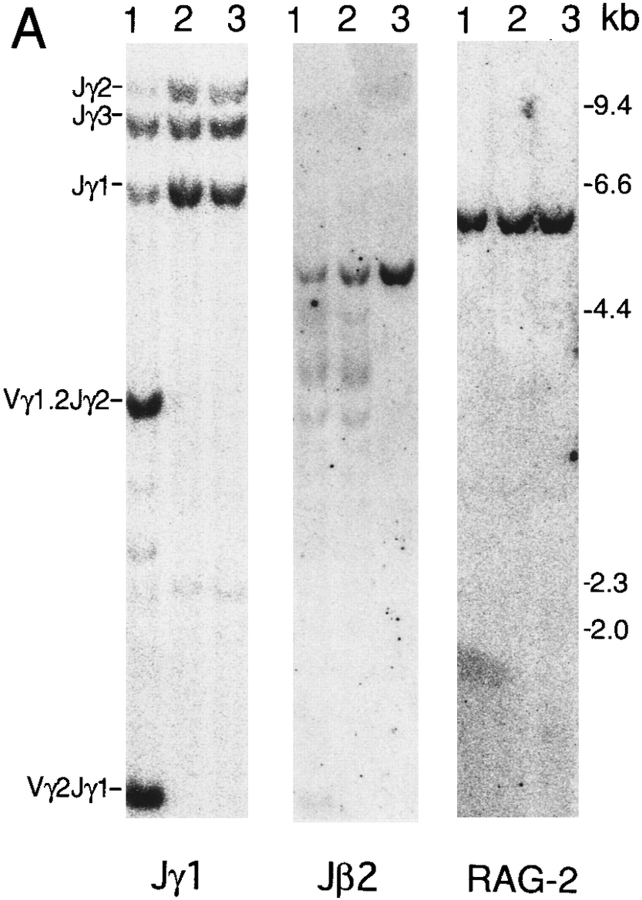
To examine whether the signal from IL-7R affects the V–J recombination, we compared the rearrangement of the TCR γ genes between the thymocytes of IL-7R +/− and −/− mice. The thymocyte DNA from 4-wk-old mice was digested with HindIII or EcoRI, and a Southern blot was hybridized with the Jγ1 probe (Fig. 1 A, left). The Jγ1 probe allows the analysis of DNA rearrangements involving not only Jγ1 but also Jγ2 and Jγ3 gene segments (15). The ES cell DNA showed a 6.6-kb Jγ1, a 9.0-kb Jγ3, and a 11.7-kb Jγ2 germline fragment. The thymocyte DNA from IL-7R +/− mice showed decreased intensity of Jγ1 and Jγ2 germline fragments compared with embryonic stem (ES) cell DNA. Furthermore, a 3.6-kb Vγ1.2–Jγ2 and a 1.4-kb Vγ2–Jγ1 fragment was clearly detected in IL-7R +/− mice. Quantification of the radioactivity revealed that 71% and 74% of Jγ1 and Jγ2 alleles, respectively, were rearranged in thymocytes. Because γδ T cells are only 0.3% of total thymocytes (12), the majority of the Vγ1.2–Jγ2 and Vγ2–Jγ1 recombined fragments are derived from αβ T cells or precursor cells. On the other hand, no fragment derived from Vγ1.2–Jγ2 or Vγ2–Jγ1 recombination was detected in IL-7R −/− mice (Fig. 1 A). This result demonstrates that Vγ1.2–Jγ2 and Vγ2–Jγ1 rearrangements are almost completely blocked in αβ T cells in IL-7R-deficient mice.
Figure 1.
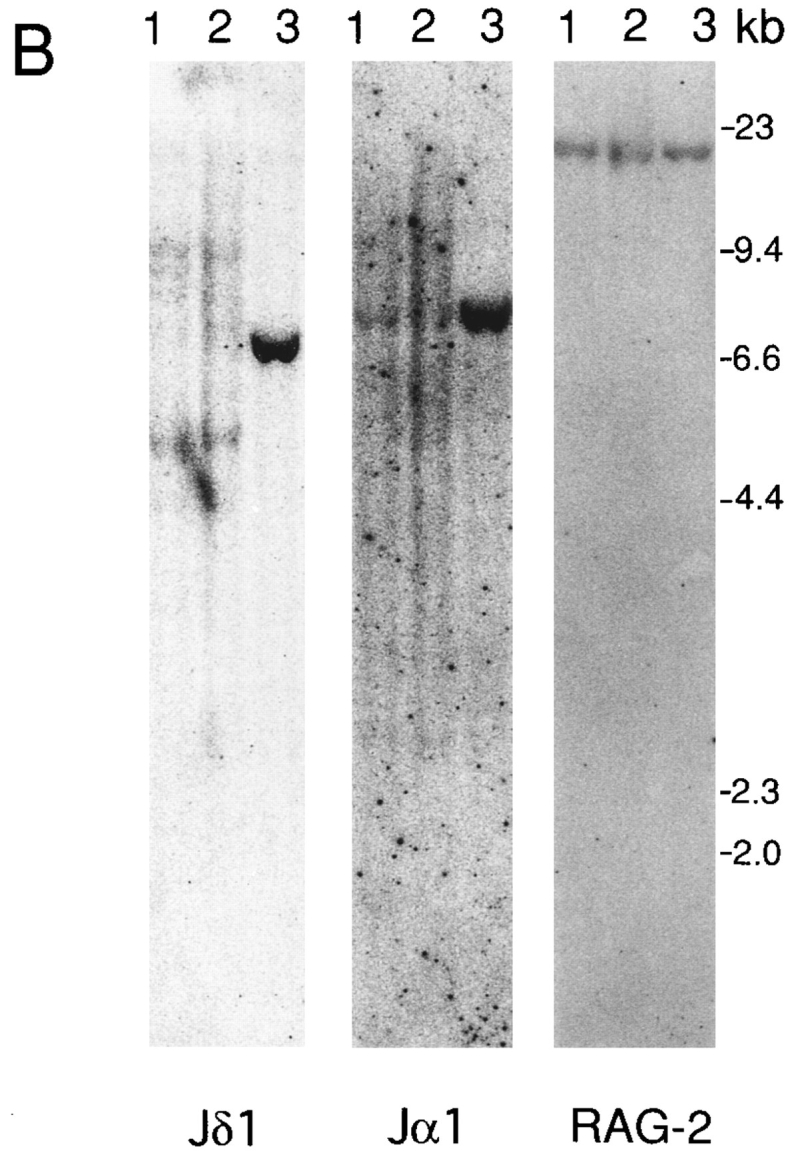
TCR gene rearrangements in the thymus of IL-7R-deficient mice. Lane 1, thymocytes from IL-7R +/− mice; lane 2, thymocytes from IL-7R −/− mice; lane 3, E14.1 ES cells. The position of HindIIIdigested phage λ DNA fragments was shown on the right. (A) Thymocyte DNA was digested with HindIII. A Southern blot was sequentially hybridized with the Jγ1 (left), the Jβ2 (middle), and the RAG-2 (right) probes. (B) Thymocyte DNA was digested with EcoRI. A Southern blot was sequentially hybridized with the Jδ1 (left), the Jα1 (middle), and the RAG-2 (right) probes.
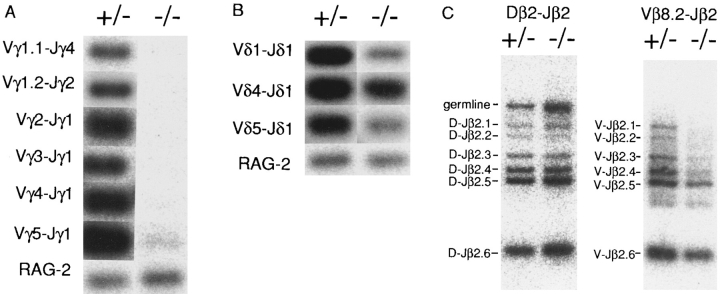
We next examined adult and fetal thymus DNA by PCR with Vγ1.1+1.2, Vγ2, Vγ3, Vγ4, Vγ5, Jγ1, Jγ2, and Jγ4 primers. Thymus DNA revealed large amounts of PCR products with all the Vγ–Jγ primer pairs in IL-7R +/− mice. On the other hand, V–J rearrangement was greatly reduced in all the TCR γ genes in IL-7R −/− thymus; the signal of Vγ5–Jγ1 product was 150-fold reduced relative to IL-7R +/− mice, and those of Vγ1.1– Jγ4, Vγ1.2–Jγ2, Vγ2–Jγ1, Vγ3–Jγ1, and Vγ4–Jγ1 products were undetectable in IL-7R −/− mice (Fig. 2 A). Amplification with RAG-2 primers produced roughly the equal amount of PCR products in both IL-7R +/− and −/− thymus, suggesting that approximately the same amount of DNA was used in this analysis. These results support the data of Southern blot analysis and suggest that the V–J recombination is almost completely blocked in IL-7R-deficient mice not only in Vγ1.2 and Vγ2 genes but also in all the other Vγ genes.
Figure 2.
TCR gene rearrangements in the thymocytes detected by PCR. Rearrangement of TCR γ (A), δ (B), and β (C) genes. The DNA from thymocytes of fetuses at day 17 of gestation (for Vγ3–Jγ1, Vγ4– Jγ1, and Vδ1–Jδ1 rearrangements) and at 4 wk old (for Vγ1.1–Jγ4, Vγ1.2–Jγ2, Vγ2– Jγ1, Vγ5–Jγ1, Vδ4–Jδ1, Vδ5– Jδ1, and all the β gene rearrangements) was amplified by PCR, and the Southern blots of the products were hybridized with oligonucleotide probes. Combination of primers used are shown on the left side (A and B). Dβ–Jβ2 and Vβ8.2–Dβ–Jβ2 rearranged fragments are shown on the left side (C).
Rearrangements of TCR α, β, and δ Genes Take Place Normally in IL-7R-deficient Mice.
We next analyzed the rearrangement of other TCR genes by Southern blot analysis. First, the Southern blot was hybridized with Jβ2 probe (see Fig. 1 A, middle). The ES cell DNA showed a 4.8-kb germline Jβ2 fragment. Thymocyte DNA showed decreased intensity of the Jβ2 germline fragment and smear patterns of Jβ2 recombined fragments in both IL-7R +/− and −/− mice. Quantification revealed that 81% and 69% of Jβ2 alleles are rearranged in IL-7R +/− and −/− thymus, respectively. Thus, the frequency of Jβ2 rearrangement is slightly decreased in IL-7R −/− mice compared with IL-7R +/− mice. Next, we hybridized a Southern blot with Jδ1 and Jα1 probes (see Fig. 1 B). A 6.8-kb Jδ1 and a 7.7-kb Jα1 germline fragment was detected in ES cell DNA. These were greatly reduced in the thymus because of deletion of the δ locus by Vα–Jα recombination in αβ T cells. Extra faint fragments and a smear pattern of Jδ recombined fragments were detected in IL-7R −/− mice as well as in IL-7R +/− mice. Thymocyte DNA from IL-7R −/− mice showed several Vδ–Dδ–Jδ and Dδ–Jδ recombined fragments at the comparable intensity with that from IL-7R +/− mice.
The rearrangement of TCR δ genes was further examined by PCR with Vδ1, Vδ4, Vδ5, and Jδ1 primers. In contrast with TCR γ genes, the signals of Vδ1–Dδ–Jδ1, Vδ4– Dδ–Jδ4, and Vδ5–Dδ–Jδ1 fragments were only slightly diminished (two- to eightfold reduction) in IL-7R −/− mice relative to IL-7R +/− mice (Fig. 2 B). Because IL-7R −/− thymus lacks γδ T cells (12), the Vδ–Jδ recombined fragments are probably derived from αβ T cells and precursor cells. Thus, the difference in the amounts of TCR δ products may be attributed to the presence and absence of γδ T cells in the thymus of IL-7R +/− and −/− mice, respectively. Collectively, TCR δ gene rearrangement seems not to be severely hampered in the IL-7R-deficient mice, supporting the data of Southern blot analysis.
Next, PCR amplification with Vβ8.2, Dβ2, and Jβ2 primers revealed six Dβ–Jβ and six Vβ–Jβ recombined fragments in both the IL-7R +/− and −/− thymus DNA (Fig. 2 C). These results demonstrate that IL-7R is not essential for both Dβ–Jβ and Vβ–Dβ–Jβ recombinations. It is recently reported that IL-7 supported Dβ to Jβ rearrangements but not Vβ to DβJβ rearrangement in fetal thymic organ culture of fetal liver precursor cells (26). However, our results do not support the notion that IL-7 may play some specific role on Dβ–Jβ recombination. All these results suggested that the rearrangements of TCR α, β, and δ genes take place normally in IL-7R-deficient mice.
Expression of Common Recombination Machinery in IL-7Rdeficient Mice.
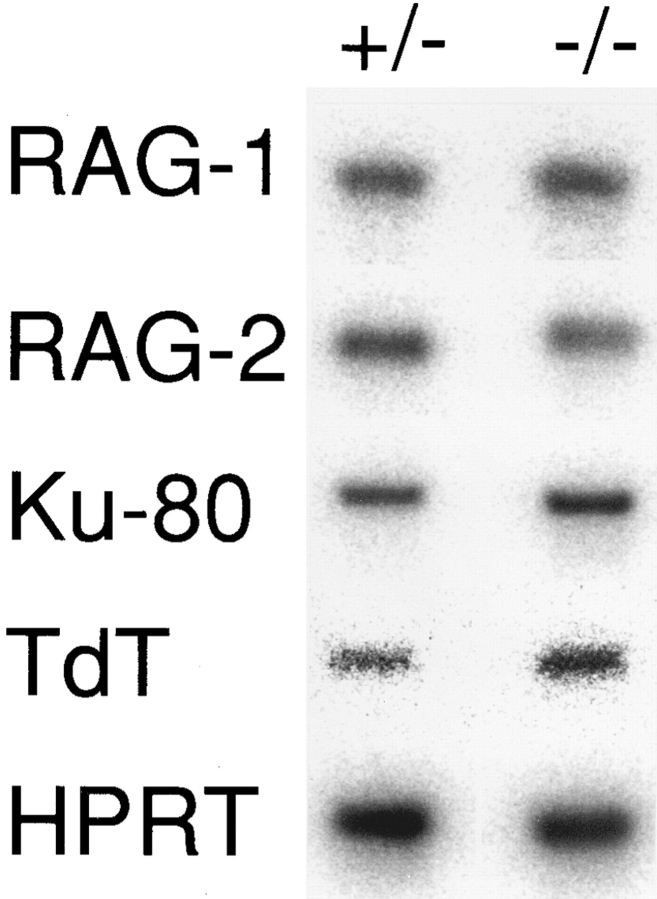
RAG-1 and RAG-2 are indispensable for V–D–J recombination, and several other gene products such as TdT, Ku p70/80 and DNA-dependent protein kinase catalytic subunit are also involved in V–D–J recombination (27). To examine whether the signal from IL-7R affects the expression of these genes, we amplified cDNA prepared from adult thymocytes of IL-7R +/− and −/− mice by PCR with RAG-1, RAG-2, TdT, and Ku-80 primers, and hybridized with each cDNA probes (Fig. 3). The levels of RAG-1, RAG-2, TdT, and Ku-80 transcripts in IL-7R −/− mice were almost comparable to those of IL-7R +/− mice. These results suggest that the mutation of IL-7R did not inhibit the expression of RAG-1, RAG-2, TdT, and Ku-80 genes.
Figure 3.
Expression of V–(D)–J recombination-associating genes in the thymus of the IL-7R-deficient mice. cDNA prepared from 4-wk-old adult thymocytes was amplified by PCR using RAG-1, RAG-2, TdT, Ku-80, and HPRT primers, and the Southern blots of PCR products were hybridized with each probe.
Discussion
TCR γ genes are frequently recombined in αβ T cells (28). More than 70% of Vγ1.2 and Vγ2 alleles are recombined in total thymocytes. In this study, we used this phenomenon to examine whether TCR γ recombination is blocked in αβ T cell precursors of IL-7R-deficient mice. IL-7R-deficient mice had no detectable TCR γ recombination by Southern blot analysis. Furthermore, the recombination of all the Vγ genes was undetectable in fetal and adult thymi by PCR analysis. Thus, we demonstrated that the signal from IL-7R is indispensable for the V–J recombination of TCR γ genes in αβ T cell precursors. And it is highly possible that the TCR γ recombination is also blocked in γδ T cell precursors as well as in αβ T cell precursors. This would be certainly one reason why IL-7Rdeficient mice lack γδ T cells.
There are three significant features in our observation. First, this blockade is specific for TCR γ genes. The recombination of TCR α, β, and δ genes are not affected. In addition, the recombination of IgH and L chain genes is probably not hampered by the mutation, because the IL-7Rdeficient mice have decreased but certain numbers of surface IgM+ B cells in the periphery (12). Second, the recombination of all the Vγ genes is blocked. In a previous report, IL-7 induced the rearrangement of Vγ2 and Vγ4, but not Vγ3 or Vγ5 genes in fetal thymic organ culture of fetal liver precursors (2). In contrast, not only Vγ2 and Vγ4 but also all the other Vγ genes in the TCR γ1, γ2, and γ4 clusters are hampered to recombine in the mutant mice. Third, TCR γ gene recombination is blocked not only in γδ but also in αβ T cell precursors. These features suggest the presence of highly specific regulation for TCR γ gene rearrangement.
To explain the specific inhibition of TCR γ recombination in the IL-7R-deficient mice, one possibility can be considered. It is to suppose that the TCR γ locus may contain a specific cis-control element(s). One possible candidate for this element is TCR γ enhancers. Recently, it was reported that IL-7 induces the phosphorylation and DNA binding activity of Stat5 protein in T cells (29). Because each TCR γ enhancer contains a Stat5 binding sequence (30, 31), Stat5 may play a role on the regulation of TCR γ recombination. It is also possible that some unknown factor(s) other than Stat5 may specifically regulate the recombination of TCR γ locus.
Acknowledgments
We thank Drs. Y. Takagaki, H. Yamagishi, O. Koiwai, and Y. Hashimoto for probes and discussion; Ms. M. Sugimori and M. Iidaka for technical assistance; Dr. S. Takeda for critically reading the manuscript; and Drs. M. Fujiwara and T. Honjo for encouragement.
Footnotes
This work was supported by a grant-in-aid from the Ministry of Education, Science, and Culture of Japan, and by the grant provided by the Ichiro Kanehara Foundation.
References
- 1.Muegge K, Vila MP, Durum SK. Interleukin-7: a cofactor for V(D)J rearrangement of the T cell receptor β gene. Science (Wash DC) 1993;261:93–95. doi: 10.1126/science.7686307. [DOI] [PubMed] [Google Scholar]
- 2.Appasamy PM, Kenniston TJ, Weng Y, Holt EC, Kost J, Chambers WH. Interleukin-7-induced expression of specific T cell receptor γ variable region genes in murine fetal liver cultures. J Exp Med. 1993;178:2201–2206. doi: 10.1084/jem.178.6.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grabstein KH, Waldschmidt TJ, Finkelman FD, Hess BW, Alpert AR, Boiani NE, Namen AE, Morrissey PJ. Inhibition of murine B and T lymphopoiesis in vivo by an anti-interleukin-7 monoclonal antibody. J Exp Med. 1993;178:257–264. doi: 10.1084/jem.178.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sudo T, Nishikawa S, Ohno N, Akiyama N, Tamakoshi M, Yoshida H, Nishikawa S. Expression and function of the interleukin-7 receptor in murine lymphocytes. Proc Natl Acad Sci USA. 1993;90:9125–9129. doi: 10.1073/pnas.90.19.9125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Freeden-Jeffry U, Vieira P, Lucian LA, McNeil T, Burdach SE, Murray R. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J Exp Med. 1995;181:1519–1526. doi: 10.1084/jem.181.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peschon JJ, Morrissey PJ, Grabstein KH, Ramsdell FJ, Maraskovsky E, Gliniak BC, Park LS, Ziegler SF, Williams DE, Ware CB, Meyer JD, Davison BL. Early lymphocyte expansion is severely impaired in interleukin-7 receptor-deficient mice. J Exp Med. 1994;180:1955–1960. doi: 10.1084/jem.180.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ikuta K, Uchida N, Friedman J, Weissman IL. Lymphocyte development from stem cells. Annu Rev Immunol. 1992;10:759–783. doi: 10.1146/annurev.iy.10.040192.003551. [DOI] [PubMed] [Google Scholar]
- 8.Haas W, Pereira P, Tonegawa S. Gamma/delta cells. Annu Rev Immunol. 1993;11:637–685. doi: 10.1146/annurev.iy.11.040193.003225. [DOI] [PubMed] [Google Scholar]
- 9.Watanabe Y, Sudo T, Minato N, Ohnishi A, Katsura Y. Interleukin 7 preferentially supports the growth of γδ T cell receptor–bearing T cells from fetal thymocytes in vitro. . Int Immunol. 1991;3:1067–1075. doi: 10.1093/intimm/3.11.1067. [DOI] [PubMed] [Google Scholar]
- 10.Matsue H, Bergstresser PR, Takashima A. Keratinocyte-derived IL-7 serves as a growth factor for dendritic epidermal T cells in mice. J Immunol. 1993;151:6012–6019. [PubMed] [Google Scholar]
- 11.Watanabe M, Ueno Y, Yajima T, Iwao Y, Tsuchiya M, Ishikawa H, Aiso S, Hibi T, Ishii H. Interleukin 7 is produced by human intestinal epithelial cells and regulates the proliferation of intestinal mucosal lymphocytes. J Clin Invest. 1995;95:2945–2953. doi: 10.1172/JCI118002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maki K, Sunaga S, Komagata Y, Kodaira Y, Mabuchi A, Karasuyama H, Yokomuro K, Miyazaki J, Ikuta K. Interleukin-7 receptor–deficient mice lack γδ T cells. Proc Natl Acad Sci USA. 1996;93:7172–7177. doi: 10.1073/pnas.93.14.7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao X, Shores EW, Hu LJ, Anver MR, Kelsall BL, Russell SM, Drago J, Noguchi M, Grinberg A, Bloom ET, Paul WE, Katz SI, Love PE, Leonard WJ. Defective lymphoid development in mice lacking expression of the common cytokine receptor γ chain. Immunity. 1995;2:223–238. doi: 10.1016/1074-7613(95)90047-0. [DOI] [PubMed] [Google Scholar]
- 14.Park SY, Saijo K, Takahashi T, Osawa M, Arase H, Hirayama N, Miyake K, Nakauchi H, Shirasawa T, Saito T. Developmental defects of lymphoid cells in Jak3 kinase-deficient mice. Immunity. 1995;3:771–782. doi: 10.1016/1074-7613(95)90066-7. [DOI] [PubMed] [Google Scholar]
- 15.Takagaki Y, Nakanishi N, Ishida I, Kanagawa O, Tonegawa S. T cell receptor-γ and -δ genes preferentially utilized by adult thymocytes for the surface expression. J Immunol. 1989;142:2112–2121. [PubMed] [Google Scholar]
- 16.Winoto A, Mjolsness S, Hood L. Genomic organization of the genes encoding mouse T-cell receptor α-chain. Nature (Lond) 1985;316:832–836. doi: 10.1038/316832a0. [DOI] [PubMed] [Google Scholar]
- 17.Takeshita S, Toda M, Yamagishi H. Excision products of the T cell receptor gene support a progressive rearrangement model of the α/δ locus. EMBO (Eur Mol Biol Organ) J. 1989;8:3261–3270. doi: 10.1002/j.1460-2075.1989.tb08486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikuta K, Hattori M, Wake K, Kano S, Honjo T, Yodoi J, Minato N. Expression and rearrangement of the α, β, and γ chain genes of the T cell receptor in cloned murine large granular lymphocyte lines: no correlation with the cytotoxic spectrum. J Exp Med. 1986;164:428–442. doi: 10.1084/jem.164.2.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science (Wash DC) 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 20.Itohara S, Mombaerts P, Lafaille J, Iacomini J, Nelson A, Clarke AR, Hooper ML, Farr A, Tonegawa S. T cell receptor δ gene mutant mice: independent generation of αβ T cells and programmed rearrangements of γδ TCR genes. Cell. 1993;72:337–348. doi: 10.1016/0092-8674(93)90112-4. [DOI] [PubMed] [Google Scholar]
- 21.Palacios R, Samaridis J. Bone marrow clones representing an intermediate stage of development between hematopoietic stem cells and pro-T-lymphocyte or proB-lymphocyte progenitors. Blood. 1993;81:1222–1238. [PubMed] [Google Scholar]
- 22.Levin SD, Anderson SJ, Forbush KA, Perlmutter RM. A dominant–negative transgene defines a role for p56lckin thymopoiesis. EMBO (Eur Mol Biol Organ) J. 1993;12:1671–1680. doi: 10.1002/j.1460-2075.1993.tb05812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson SJ, Abraham KM, Nakayama T, Singer A, Perlmutter RM. Inhibition of T-cell receptor β-chain gene rearrangement by overexpression of the non-receptor protein tyrosine kinase p56l ck . EMBO (Eur Mol Biol Organ) J. 1992;11:4877–4886. doi: 10.1002/j.1460-2075.1992.tb05594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol– chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 25.Koiwai O, Yokota T, Kageyama T, Hirose T, Yoshida S, Arai K. Isolation and characterization of bovine and mouse terminal deoxynucleotidyltransferase cDNAs expressible in mammalian cells. Nucleic Acids Res. 1986;14:5777–5792. doi: 10.1093/nar/14.14.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsuda S, Rieke S, Hashimoto Y, Nakauchi H, Takahama Y. IL-7 supports D–J but not V–DJ rearrangement of TCR-β gene in fetal liver progenitor cells. J Immunol. 1996;156:3233–3242. [PubMed] [Google Scholar]
- 27.Lin WC, Desiderio S. V(D)J recombination and the cell cycle. Immunol Today. 1995;16:279–289. doi: 10.1016/0167-5699(95)80182-0. [DOI] [PubMed] [Google Scholar]
- 28.Heilig JS, Tonegawa S. T-cell γ gene is allelically but not isotypically excluded and is not required in known functional T-cell subsets. Proc Natl Acad Sci USA. 1987;84:8070–8074. doi: 10.1073/pnas.84.22.8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foxwell BM, Beadling C, Guschin D, Kerr I, Cantrell D. Interleukin-7 can induce the activation of Jak 1, Jak 3 and STAT 5 proteins in murine T cells. Eur J Immunol. 1995;25:3041–3046. doi: 10.1002/eji.1830251109. [DOI] [PubMed] [Google Scholar]
- 30.Vernooij BT, Lenstra JA, Wang K, Hood L. Organization of the murine T-cell receptor γ locus. Genomics. 1993;17:566–574. doi: 10.1006/geno.1993.1373. [DOI] [PubMed] [Google Scholar]
- 31.Ihle JN. STATs: signal transducers and activators of transcription. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]