Abstract
Major histocompatibility complex (MHC) class II–positive cell lines which lack HLA-DM expression accumulate class II molecules associated with residual invariant (I) chain fragments (class II–associated invariant chain peptides [CLIP]). In vitro, HLA-DM catalyzes CLIP dissociation from class II–CLIP complexes, promoting binding of antigenic peptides. Here the physical interaction of HLA-DM with HLA-DR molecules was investigated. HLA-DM complexes with class II molecules were detectable transiently in cells, peaking at the time when the class II molecules entered the MHC class II compartment. HLA-DR αβ dimers newly released from I chain, and those associated with I chain fragments, were found to associate with HLA-DM in vivo. Mature, peptide-loaded DR molecules also associated at a low level. These same species, but not DR-I chain complexes, were also shown to bind to purified HLA-DM molecules in vitro. HLA-DM interaction was quantitatively superior with DR molecules isolated in association with CLIP. DM-DR complexes generated by incubating HLA-DM with purified DR αβCLIP contained virtually no associated CLIP, suggesting that this superior interaction reflects a prolonged HLA-DM association with empty class II dimers after CLIP dissociation. Incubation of peptide-free αβ dimers in the presence of HLA-DM was found to prolong their ability to bind subsequently added antigenic peptides. Stabilization of empty class II molecules may be an important property of HLA-DM in facilitating antigen processing.
The recognition of antigen by CD4+ T helper cells requires the presentation of short peptides displayed in the binding groove of MHC class II molecules (1, 2). MHC class II molecules are cell surface expressed heterodimeric (αβ) glycoproteins. Newly synthesized class II α and β chains assemble in the ER with a third transmembrane glycoprotein, the invariant (I) chain (3, 4). With the aid of the molecular chaperone calnexin, three αβ dimers bind sequentially to a trimer of the I chain in the ER (5, 6). The nonameric complex moves through the Golgi apparatus and is sorted by signals in the cytoplasmic domain of the I chain (7) and the class II β chain (8) to endosomal compartments with late endosomal (class II–containing vesicles, CIIV; 9) or lysosome-like characteristics (MHC class II compartment [MIIC]1; 10–13) where the I chain is proteolytically cleaved by aspartic and cysteine proteases (14–17). The released αβ dimers are loaded with peptides derived from internalized pathogen-derived or endogenous proteins present in the endocytic system and transported to the cell surface.
The expression of HLA-DM, encoded by MHC-linked genes (18, 19), is required for class II–restricted processing and presentation of most protein antigens but not for the presentation of exogenously added antigenic peptides (reviewed in reference 20). Class II molecules in DM-negative B-lymphoblastoid cell lines (B-LCL) are expressed on the cell surface at wild-type levels but are associated with a set of peptides derived from residues 81-104 of the I chain (class II–associated invariant chain peptides, or CLIP) instead of antigenic peptides (21, 22). Similar complexes accumulate in class II–positive cell lineages in mice in which the Ma (DMA) gene is disrupted (23–25). The ability to isolate αβCLIP complexes from wild-type APCs (2), their transient appearance during pulse–chase analysis of class II transport (26–27), and the proteolytic generation of αβCLIP complexes from αβI in vitro (27), all indicate that αβCLIP complexes are an intermediate in the class II processing pathway and that CLIP probably represents the end product of I chain proteolysis. Recent x-ray crystallographic analysis of HLA-DR3-CLIP complexes has demonstrated that CLIP binds in the antigen binding groove of the class II molecules (28) indicating that CLIP must first be removed before endocytically generated peptides can bind.
Recently, we and others have shown that CLIP removal and peptide loading are directly catalyzed by DM in vitro (29–31), suggesting that DM also removes CLIP from αβCLIP complexes in vivo. For DM to catalyze the release of CLIP, it seemed likely that a DM–class II interaction must occur and, indeed, Sanderson et al. (32) have shown by coprecipitation that DM and DR associate under steady state conditions in vivo. The association is favored by low pH in the nonionic detergent digitonin, and occurs in dense compartments, probably in MIICs. In this study, using biosynthetic labeling and coprecipitation of DR with DM, we show that in vivo the DM-DR association is transient, and that DM associates with peptide loaded αβ dimers, extending the initial observations reported by Sanderson et al. (32). Additionally, we examine the in vitro interaction of affinity-purified DM with DR molecules associated with I chain, fragments of I chain, CLIP, or the normal complement of peptides expressed in wild-type cells. The results indicate a degree of specificity for αβCLIP in the interaction, and suggest that one function of HLA-DM is to prolong the survival of empty class II molecules in the MIIC, a chaperone-like function which is likely to be important in antigen processing.
Materials and Methods
Cell Lines.
The EBV transformed B-lymphoblastoid cell line (B-LCL) Pala (HLA-DR3.Dw17) and the T × B hybrid cell line .174 × CEM.T2 (33) transfected with HLA-DR3 (T2.DR3; 34) were maintained in Iscove's DMEM (GIBCO BRL, Gaithersburg, MD) with 5% calf serum (GIBCO BRL) in the presence of 5% CO2 at 37°C. The derivation of the mutant B-LCL 10.24.6 (HLA-DR3, -DRw52a, -DQ2, -DP4.1) and the progenitor BLCL 8.1.6 have been described (35). These cells were maintained in RPMI (GIBCO BRL) supplemented with 1% glutamine (GIBCO BRL) and 10% calf serum.
Antibodies and Peptide.
The hybridoma cell lines L243 (antiHLA-DR αβ; 36), DA6.147 (anti-HLA-DR α chain; 37), GAP.A3 (anti-HLA-A3; 38), W6/32 (anti-HLA-class I; 39), HB10A (anti-HLA-DR β chain; 40), PIN.1 (anti-I chain NH2terminal; 41), and CerCLIP.1 (anti-CLIP24; 42), have been previously described. Rabbit anti-DM serum was a generous gift of Dr. Hans Zweerink (Merck Research Laboratories, Rahway, NJ) and was generated against recombinant, soluble DM, (31). Rabbit anti-C23V serum, specific for an influenza hemagglutinin peptide, was generated in our laboratory and has been described (42).
The DR3-restricted MOMP peptide (QASLALSYRLNMFTP; 21) is derived from residues 251-265 of the major outer membrane protein of Chlamydia trachomatis and was synthesized by the Keck Foundation Biotechnology Resource Laboratory (Yale University). The peptide was purified by RP-HPLC and laser desorption mass spectrometry indicated that it was a single species.
Metabolic Labeling.
Cells for pulse labeling were washed once with methionine and cysteine-free DMEM supplemented with 3% dialyzed FBS, 15 mM Hepes, 2 mM glutamine, and 1 mM sodium pyruvate (labeling medium; all from GIBCO BRL) and preincubated at 3 × 106 cells/ml in labeling medium for 60 min at 37°C. Cells were pelleted and resuspended at 1 × 107 cells/ml in fresh labeling medium plus 1 mCi/ml of l-[35S]methionine in vitro cell labeling mix (Amersham Corp., Arlington Heights, IL). After incubation at 37°C for 15 min to 6 h, the pulsed cells were pelleted, resuspended at 1 × 106 cells/ml with IDMEM/5% calf serum/5% FBS containing a 15-fold excess of cold methionine and cysteine, and then chased for up to 18 h at 37°C. At each chase point, the cells were diluted into ice-cold serum-free IDMEM, pelleted by centrifugation, and either lysed immediately for immunoprecipitation and affinity purification or stored at −20°C until ready for use.
Immunoprecipitations and Endo H Digestion.
Radiolabeled cell pellets were extracted for 30 min on ice in 20 mM Tris, 130 mM NaCl (pH 7.4) (TBS), containing 1% CHAPS (Pierce, Rockford, IL), or 1% CHAPS plus 1% Triton X-100 (Sigma Chem. Co., St. Louis, MO), 0.5 mM PMSF, 0.1 mM TLCK, and 5 mM iodoacetamide (Sigma) at 5 × 105–2 × 106 cells/ml. After the removal of nuclear material by centrifugation, lysates were precleared with 2 μl normal rabbit serum and 75 μl Zysorbin (Zymed Laboratories, South San Francisco, CA) per ml of cell lysate for 1 h at 4°C before incubation with specific antibody (2–3 μl ascites or rabbit antiserum, 100 μl tissue culture supernatant) and protein A–Sepharose (Sigma) or protein G–Sepharose (Pharmacia, Piscataway, NJ). For anti class II or I chain immunoprecipitations 5 × 105 cell equivalents were used. For anti DM immunoprecipitation 2 × 106 cell equivalents were used. Sepharose pellets were washed three times with TBS plus 1% CHAPS or 10 mM Tris, pH 8.0, 300 mM NaCl, 0.1% SDS, 0.05% Triton X-100, or TBS plus 0.1% Triton-100, and stored at −20°C. For reprecipitation experiments, 1 ml of TBS containing 0.5% deoxycholate (DOC; Sigma) was added to the DM precipitates and they were incubated for 3 h at 4°C. After centrifugation, the released DR was immunoprecipitated from the DOC supernatants as described above. For Endo H digestion, beads were boiled for 5 min in 20 μl of 0.06 M NaPO4, 0.3% SDS, 0.06% NaN3, diluted with 40 μl H2O, and the supernatants were divided into two aliquots. To one aliquot, 2 mU recombinant Endo H (Boehringer Mannheim, Indianapolis, Indiana) was added and both aliquots were incubated overnight at 37°C.
For SDS-PAGE analysis of precipitates, reducing or non-reducing Laemmli sample buffer was added and the samples were either boiled for 5 min or incubated at room temperature for 30 min before analysis by SDS-PAGE (10.5–12%). Gels were treated for fluorography with 125 mM salicylate, dried and exposed to film. Bands were quantitated with a Bio-Rad GS-525 Molecular Imager, subtracting the background obtained by integrating a blank area of the appropriate size on each gel.
Affinity Purification of Radiolabeled Class II Complexes and HLADM.
Affinity purification of αβI, αβLIP, αβCLIP, and αβ peptide complexes from radiolabeled cell lysates were as previously described (14, 29, 43). The affinity purification of HLADM from wild-type B-LCLs has also been described (29).
SDS Stability Assay.
Affinity-purified HLA-DM (2 ng) was incubated with 500 nM MOMP peptide and ∼10,000 CPM of radiolabeled mutant (10.24.6-derived) or wild-type αβCLIP complexes in TBS (pH 8), 1% CHAPS. The pH was adjusted to 5.0 by the addition of 1 M acetic acid and the samples were incubated at 37°C for 0–120 min. After neutralization by the addition of 1 M Tris, 10× non-reducing Laemmli sample buffer was added and the samples were separated by 10.5% SDS-PAGE.
In Vitro Association Assay.
Affinity–purified, radiolabeled αβI, αβLIP/SLIP, αβCLIP, αβ peptide, or 10.24.6 αβCLIP complexes (∼40,000 CPM) in TBS (pH 8), 1% CHAPS were incubated in the absence or presence of either 50 ng affinity-purified DM or decreasing amounts of DM (50 ng to 0.156 ng) and the pH was adjusted to 5.0 by the addition of 1 M acetic acid. After an incubation at 37°C for 20–30 min, the samples were placed on ice and 500 μl of 50 mM sodium acetate, 150 mM NaCl (pH 6.0), 1% CHAPS and 2 μl anti-DM or anti-C23V and 25 μl protein A–Sepharose were added. The samples were incubated for 30 min at 4°C with rotating, washed three times with 50 mM sodium acetate, 150 mM NaCl (pH 6.0), 1% CHAPS, analyzed by SDS-PAGE and quantitated by image analysis.
In Vitro Stabilization Assay.
Radiolabeled DR3 αβCLIP complexes (10,000 cpm; 25 nM) were incubated in the presence or absence of affinity-purified HLA-DM (25 nM) in TBS (pH 8.0), 1% octyl glucoside. The pH was adjusted to pH 4.5 and the samples were incubated at 37°C for 0 to 10 h. At various time points, MOMP peptide was added to a concentration of 1 μM and the samples were incubated for an additional 45 min at 37°C. After neutralization by the addition of 1 M Tris, 10× non-reducing Laemmli sample buffer was added and the samples were separated by 10.5% SDS-PAGE and analyzed by image analysis.
Results
HLA-DM and -DR Associate In Vivo.
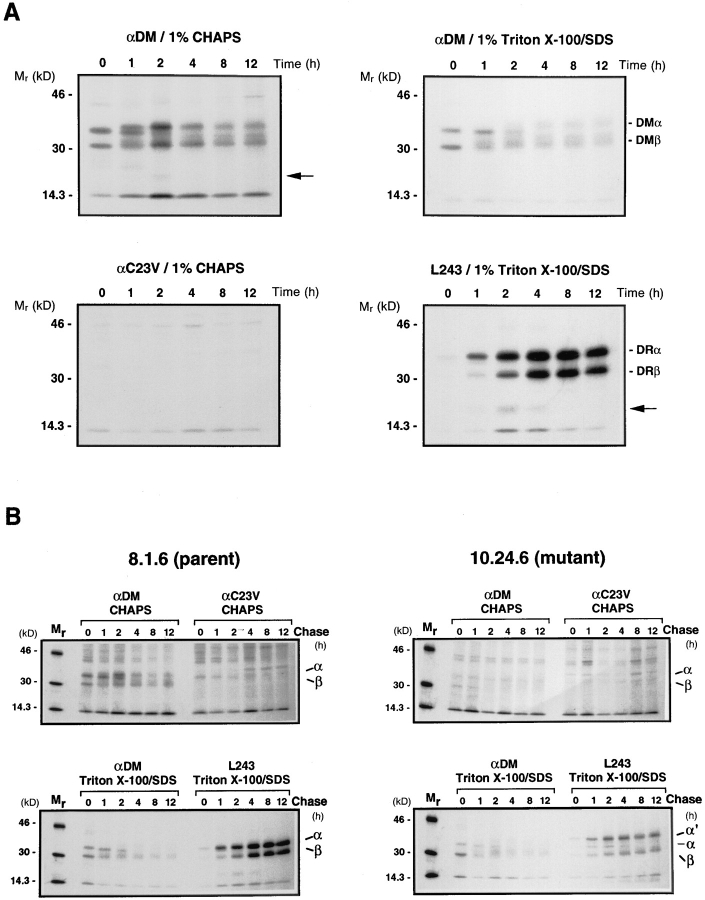
HLA-DM has been shown to mediate the exchange of CLIP for antigenic peptides under acidic conditions in vitro (29–31). For DM to catalyze the release of CLIP, it seemed likely that a direct physical interaction between DM and class II molecules must occur. HLA-DM is a resident of the MIIC (44–47), and class II molecules traffic through this compartment to be loaded with antigenic peptides by DM before subsequent arrival at the cell surface. Therefore, any interaction between DM and DR molecules should occur in the MIIC and be transient in nature. Sanderson et. al. (32), have demonstrated direct DM-DR association under steady-state conditions in vivo. To examine the kinetics of DM-DR association, wild-type Pala cells were pulsed for 15 min with [35S]methionine and chased in the presence of 15-fold excess methionine and cysteine for up to 12 h (Fig. 1 A). At each time point, radiolabeled cells were lysed in 1% CHAPS and split into four aliquots. One aliquot was immunoprecipitated with a polyclonal rabbit antiserum to the DM heterodimer (anti-DM) and another with a negative control rabbit antiserum to an influenza hemagglutinin peptide (anti-C23V). DM and DR have similar mobilities on SDSPAGE, however their processing patterns are somewhat different. DM-DR association has been shown to be sensitive to the detergent used for solubilization of the cells (32; unpublished data). To determine the exact position and processing pattern of DM and DR on SDS-PAGE, 1% Triton X-100 was added to the other two aliquots of cell lysate to dissociate the DM-DR complexes. Immunoprecipitation was performed with anti-DM or with the DRspecific mAb, L243, followed by extensive washing with a buffer containing Triton X-100 and SDS (see Materials and Methods) to ensure any remaining DM-DR complexes were dissociated. SDS-PAGE analysis (Fig. 1 A, lower right) shows that in Pala cells L243-reactive αβ dimers were initially generated between 1 and 2 h of chase and that probable invariant chain fragments (26; indicated on the right hand side with an arrow) were coprecipitated with the DR molecules early in the chase period (1 to 4 h). Immunoprecipitation with anti-DM serum (Fig. 1 A; upper right) revealed the position of the DMα and DMβ chains in the absence of any associated DR molecules. DM αβ dimers were observed immediately after the pulse and remained throughout the chase period, consistent with previous reports (42). Comparison of the anti-DM and L243 immunoprecipitates in Triton X-100 clearly demonstrates that the DM and DR processing patterns on SDS-PAGE are very different. SDS-PAGE analysis of an anti-DM immunoprecipitate in the detergent CHAPS (Fig. 1A; upper left) showed that after the pulse point, only the DM αβ dimer was precipitated. However, after 1 h of chase additional species with the same mobilities as DRα and β subunits coprecipitated with the DM dimers. The coprecipitated species peaked at 2 h and then decreased with time, with some remaining associated even at the 12 h time point. Additionally, fragments of the invariant chain (marked with an arrow) were coprecipitated with the DM complex at 2 h indicating that DM can associate with putative DR molecules still complexed with these fragments, suggesting that HLA-DM is associated with the class II-I chain complex during degradation of the I chain.
Figure 1.
(A) Kinetics of HLADM and HLA-DR association in wild-type Pala cells. Cells were pulse labeled with [35S]methionine for 15 min and chased in the presence of an excess of methionine and cysteine for the indicated periods of time (h). Class II and DM molecules were immunoprecipitated at each time point from CHAPS or Triton X-100 solubilized lysates (as indicated above each gel) with the DR-specific mAb L243 or with a polyclonal rabbit antiserum specific for the DM heterodimer (αDM) and analyzed by SDSPAGE (10.5%). Control precipitates were performed from CHAPS solubilized lysates with an antiserum specific for an influenza hemagglutinin peptide (αC23V). The bands corresponding to the DMα, DMβ, DRα and DRβ proteins are marked on the right and an I degradation fragment is indicated by a solid arrow head. (B) HLADM does not associate with HLA-DR in 10.24.6 cells expressing a mutant DRα chain containing an extra N-linked glycan. Mutant 10.24.6 and wild-type 8.1.6 cells were pulsed labeled, chased and immunoprecipitated as in A. The positions of the individual class II and DMα and β chains are indicated on the right and α′ marks the position of the mutant DRα chain expressed by 10.24.6 cells.
DM-positive 10.24.6 cells express a mutant DR α chain containing a proline to serine substitution at position 96 that results in the addition of an extra N-linked glycan. In this cell line DR3 αβ dimers accumulate as αβCLIP complexes (35). Purified αβCLIP complexes from 10.24.6 cannot be loaded with peptide by recombinant soluble DM in vitro (31) or by affinity-purified native DM (see below), and steady-state DM-DR association is greatly reduced in this cell line (32). To confirm that the putative DR-DM interaction seen in wild-type Pala cells was specific, we repeated the pulse-chase experiment with the 10.24.6 cell line and the matched parental cell line, 8.1.6 (35). The results (Fig. 1 B, upper left) showed that in the wild-type cell line 8.1.6, DR was coprecipitated with DM and that this association was strongest at 1–2 h of chase, similar to what was observed in Pala cells (Fig. 1 A). In 10.24.6 cells, antiDM and L243 (anti-DR) precipitations in Triton X-100 clearly showed that this cell line expresses both DM and DR (Fig. 1 B lower right). The reduced mobility of the mutant DRα chain (α′) caused by the additional N-linked glycan is readily apparent. None of the mutant DR was coprecipitated with DM in CHAPS (Fig. 1 B, upper right) even upon prolonged autoradiographic exposure (data not shown).
DM Associates with DR in a Post-Golgi Compartment.
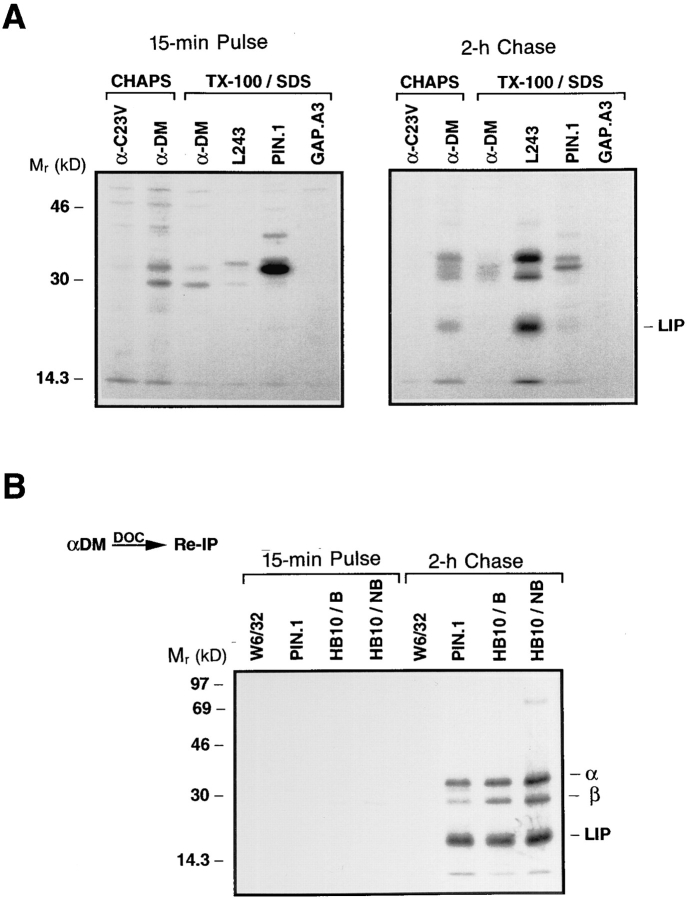
Pulsechase analysis (Fig. 1 A) showed that DM-DR association was maximal after 1–2 h of chase, the time at which the αβI complex is delivered into the MIIC and the invariant chain is being degraded in this cell line (46; unpublished results). Therefore, the interaction must occur in a post-Golgi compartment and the DM-associated DR should be resistant to Endo H digestion. To determine this, wild-type Pala cells were pulse-labeled for 15 min with [35S]methionine and chased in the presence of an excess of methionine and cysteine. After lysis of the cells in CHAPS, the DM–class II complexes were immunoprecipitated with anti-DM. The coprecipitated DR was eluted from the DM immunoprecipitates with 0.5% DOC and reprecipitated from supernatants with a DR β chain–specific mAb, HB10A or with a negative control antibody to HLA class I molecules, W6/ 32. Duplicate HB10A and W6/32 precipitates were treated with Endo H or mock-treated overnight and analyzed by SDS-PAGE. No DM associated DR was detected early in transport since the faint Endo H–sensitive bands present immediately after the pulse in the HB10A precipitates (Fig. 2 A) were also present in the negative control precipitates (Fig. 2 B). As observed in Fig. 1, DM-associated DR was first detected after 1 h of chase and the associated DR was resistant to Endo H digestion (Fig. 2 A, note that one of the DRα chain N-linked glycans remained in the high mannose form upon maturation; 48). These data indicate that DM and DR are not associated in the ER and that DM-DR must interact after the DR molecules have traversed the medial Golgi. No HLA class I molecules were coprecipitated with DM, verifying the specificity of the DR-DM association (Fig. 2 B).
Figure 2.
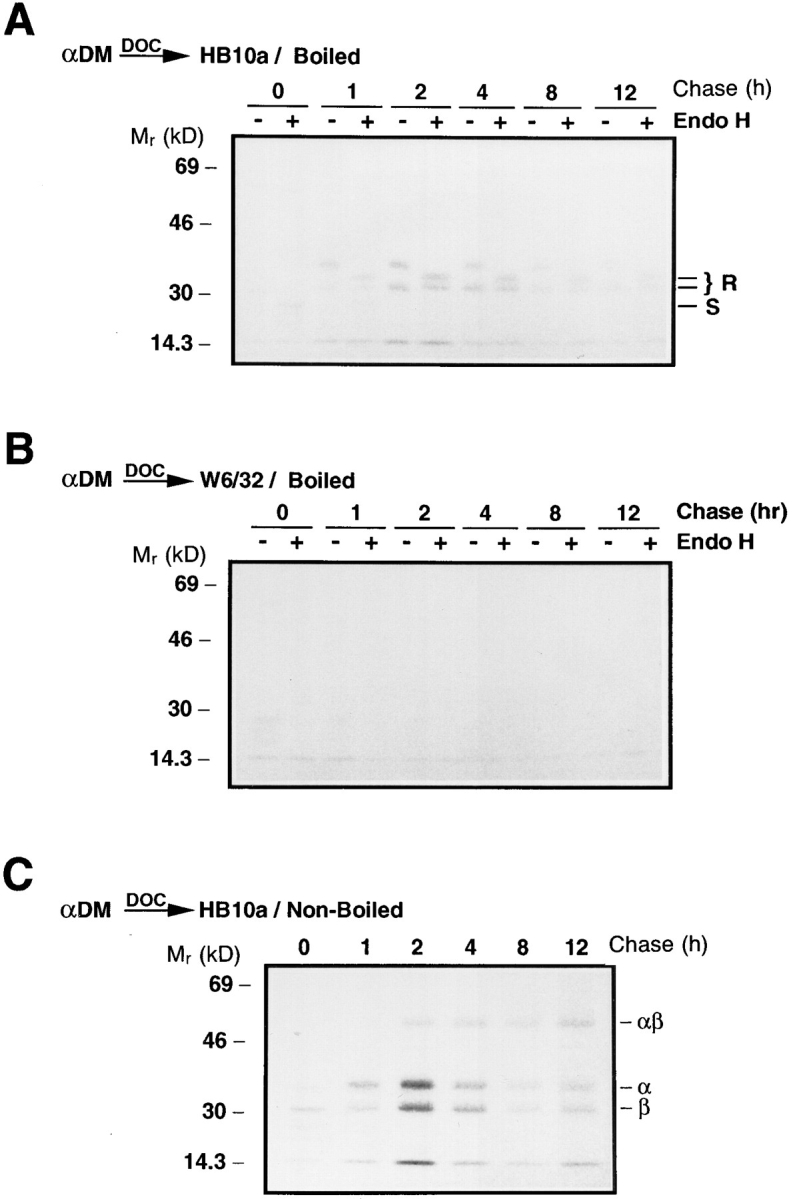
(A and B) HLA-DM associates with DR in a post-Golgi compartment. Pala cells were pulsed labeled with [35S]methionine for 15 min and chased for the indicated periods of time (h). DM-DR complexes were immunoprecipitated at each time point from CHAPS solubilized lysates with anti-DM (αDM). DM-associated class II molecules were eluted with 0.5% DOC and the supernatants reprecipitated with DR β chain-specific mAb HB10A (A) or control mAb W6/32 (B). The precipitates were either treated with Endo H or mock treated and analyzed by SDS-PAGE (10.5%). Locations of Endo H resistant (R) and sensitive (S) DR α and β bands are indicated on the right. (C) HLA-DM associates with SDS-stable αβ dimers. Pala cells were pulse labeled, immunoprecipitated and re-precipitated with mAb HB10A as in A. After the addition of Laemmli sample buffer, the precipitates were incubated at room temperature for 30 min and analyzed by SDS-PAGE (10.5%). The positions of the individual α and β chains and the αβ dimers are indicated on the right.
DM can be coprecipitated by the DR3 conformational specific mAb 16.23 (32). This antibody is thought to recognize only peptide loaded DR3 molecules and does not bind αβI or αβCLIP complexes (49). Sanderson et al. (32), suggest that because DM can be efficiently coprecipitated by mAb 16.23, that the DM is able to associate with mature, peptide-loaded DR3 molecules. Since the exact specificity of mAb 16.23 is not known, the DM associated DR they observed may actually be empty DR3 molecules. SDS-stability of class II molecules has been shown to correlate with peptide binding (50). To determine if DM can associate with peptide-loaded DR molecules, Pala cells were pulsed with [35S]methionine, chased for various times, extracted in CHAPS, and immunoprecipitated with antiDM. After release of the DM-associated DR by 0.5% DOC, the DR was reprecipitated with HB10A as described above. The HB10A immunoprecipitates were incubated in SDS-PAGE sample buffer at room temperature for 30 min and analyzed by SDS-PAGE for SDS-stable, peptide-loaded αβ dimers. The peak of class II association with DM was again at 2 h, and at this time the majority is clearly unstable. However, stable αβ dimers were clearly present, indicating that DM can associate with mature, peptide-loaded αβ dimers (Fig. 2 C ). This result is reproducible and does not reflect an artefactual, post-solubilization association of DM and mature DR molecules, because coextraction under the same conditions of labeled T2.DR3 cells mixed with unlabeled T2.DM cells did not generate such complexes (data not shown). Although the relative amount of the total DR that was SDS-stable increased with time, the majority of the DM-associated DR was not stable in SDS. This suggests that most of the peptide loaded DR dissociated from DM during the chase.
DM Associates with αβLIP and αβSLIP Complexes In Vivo.
We have shown that DM is able to catalyze in vitro the removal of LIP, a 21-kD NH2-terminal fragment of the I chain that accumulates in the presence of the protease inhibitor leupeptin, from αβLIP complexes (29). This suggested that DM can interact with class II complexed with fragments of the I chain. Additionally, pulse–chase analysis of DM in the detergent CHAPS (Fig. 1 A) demonstrated a band at 2 h that coprecipitated with DM and probably is a fragment of the I chain. To directly examine if DM can interact with DR complexed with I chain fragments, Pala cells were pulsed with [35S]methionine for 15 min and chased for 2 h in the presence of the protease inhibitor leupeptin to accumulate αβLIP (14) and αβSLIP (15) complexes. Radiolabeled cells were lysed in CHAPS, immunoprecipitated with anti-DM or anti-C23V (negative control), or lysed in CHAPS plus Triton X-100 and immunoprecipitated with anti-DM, the DR specific mAb L243, the I chain specific mAb PIN.1, or the HLA-A3 specific mAb, GAP.A3 (negative control) and analyzed by SDS-PAGE. Immediately after the pulse, as expected, no DM-associated DR was observed and no αβLIP and αβSLIP had yet been generated (Fig. 3 A). After a 2-h chase, the mAb L243 precipitated DR αβ in addition to lower molecular weight fragments of the I chain, LIP and SLIP (which runs on the dye front of the gel). Immunoprecipitation with anti-DM showed that αβLIP and αβSLIP complexes were coprecipitated with DM in the detergent CHAPS but not in Triton X-100 (Fig. 3 A), as observed in Fig. 1. These results show that DM can associate with DR complexed with I chain fragments.
Figure 3.
HLA-DM associates with αβLIP and αβSLIP complexes. (A) Pala cells were pulse labeled for 15 min with [35S]methionine and chased for 2 h in the presence of an excess of methionine and cysteine. Class II, DM and I chain molecules were immunoprecipitated from CHAPS solubilized lysates with anti-DM (αDM) or control anti-C23V (αC23V) or from Triton X-100 solubilized lysates with anti-DM, DR-specific mAb L243, I chain–specific mAb PIN.1 or control mAb GAP.A3 and analyzed by SDS-PAGE (12%). (B) Wild-type Pala cells were pulse labeled and chased as in A. DM-associated class II and class II–I chain complexes were eluted with 0.5% DOC, the supernatants reprecipitated with mAbs PIN.1, HB10A, and W6/32 (control) and analyzed by SDS-PAGE (12%). The positions of the individual DR α and β chains and LIP are indicated on the right. The right lane shows the HB10A-precipitated material run without boiling the sample.
Previous in vivo analysis has shown that DM coprecipitated with full-length I chain in the ER and with I chain fragments (including LIP) in dense Percoll gradient fractions (32). However, these experiments were performed using immunoprecipitation with an antibody specific for the NH2-terminal region of the I chain (VICY1) followed by Western blotting, and thus it was impossible to distinguish between direct DM interaction with the I chain or association with the I chain via its interaction with class II molecules. A similar problem exists in the experiment shown in Fig. 3 A. An experiment was designed to determine whether the DM-associated I chain fragments seen in Fig. 3 A were free or complexed to class II molecules. Pala cells were pulsed for 15 min with [35S]methionine, chased for 2 h in the presence of leupeptin, lysed in CHAPS and immunoprecipitated with anti-DM. The DM-associated material was eluted by the addition of 0.5% DOC, which should not dissociate αβI or αβ complexed to fragments of the I chain. The eluted complexes were reprecipitated from the DOC supernatant with mAb PIN.1 (anti-I chain), mAb HB10A (anti-DR β chain) or a negative control mAb W6/32 (anti-class I). SDS-PAGE analysis of the immunoprecipitates is shown in Fig. 3 B. Immunoprecipitation with the class II specific mAb HB10A showed that after a 15-min pulse, no class II was associated with DM and reprecipitation with an antibody specific for the I chain showed that very little, if any, I chain was associated with DM. After 2 h of chase, re-precipitation of DM-associated class II with mAb HB10A showed that LIP was clearly present, demonstrating that DM interacts with αβLIP complexes. Reprecipitation with the I chain specific mAb PIN.1 also precipitated αβLIP complexes. Because HLADM can bind to αβCLIP and αβ peptide complexes (see below), it seems likely that DM associates with LIP via an interaction with the DR molecule and not directly with the LIP fragment itself.
Mutant αβCLIP Complexes Cannot Be Loaded by Affinitypurified HLA-DM.
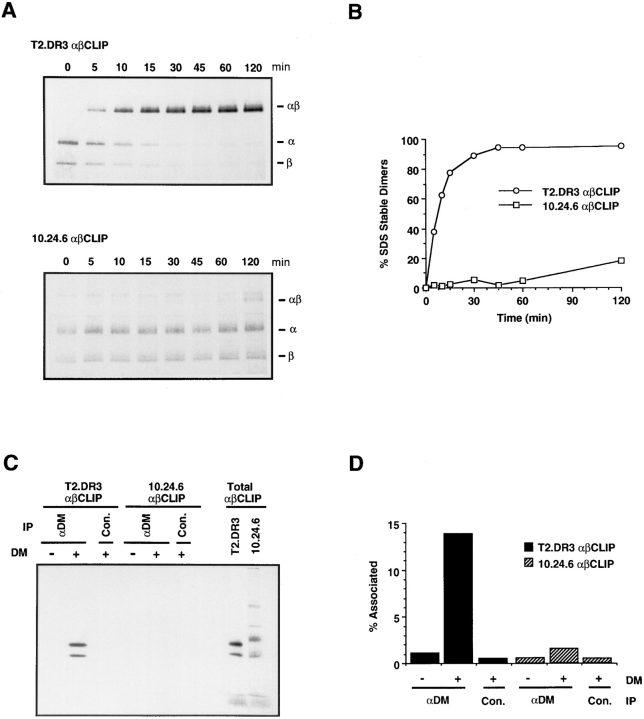
Class II molecules containing a mutant DR α chain in the DM positive 10.24.6 cell line are unable to associate with DM and therefore arrive at the cell surface complexed with CLIP (Fig. 1 B; 32, 35). To determine if 10.24.6-derived αβCLIP complexes could be loaded in vitro by native DM we compared the rates at which mutant and wild-type αβCLIP complexes could be converted to SDS-stable dimers. T2.DR3 and 10.24.6 cells were pulsed with [35S]methionine for 4 h and chased in the presence of a 15-fold excess of methionine and cysteine for 16 h to ensure degradation of the I chain and maturation of the αβCLIP complexes. Mutant and wild-type αβCLIP complexes were affinity-purified from radiolabeled cell lysates using a CerCLIP.1 antibody column. Radiolabeled wildtype and mutant DR3 αβCLIP complexes and the DR3specific peptide MOMP were added to affinity-purified DM and incubated at pH 5 for periods up to 120 min at 37°C (29). After neutralization, the samples were analyzed by SDS-PAGE. The kinetics of DM-induced dimer formation for wild-type αβCLIP complexes showed that SDSstable αβ dimers were detected after only 5 min of incubation at 37°C and the formation of dimers proceeded rapidly for the first 30–45 min of incubation before leveling off (Fig. 4, A and B). In contrast, αβCLIP complexes purified from 10.24.6 molecules were inefficiently loaded by affinity-purified DM. After 120 min only 18% of the mutant αβCLIP complexes were converted to SDS-stable dimers compared to 95% dimers for wild-type αβCLIP. These results show that native DM is unable to efficiently load mutant αβCLIP complexes from 10.24.6 cells and confirm the results of Sloan et al. (31), who demonstrated that the mutant αβCLIP complexes cannot be loaded in vitro by recombinant, soluble DM.
Figure 4.
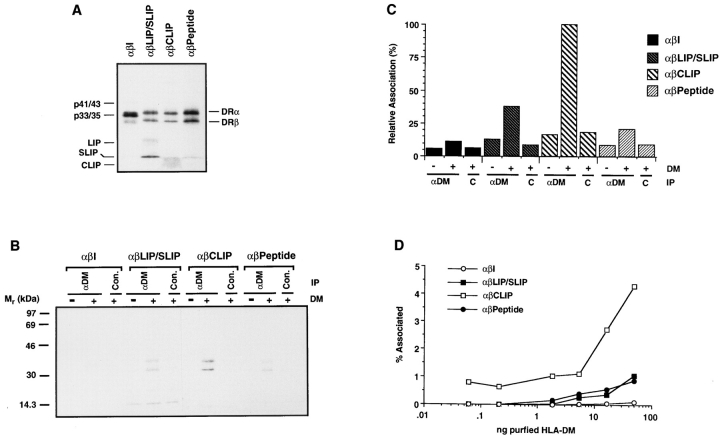
(A and B) HLADR3 αβCLIP complexes from 10.24.6 cells cannot be loaded by affinity-purified HLA-DM in vitro. Purified HLA-DR3 αβCLIP complexes from T2.DR3 and mutant 10.24.6 cells were incubated with the DR3-restricted MOMP peptide and 2 ng of affinity-purified HLA-DM at 37°C for the indicated times at pH 5.0. After neutralization, samples were analyzed by SDS-PAGE (10.5%) (A) and the percent SDS-stable dimers formed at each time point quantitated by image analysis (B). The positions of the individual α and β chains and αβ dimers are indicated on the right. (C and D) HLA-DM associates with αβCLIP complexes in vitro. Radiolabeled αβCLIP complexes from T2.DR3 and 10.24.6 cells were incubated in the presence (50 ng) or absence of affinitypurified HLA-DM at 37°C for 30 min at pH 5.0. The pH was adjusted to 6.0 and the reaction mixtures were immunoprecipitated with anti-DM (αDM) or control anti-C23V (con.) and analyzed by SDS-PAGE (10.5%) (C) and quantitated by image analysis (D). The lanes labeled Total αβCLIP represent onetenth the amount of radiolabeled αβCLIP used in the reaction mixture and was used to calculate the percentage DM associated class II (% Associated). IP in C, indicates the antibody used for immunoprecipitation.
DM Associates with αβCLIP Complexes In Vitro.
DM is able to catalyze the removal of CLIP from αβCLIP complexes and exchange it for antigenic peptides in vitro (29–31). This suggests that DM and DR must associate during the in vitro reaction. To look for such an interaction, radiolabeled, CerCLIP.1-purified normal αβCLIP from T2.DR3 or mutant αβCLIP from 10.24.6 were added to affinitypurified DM and incubated at pH 5.0 for 20–30 min at 37°C. After adjusting the pH to 6.0, the reaction mixtures were immunoprecipitated with anti-DM or anti-C23V, which served as a negative control antibody, and analyzed by SDS-PAGE. When affinity-purified DM and wild-type DR αβCLIP complexes were incubated together, antiDM but not anti-C23V coprecipitated the radiolabeled DR (Fig. 4 C). However, DM failed to form a detectable complex with mutant αβCLIP, since immunoprecipitation with anti-DM did not coprecipitate radiolabeled mutant DR. Image analysis (Fig. 4 D) indicated that 14% of the α and β chains from wild-type αβCLIP was coprecipitated with DM, whereas less than 4% of those of the mutant αβCLIP complexes were coprecipitated (Fig 4 D). These results show that DM and DR αβCLIP complexes do interact in vitro and that no additional components are necessary for the interaction. CLIP, which runs below the dye front on SDS-PAGE (29), is conspicuously absent from the class II complex coprecipitated by anti-DM in Fig. 4 C, even though it can readily be seen in the initial αβCLIP complex (second lane from the right). DM appears to be associated with empty class II αβ dimers in this experiment, because no peptides were added to the reaction mixture. CLIP is consistently absent from or severely reduced in complexes formed by incubating DM with αβCLIP.
DM Stabilizes Empty Class II Molecules.
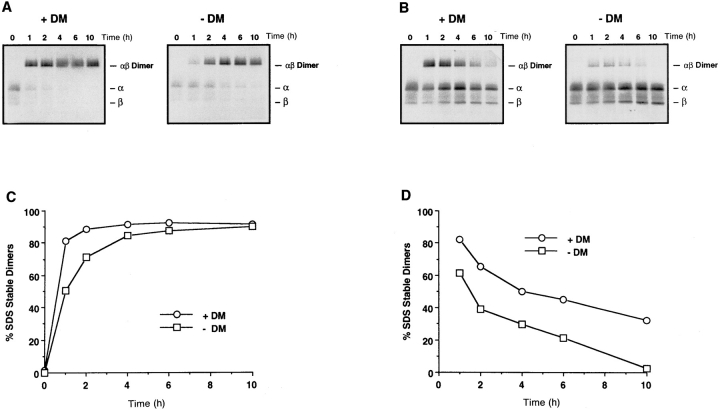
Empty class II αβ dimers have been shown to aggregate and lose their ability to bind antigenic peptides (51–53). The absence of CLIP from class II complexes coprecipitated with DM (Fig. 4 C) indicated that DM continues to interact with empty class II molecules after CLIP dissociation. Such an association might stabilize the αβ dimers and prevent them from aggregating and losing the capacity to bind antigenic peptides. To test this hypothesis, we took advantage of the ability of the detergent octyl glucoside to induce CLIP release from DR3 αβCLIP complexes (21, 27). Radiolabeled DR3 αβCLIP complexes were incubated in 1% octyl glucoside at pH 4.5 with the DR3-specific MOMP peptide in the presence and absence of affinity-purified DM. After acidification and incubation for 0 to 10 hr at 37°C, the samples were neutralized, analyzed by SDS-PAGE and quantitated by image analysis. The results showed that the conversion of class II αβCLIP complexes to αβ peptide complexes in the detergent octyl glucoside was almost as efficient in the absence of DM as in its presence. By 6 h, the percentage of SDS-stable dimers was virtually identical (∼90%; Fig. 5, A and B). This demonstrates that octyl glucoside can induce CLIP release from class II αβCLIP complexes almost as efficiently as DM under these experimental conditions. To determine whether DM could stabilize empty class II molecules, radiolabeled αβCLIP complexes were incubated without peptide in 1% octyl glucoside in the presence and absence of DM at pH 4.5 for 0–10 h. At each time point, MOMP peptide was added to 1 μM and the samples were incubated for an additional 45 min at 37°C to allow surviving, functional class II molecules to bind peptide. After neutralization, the samples were analyzed by SDS-PAGE and quantitated by image analysis. The results showed that after only 1 h of incubation in the absence of peptide more peptide-receptive class II molecules survived in the presence of DM (Fig. 5 C). With increasing incubation time, the amount of peptide-receptive class II progressively decreased both with and without DM. However, at all time points the presence of HLA-DM resulted in increased peptide loading. After 10 h of incubation no peptide-receptive class II molecules were present in the absence of DM, whereas ∼35% of class II molecules retained their peptide binding function when DM was present (Fig. 5, C and D). Control proteins at the same or 10-fold higher concentrations failed to preserve peptidereceptive class II molecules demonstrating that this function is a specific property of HLA-DM (data not shown).
Figure 5.
Stabilization of empty class II molecules by HLA-DM. (A and B) Loading of class II αβCLIP complexes in octyl glucoside versus loading in octyl glucoside and DM. Purified HLA-DR3 αβCLIP complexes (25 nM) from T2.DR3 cells were incubated in 1% octyl glucoside with the DR3-specific MOMP peptide in the presence or absence of 25 nM affinity-purified HLA-DM at 37°C for the indicated times at pH 4.5. After neutralization, samples were analyzed by SDS-PAGE (A) and the percent SDS-stable dimers formed in the presence and absence of HLA-DM at each time point were quantitated by image analysis (B). The positions of the individual α and β chains and the αβ dimers are indicated on the right. (C and D) HLA-DM stabilizes empty class II molecules. Radiolabeled DR3 αβCLIP complexes (25 nM) were incubated at 37°C in 1% octyl glucoside in the presence and absence of HLA-DM (25 nM) for the indicated times at pH 4.5. At each time point, 1 μM MOMP peptide was added and the samples were incubated at 37°C for an additional 45 min. After neutralization, samples were analyzed by SDS-PAGE (C). To quantitate the results in panel C, the amount of SDSstable dimers at each time point is represented in panel D as the percentage of the maximal available SDS-stable dimers generated at the same time point in the presence and absence of DM but with MOMP continuously present (B).
DM Interacts In Vitro with αβ Dimers Associated with I Chain Fragments or Endogenous Peptides but not αβI Complexes.
During transport, maturing class II complexes have many opportunities to interact with DM. In vivo pulsechase (Figs. 1–3) and steady-state Western blotting experiments (32) showed that DM does not coprecipitate with αβI complexes, but can be coprecipitated with αβ complexed to I chain fragments (LIP) and with peptide-loaded αβ dimers. Lack of association with αβI, particularly, could either be a reflection of limiting amounts of DM in the ER and later αβI-containing compartments or of an inability of DM to interact. To examine this question, [35S]methionine-labeled αβI, αβLIP/SLIP and mature αβ dimers were purified from wild type cells. SDS-PAGE of the purified αβI, αβLIP/SLIP, αβCLIP, and αβ peptide complexes used for the in vitro association assays are shown in Fig. 6 A. The loaded samples represent one-tenth the amount of each complex used for the association assay. In this particular experiment, the SLIP fragment (15) was unusually well represented in the class II molecules purified from leupeptin-treated Pala cells. For association experiments, the various complexes were incubated in the presence and absence of affinity-purified DM at pH 5.0 for 30 min at 37°C, immunoprecipitated with anti-DM or antiC23V (negative control) and analyzed by SDS-PAGE (Fig. 6 B). The results showed that DM interacted detectably with αβLIP/SLIP, αβCLIP, and αβ peptide complexes, but not with αβI. In other experiments we were unable to detect association of DM with affinity-purified I chain trimers (data not shown). Image analysis of the co-association experiment (Fig. 6 C) suggested that DM interacted better with αβCLIP complexes than with αβLIP/SLIP or αβ peptide complexes. To better quantitate the interactions, titrated amounts of purified DM were added to each of the purified class II complexes and the percentage of each class II complex coprecipitated with DM was determined for each concentration. The results were quantitated by image analysis after SDS-PAGE (Fig. 6 D). DM clearly associated better with class II molecules when αβCLIP was used than when αβ dimers complexed with larger I chain fragments or αβ peptide complexes were used. These results probably reflect prolonged association with empty αβ dimers after CLIP dissociation (see Fig. 5). Interaction with αβI was essentially undetectable.
Figure 6.
HLA-DM interacts in vitro with αβ dimers associated with I chain fragments or endogenous peptides but not αβI. (A) SDS-PAGE (12%) analysis of radiolabeled, purified αβI, αβLIP/SLIP, αβCLIP, and αβ peptide complexes used for the experiments shown in B and D. The positions of the individual DR α and β chains are indicated on the right and the positions of the I chain (p41/43 and p33/35), LIP, SLIP and CLIP are indicated on the left. (B and C) Radiolabeled complexes (as indicated) were incubated in the presence and absence of purified DM at 37°C for 30 min at pH 5.0, the pH was adjusted to 6.0 and the reaction mixtures were immunoprecipitated with anti-DM (αDM) or control anti-C23V (con.). After analysis of the precipitates by SDS-PAGE (12%) (B), the results were quantitated by image analysis (C) to determine the relative percentage each DR complex that remained associated with DM. (D) Purified complexes were incubated with various concentrations of affinity-purified HLA-DM as indicated (50 ng to 0.156 ng) at 37°C for 30 min at pH 5.0 and the reaction mixtures were immunoprecipitated with anti-DM. After analysis of the precipitates by SDSPAGE, the percentage of each class II complex coprecipitated with DM was determined for each concentration by image analysis.
Discussion
Upon arrival in the MIIC, the I chain component of αβI complexes is degraded, leaving CLIP as a residual fragment bound to class II αβ dimers. Through a process mediated by HLA-DM, CLIP is exchanged for endocytically generated peptides and the mature class II molecules are transported to the cell surface (29–31). This process implies a physical interaction between class II molecules and HLADM. In this paper, we have confirmed that such an interaction, first described by Sanderson et al. (32), occurs in vivo, and have complemented this finding by analyzing the specificity of the interaction for different class II complexes in vitro. In addition, we have shown that HLA-DM associates with empty class II molecules after CLIP dissociation, and that this association increases the stability of the peptidefree αβ dimers, prolonging their ability to bind peptides.
HLA-DM is a resident protein of MIICs (44–47). Not surprisingly, therefore, the interaction between class II molecules and HLA-DM is readily detectable at steady state in dense Percoll fractions (32). Examination of the kinetics of the interaction by pulse-chase analysis (Fig. 1), shows that it is transient, peaking at the time when, based on our previous experiments, HLA-DR molecules arrive in MIICs (46; unpublished data). Consistent with this, DM association is not seen until the class II molecules are resistant to Endo H digestion (Fig. 2). Additionally, in MIICs purified by a combination of Percoll gradient separation and immunoisolation using an antibody to the DMβ cytoplasmic domain, we have also been able to observe DM-DR association after CHAPS solubilization (Hammond, C., unpublished observations).
There are two possible, non-mutually exclusive, explanations for the peak of DM-DR association occurring in MIICs. First, this is believed to be where αβCLIP is generated and αβCLIP could be the optimal form of class II for DM binding. Second, the interaction could peak in MIICs because this is the organelle where DM is maximally concentrated. At one extreme, all the forms of class II (αβI through αβ with endocytically acquired peptides) could bind equally well to DM, but because DM is encountered primarily in MIICs, only the later forms would be found to associate in vivo. Some evidence for this is seen in Fig. 1, where what appears to be an I chain fragment is coprecipitated with DM, consistent with the idea that αβ dimers with residual I chain fragments can bind. If complexes of class II with I chain fragments are deliberately induced by leupeptin treatment, their in vivo association with DM is even more dramatically evident (Fig. 3; 32). Under such conditions, these complexes accumulate in MIICs. In addition, it is clear that SDS-stable dimers, presumably loaded with peptides other than CLIP, can associate with DM (Fig. 2 C). At later times in the chase these may represent an internalized subset of surface class II molecules. They do not reflect an artefactual, post-solubilization association of DM and mature DR molecules, because coextraction under the same conditions of labeled T2.DR3 cells mixed with unlabeled T2.DM cells did not result in the formation of such complexes (data not shown).
To properly determine the specificity of DM for the different forms of maturing class II molecules, the development of an in vitro binding assay was essential. Such an assay also allows one to differentiate between a binding interaction between class II and DM and the functional activity of DM. We previously found that αβLIP and αβCLIP were substrates for DM-mediated peptide loading, whereas αβI and mature αβ peptide complexes from wild-type DR3-positive Pala cells were not (29). The experiments in Figs. 4 and 5 show that DR3 αβCLIP, αβLIP/SLIP, and αβ peptide complexes can all associate with HLA-DM under the in vitro conditions used, whereas αβI complexes do not. The αβCLIP complexes associate best although mutant αβCLIP complexes from the 10.24.6 cell line, which fail to bind antigenic peptides when incubated with DM (31; Fig. 4, A and B), fail to associate with HLA-DM both in vivo (32; Fig. 1 B) and in vitro (Fig. 4, C and D). Thus, binding and functional susceptibility correlate well, with the exception of mature αβ peptide complexes which bind to HLA-DM but exchange peptides poorly (29). CLIP-containing αβ dimers from the cell line 10.24.6 presumably do not bind DM because of steric interference by the additional N-linked glycan on the mutant α chain (35). Similarly, αβI complexes may not bind to DM because the COOH-terminal region of the I chain, which has been postulated to interact directly with the αβ dimer (54, 55) also sterically interferes with the interaction.
The in vitro association of DM with class II molecules is relatively inefficient. In the experiments shown in Figs. 4 and 6, DM was added in a large excess over DR and in the best experiments only ∼15% of the class II (in this case, αβCLIP) added is recoverable as DM-associated material. This inefficiency is probably attributable to several limitations of the in vitro system. First, it has been shown that DM-DR complexes are more efficiently isolated from intact cells at pH 5.0 than at pH 7.0 (32). The use of immunoprecipitation to recover DM-DR complexes required increasing the pH from 5.0 to 6.0, a pH at which the DM-DR complex may not be particularly stable. Second, DM and DR molecules are present in vivo in a membrane-bound form and are concentrated in MIICs. These conditions are not met in vitro. Additionally, there could be other proteins that stabilize the DM-DR interaction in vivo which are absent from our assay. However, we think this is unlikely since purified, soluble recombinant DM appears to be fully functional in vitro. DM-DR association in vitro may be inefficient simply because the complex is in equilibrium with free class II and DM, so that any time only a fraction of the DM and DR are associated.
The substoichiometric levels of DM compared to class II in vivo (29, 42) suggest that DM acts catalytically on multiple class II molecules. One way to ensure that DM interacts with many class II molecules would be to regulate DMclass II association by equilibrium. It has been demonstrated that in clones of wild-type human Swei cells transfected with I-Ab, low DM expression results in more αβCLIP and less αβ peptide being expressed at the cell surface (56). Ramachandra et al. (56), use this data to argue that there must be a quasi-stoichiometric ratio of class II to DM for efficient peptide loading, and that DM may not be functioning catalytically. However, if class II moves through the MIIC at the same rate, less DM could still result in less peptide loading of αβCLIP complexes.
Previously, we reported that DM does not associate with I chain in the detergent Triton X-100 (42), and in vitro association experiments in which affinity-purified I chain trimers were incubated with affinity-purified DM in the detergent CHAPS also failed to demonstrate an interaction (data not shown). However, Karlsson and co-workers have provided good evidence in the mouse system that DM and the I chain can associate in vivo (57). Removal of the cytoplasmic tail of the mouse DM β chain and coexpression with the α chain resulted in cell surface expression of mouse DM (57, 58). When the transfected molecule was coexpressed with I chain, it was redistributed to MIIC, showing that the two proteins interact (57). Additionally, association between DM and I chain has been detected by coprecipitation experiments in the detergent digitonin (32, 47, 57). However, Sanderson et al. (32), demonstrated that the DM-associated I chain complex never became Endo H–resistant, indicating that the complex failed to leave the ER, a result which we have confirmed (data not shown). Such a complex may result from mis-assembly in the ER. Pulse–chase analysis of DM in CHAPS (Fig. 1 A) showed that fragments of the I chain are coprecipitated with DM. However, elution of the DM associated proteins followed by reprecipitation indicated that the majority, and perhaps all, of the I chain fragments are still complexed to class II molecules (Fig 3; unpublished results). Overall, the data argue against a functionally important direct interaction between DM and the I chain, at least in the human system.
What is the physiological substrate for DM in wild-type cells? Recent in vitro experiments have implicated αβCLIP as the DM substrate although some other peptides can also be removed from αβ dimers by DM (31), and αβLIP can serve as an in vitro substrate (29). The data presented here are consistent with the hypothesis that αβCLIP is the preferred substrate. First, the level of class II association with DM in vitro is much higher when αβCLIP complexes, rather than αβLIP/SLIP or αβ peptide complexes, are used (Fig. 6). Second, when αβCLIP is incubated with DM, virtually no CLIP is visible in the DM-class II complex formed (e.g., Fig. 4 C). This is also true for DM-DR complexes isolated from intact cells. For example, in Fig. 1 A, upper left, no CLIP is visible below the dye front where it normally runs, even at the peak of DM-DR association and even upon over-exposure of the gels (data not shown). A reasonable hypothesis to explain this is that CLIP release is so readily mediated by DM that it does not remain associated for any significant period after the complex forms. Thus, the higher level of α and β subunit association with DM seen when αβCLIP is added, compared to when αβLIP/SLIP or αβ peptide is added, could be because DM actually has a higher affinity for empty class II molecules than for class II molecules occupied with I chain fragments or peptides.
Preferred association of HLA-DM with empty class II molecules could in part account for its ability to catalyze peptide loading. HLA-DM molecules may bind with αβ dimers associated with CLIP, other peptides or I chain fragments. This interaction might favor a conformation (or stabilize a transition state; 28) which has a significantly reduced affinity for CLIP and certain other peptides, but not for the majority of bound peptides. After dissociation of CLIP, stabilization of the empty molecules by DM association (Fig. 5) could enhance their ability to associate with other, endocytically generated, peptides. The COOH-terminal region of the I chain has been reported to stabilize empty recombinant DR1 molecules, which normally tend to aggregate (51), thus enhancing in vitro peptide binding (54). We previously observed that DM was superior to octyl glucoside in facilitating peptide binding to DR3 αβCLIP complexes, even though octyl glucoside was better at inducing CLIP dissociation (29), a result now explained by the stabilization of empty αβ dimers by DM association (Fig. 5). This chaperone-like function of HLA-DM could also explain the finding that it can enhance antigen processing involving class II alleles with a low affinity for CLIP, for example, I-Ak (34). After spontaneous release of CLIP from I-Ak molecules in vivo, empty I-Ak molecules may be better preserved in MIICs by interaction with DM, enhancing their ability to generate functional I-Ak-peptide complexes. Ultimately, these ideas can best be tested by isolating and characterizing DM–DR complexes. Such experiments are the next challenge in understanding the catalytic function of HLA-DM.
Acknowledgments
We would like to thank Dr. Hans Zweerink who kindly supplied the anti-DM antiserum, Dr. Betsy Mellins for providing us with the 10.24.6 and 8.1.6 cell lines, Dr. Derek Sant'Angelo for critical reading of the manuscript, and N. Dometios for expert preparation of the manuscript.
Footnotes
This work was supported by National Institutes of Health grant AI 23081, a gift from Pfizer Inc., and the Howard Hughes Medical Research Institute. L.K. Denzin is supported by a fellowship from the Patrick and Catherine Weldon Donaghue Medical Research Foundation.
1 Abbreviations used in this paper: B-LCL, B-lymphoblastoid; CIIV, class II– containing vesicles; CLIP, class II–associated invariant chain peptides; DOC, deoxycholate; MIIC, MHC class II compartment.
References
- 1.Unanue ER. Antigen-presenting function of the macrophage. Ann Rev Immunol. 1984;2:395–428. doi: 10.1146/annurev.iy.02.040184.002143. [DOI] [PubMed] [Google Scholar]
- 2.Cresswell, P. 1994. Assembly, transport and function of MHC class II molecules. Annu. Rev. Immunol 12:259–293. [DOI] [PubMed]
- 3.Jones PP, Murphy DB, Hewgill D, McDevitt HO. Detection of a common polypeptide chain in I-A and I-E sub-region immunoprecipitates. Mol Immunol. 1979;16:51–60. doi: 10.1016/0161-5890(79)90027-0. [DOI] [PubMed] [Google Scholar]
- 4.Machamer CE, Cresswell P. Biosynthesis and glycosylation of the invariant chain associated with HLA-DR antigens. J Immunol. 1982;129:2564–2569. [PubMed] [Google Scholar]
- 5.Anderson KS, Cresswell P. A role for calnexin (IP90) in the assembly of class II MHC molecules. EMBO (Eur Mol Biol Organ) J. 1994;13:675–682. doi: 10.1002/j.1460-2075.1994.tb06306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roche PA, Marks MS, Cresswell P. Formation of a nine-subunit complex by HLA class II glycoproteins and the invariant chain. Nature (Lond) 1991;354:392–394. doi: 10.1038/354392a0. [DOI] [PubMed] [Google Scholar]
- 7.Bakke O, Dobberstein B. MHC class II-associated invariant chain contains a sorting signal for endosomal compartments. Cell. 1990;63:707–716. doi: 10.1016/0092-8674(90)90137-4. [DOI] [PubMed] [Google Scholar]
- 8.Chervonsky AV, Gordon L, Sant AJ. A segment of the MHC Class II β chain plays a critical role in targeting class II molecules to the endocytic pathway. Int Immunol. 1994;6:1973–1982. doi: 10.1093/intimm/6.7.973. [DOI] [PubMed] [Google Scholar]
- 9.Amigorena S, Drake JR, Webster P, Mellman I. Transient accumulation of new class II MHC molecules in a novel endocytic compartment in B lymphocytes. Nature (Lond) 1994;369:113–119. doi: 10.1038/369113a0. [DOI] [PubMed] [Google Scholar]
- 10.Peters PJ, Neefjes JJ, Oorschot V, Ploegh HL, Geuze HJ. Segregation of MHC class II molecules from MHC class I molecules in the Golgi complex for transport to lysosomal compartments. Nature (Lond) 1991;349:669–676. doi: 10.1038/349669a0. [DOI] [PubMed] [Google Scholar]
- 11.Qui Y, Xu X, Wandinger-Ness A, Dalke DP, Pierce SK. Separation of subcellular compartments containing distinct functional forms of MHC class II. J Cell Biol. 1994;12:595–605. doi: 10.1083/jcb.125.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tulp A, Verwoerd D, Dobberstein B, Ploegh HL, Pieters J. Isolation and characterization of the intracellular MHC class II compartment. Nature (Lond) 1994;369:120–126. doi: 10.1038/369120a0. [DOI] [PubMed] [Google Scholar]
- 13.West MA, Lucocq JM, Watts C. Antigen processing and class II MHC peptide-loading compartments in human B-lymphoblastoid cells. Nature (Lond) 1994;369:147–150. doi: 10.1038/369147a0. [DOI] [PubMed] [Google Scholar]
- 14.Blum JS, Cresswell P. Role for intracellular proteases in the processing and transport of class II HLA antigens. Proc Natl Acad Sci USA. 1988;85:3975–3979. doi: 10.1073/pnas.85.11.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maric MA, Taylor MD, Blum JS. Endosomal aspartic proteinases are required for invariant chain processing. Proc Natl Acad Sci USA. 1994;91:2171–2175. doi: 10.1073/pnas.91.6.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neefjes JJ, Ploegh HL. Inhibition of endosomal proteolytic actitity by leupeptin blocks surface expression of MHC class II molecules and their conversion to SDS resistant αβ heterodimers in endosomes. EMBO (Eur Mol Biol Organ) J. 1992;11:411–416. doi: 10.1002/j.1460-2075.1992.tb05069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riese RJ, Wolf PR, Brömme D, Natkin LR, Villadangos JA, Ploegh HL, Chapman HA. Essential role for cathepsin S in MHC class II-associated invariant chain processing and peptide loading. Immunity. 1996;4:357–366. doi: 10.1016/s1074-7613(00)80249-6. [DOI] [PubMed] [Google Scholar]
- 18.Kelly AP, Monaco JJ, Cho S, Trowsdale J. A new human HLA class II-related locus, DM. Nature (Lond) 1991;353:571–573. doi: 10.1038/353571a0. [DOI] [PubMed] [Google Scholar]
- 19.Cho S, Attaya M, Monaco JJ. New class II-like genes in the murine MHC. Nature (Lond) 1991;353:573–576. doi: 10.1038/353573a0. [DOI] [PubMed] [Google Scholar]
- 20.Busch R, Mellins ED. Developing and shedding inhibitions: how MHC class II molecules reach maturity. Curr Opin Immunol. 1996;8:51–58. doi: 10.1016/s0952-7915(96)80105-1. [DOI] [PubMed] [Google Scholar]
- 21.Riberdy JM, Newcomb JR, Surman MJ, Barbosa JA, Cresswell P. HLA-DR molecules from an antigen-processing mutant cell line are associated with invariant chain peptides. Nature (Lond) 1992;360:474–477. doi: 10.1038/360474a0. [DOI] [PubMed] [Google Scholar]
- 22.Sette A, Ceman S, Kubo RT, Sakaguch K, Appella E, Hunt DF, Davis TA, Michel H, Shabanowitz J, Rudersdorf R, et al. Invariant chain peptides in most HLA-DR molecules of an antigen-processing mutant. Science (Wash DC) 1992;258:1801–1804. doi: 10.1126/science.1465617. [DOI] [PubMed] [Google Scholar]
- 23.Martin WD, Hicks GG, Mendiratta SK, Leva HI, Ruley HE, Van Kaer L. H2-M mutant mice are defective in the peptide loading of class II molecules, antigen presentation, and T cell repertoire selection. Cell. 1996;84:534–550. doi: 10.1016/s0092-8674(00)81030-2. [DOI] [PubMed] [Google Scholar]
- 24.Miyazaki T, Wolf P, Tourne S, Waltzinger C, Dierich A, Barois N, Ploegh H, Benoist C, Mathis D. Mice lacking H2-M complexes, enigmatic elements of the MHC class II peptide-loading pathway. Cell. 1996;84:531–541. doi: 10.1016/s0092-8674(00)81029-6. [DOI] [PubMed] [Google Scholar]
- 25.Fung-Leung W, Surh CD, Liljedahl M, Pang J, Leturcq D, Peterson PA, Webb SR, Karlsson L. Antigen presentation and T cell development in H2-M deficient mice. Science (Wash DC) 1996;271:1278–1281. doi: 10.1126/science.271.5253.1278. [DOI] [PubMed] [Google Scholar]
- 26.Riberdy JM, Avva RR, Geuze HJ, Cresswell P. Transport and intracellular distribution of MHC class II molecules and associated invariant chain in normal and antigen-processing mutant cell lines. J Cell Biol. 1994;125:1225–1237. doi: 10.1083/jcb.125.6.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Avva RR, Cresswell P. In vivo and in vitro formation and dissociation of HLA-DR complexes with invariant chain-derived peptides. Immunity. 1994;1:63–74. doi: 10.1016/s1074-7613(94)80018-9. [DOI] [PubMed] [Google Scholar]
- 28.Ghosh P, Amaya M, Mellins E, Wiley DC. The structure of an intermediate in class II MHC maturation: CLIP bound to HLA-DR3. Nature (Lond) 1995;378:457–462. doi: 10.1038/378457a0. [DOI] [PubMed] [Google Scholar]
- 29.Denzin LK, Cresswell P. HLA-DM induces CLIP dissociation from MHC class II αβ dimers and facilitates peptide loading. Cell. 1995;82:155–165. doi: 10.1016/0092-8674(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 30.Sherman MA, Weber DA, Jensen PE. DM enhances peptide binding to class II MHC by release of invariant chain-derived peptides. Immunity. 1995;3:197–205. doi: 10.1016/1074-7613(95)90089-6. [DOI] [PubMed] [Google Scholar]
- 31.Sloan VS, Cameron P, Porter G, Gammon M, Amaya M, Mellins E, Zaller DM. Mediation by HLA-DM of dissociation of peptides from HLA-DR molecules. Nature (Lond) 1995;375:802–806. doi: 10.1038/375802a0. [DOI] [PubMed] [Google Scholar]
- 32.Sanderson F, Thomas C, Neefjes J, Trowsdale J. Association between HLA-DM and HLA-DR in vivo. Immunity. 1996;4:87–96. doi: 10.1016/s1074-7613(00)80301-5. [DOI] [PubMed] [Google Scholar]
- 33.Salter RD, Howell DN, Cresswell P. Genes regulating HLA class I antigen expression in T-B lymphoblast hybrids. Immunogenetics. 1985;21:235–246. doi: 10.1007/BF00375376. [DOI] [PubMed] [Google Scholar]
- 34.Riberdy JM, Cresswell P. The antigen-processing mutant T2 suggests a role for MHC-linked genes in class II antigen presentation. J Immunol. 1992;148:2586–2590. [PubMed] [Google Scholar]
- 35.Mellins E, Cameron P, Amaya M, Goodman S, Pious D, Smith L, Arp L. A mutant human histocompatibility leukocyte antigen DR molecule associated with invariant chain peptides. J Exp Med. 1994;179:541–549. doi: 10.1084/jem.179.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lampson LA, Levy R. Two populations of Ialike molecules on a human cell line. J Immunol. 1980;125:293–299. [PubMed] [Google Scholar]
- 37.Guy K, van Heyningen V, Cohen BB, Deane DL, Steel CM. Differential expression and serologically distinct subpopulations of human Ia antigens detected with monoclonal antibodies to Ia alpha and beta chains. Eur J Immunol. 1982;12:942–948. doi: 10.1002/eji.1830121109. [DOI] [PubMed] [Google Scholar]
- 38.Berger AE, Davis JE, Cresswell P. Monoclonal antibody to HLA-A3. Hybridoma. 1982;1:87–90. doi: 10.1089/hyb.1.1982.1.87. [DOI] [PubMed] [Google Scholar]
- 39.Parham P, Barnstable CJ, Bodmer WF. Use of a monoclonal antibody (w6/32) in structural studies of HLAA,B,C antigens. J Immunol. 1979;123:342–349. [PubMed] [Google Scholar]
- 40.Clark, E.A., and T. Yakoshi. 1984. In Leukocyte Typing. A. Bernard, L. Baunsell, J. Dausset, and S. Schlossman, editors. Springer, Heidelberg. 195–211.
- 41.Lamb CA, Cresswell P. Assembly and transport properties of invariant chain trimers and HLA-DR-invariant chain complexes. J Immunol. 1992;148:3478–3482. [PubMed] [Google Scholar]
- 42.Denzin LK, Robbins NF, Carboy-Newcomb C, Cresswell P. Assembly and intracellular transport of HLA-DM and correction of the class II antigen-processing defect in T2 cells. Immunity. 1994;1:595–606. doi: 10.1016/1074-7613(94)90049-3. [DOI] [PubMed] [Google Scholar]
- 43.Machamer CE, Cresswell P. Monensin prevents terminal glycosylation of the N- and O-linked oligosaccharides of the HLA-DR-associated invariant chain and inhibits its dissociation from the α-β chain complex. Proc Natl Acad Sci USA. 1984;81:1287–1291. doi: 10.1073/pnas.81.5.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanderson F, Kleijmeer MJ, Kelly A, Verwoerd D, Tulp A, Neefjes JJ, Geuze HJ, Trowsdale J. Accumulation of HLA-DM, a regulator of antigen presentation, in MHC class II compartments. Science (Wash DC) 1994;266:1566–1569. doi: 10.1126/science.7985027. [DOI] [PubMed] [Google Scholar]
- 45.Nijman HW, Kleijmeer MJ, Ossevoort MA, Oorschot VM, Vierboom MP, van de Keur M, Kenemans P, Kast WM, Geuze HJ, Melief CJ. Antigen capture and major histocompatibility class II compartments of freshly isolated and cultured human blood dendritic cells. J Exp Med. 1995;182:163–174. doi: 10.1084/jem.182.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robbins NF, Hammond C, Denzin LK, Pan M, Cresswell P. Trafficking of MHC class II molecules through intracellular compartments containing HLA-DM. Human Immunol. 1995;45:13–23. doi: 10.1016/0198-8859(95)00152-2. [DOI] [PubMed] [Google Scholar]
- 47.Karlsson L, Peleraux A, Lindstedt R, Liljedahl M, Peterson PA. Reconstitution of an operational MHC class II compartment in nonantigen-presenting cells. Science (Wash DC) 1994;266:1569–1572. doi: 10.1126/science.7985028. [DOI] [PubMed] [Google Scholar]
- 48.Shackelford DA, Strominger JL. Analysis of the oligosaccharides on the HLA-DR and DC1 B cell antigens. J Immunol. 1993;130:274–282. [PubMed] [Google Scholar]
- 49.Mellins E, Kempin S, Smith L, Monji T, Pious D. A gene required for class II–restricted antigen presentation maps to the major histocompatibility complex. J Exp Med. 1991;174:1607–1615. doi: 10.1084/jem.174.6.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Germain RN, Hendrix LR. MHC class II structure, occupancy and surface expression determined by postendoplasmic reticulum antigen binding. Nature (Lond) 1991;353:134–139. doi: 10.1038/353134a0. [DOI] [PubMed] [Google Scholar]
- 51.Stern LJ, Wiley DC. The human class II MHC protein HLA-DR1 assembles as empty αβ heterodimers in the absence of antigenic peptide. Cell. 1992;68:465–477. doi: 10.1016/0092-8674(92)90184-e. [DOI] [PubMed] [Google Scholar]
- 52.Germain R, Rinker AG., Jr Peptide binding inhibits protein aggregation of invariant-chain free class II dimers and promotes selective cell surface expression of occupied molecules. Nature (Lond) 1993;363:725–728. doi: 10.1038/363725a0. [DOI] [PubMed] [Google Scholar]
- 53.Sadegh-Nasseri S, Stern LJ, Wiley DC, Germain RN. MHC class II function preserved by low-affinity peptide interactions preceding stable binding. Nature (Lond) 1994;370:647–650. doi: 10.1038/370647a0. [DOI] [PubMed] [Google Scholar]
- 54.Park A-J, Sadegh-Nasseri S, Wiley DC. Invariant chain made in Escherichia coli has an exposed N-terminal segment that blocks antigen binding to HLA-DR1 and a trimeric C-terminal segment that binds empty HLA-DR1. Proc Natl Acad Sci USA. 1995;92:11289–11293. doi: 10.1073/pnas.92.24.11289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Romagnoli P, Germain RN. The CLIP region of invariant chain plays a critical role in regulating major histocompatibility complex class II folding, transport, and occupancy. J Exp Med. 1994;180:1107–1113. doi: 10.1084/jem.180.3.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramachandra L, Kovats S, Eastman A, Rudensky AY. Variation in HLA-DM expression influences conversion of MHC class II αβ:class II-associated invariant chain peptide complexes to mature peptide bound class II αβ dimers in a normal B cell line. J Immunol. 1996;156:2196–2204. [PubMed] [Google Scholar]
- 57.Lindstedt R, Liljedahl M, Péléraux A, Peterson PA, Karlsson L. The MHC class II molecule H2-M is targeted to an endosomal compartment by a tyrosine-based targeting motif. Immunity. 1995;3:361–572. doi: 10.1016/1074-7613(95)90127-2. [DOI] [PubMed] [Google Scholar]
- 58.Marks MS, Roche PA, van Donselaar E, Woodruff L, Peters PJ, Bonifacino JS. A lysosomal targeting signal in the cytoplasmic tail of the β chain directs HLA-DM to MHC class II compartments. J Cell Biol. 1995;131:351–369. doi: 10.1083/jcb.131.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]