Abstract
Telomerase is a ribonucleoprotein reverse transcriptase that synthesizes telomeric DNA. A pseudoknot structure is phylogenetically conserved within the RNA component of telomerase in all ciliated protozoans examined. Here, we report that disruptions of the pseudoknot base pairing within the telomerase RNA from Tetrahymena thermophila prevent the stable assembly in vivo of an active telomerase. Restoring the base-pairing potential of the pseudoknot by compensatory changes restores telomerase activity to essentially wild-type levels. Therefore, the pseudoknot topology rather than sequence is critical for an active telomerase. Furthermore, we show that disruption of the pseudoknot prevents the association of the RNA with the reverse transcriptase protein subunit of telomerase. Thus, we provide an example of a structural motif within the telomerase RNA that is required for telomerase function and identify the domain that is required for telomerase complex formation. Hence, we identify a biological role for a pseudoknot: promoting the stable assembly of a catalytically active ribonucleoprotein.
Telomeres are the specialized DNA–protein structures at the ends of eukaryotic chromosomes (1, 2). A minimal telomeric DNA length is required for chromosome stability and cellular viability. Failure to maintain telomere length leads to replicative senescence in Tetrahymena, yeast, and mammalian cells (3). Telomeric DNA is usually composed of short tandem repeats that are synthesized by the ribonucleoprotein (RNP) telomerase (4). Activation of telomerase is characteristic of most cell lines and tumors, and ectopic expression of telomerase allows certain human primary cell lines in culture to bypass senescence and crisis and to continue proliferation (5, 6).
Known essential core components of telomerase include the telomerase RNA (TER; ref. 7), containing a short template sequence that is copied into telomeric DNA (8), and a protein reverse transcriptase (RT) subunit (TERT; refs. 9–11). Telomerase is unique in that it carries its own template as an intrinsic part of the enzyme. TERT has been classified into its own discrete phylogenetic subgroup that seems to have ancient origins and evolutionarily seems most related to the non-long-terminal-repeat family of RTs (12). Though TERT contains all the conserved amino acid motifs characteristic of other RTs, telomerase is unusual for an RT in that specific mutations in TER, within and outside the template domain, can dramatically affect the enzymatic activity, fidelity, and processivity of telomerase (13–15). These findings suggest that TER evolved in a close functional relationship with TERT and might have roles other than providing the template. In addition to TERT, other proteins have been found that associate with TER in yeasts, ciliates, and mammals, but their functions are unknown (16–19). In Tetrahymena thermophila, TERT is a 133-kDa protein (p133; ref. 20), and the other known TER-associated proteins are 80 kDa (p80) and 95 kDa (p95; ref. 16).
The relatively small (≈150–200 nt) TERs of ciliated protozoa contain a well defined conserved core structure that includes a pseudoknot positioned downstream of the template domain (ref. 21; Fig. 1A). Whether the much larger TERs of mammals (≈400 nt) or yeasts (up to ≈1,300 nt) contain a comparable pseudoknot is not known (4). RNA pseudoknots are found within many classes of RNAs and play critical roles in various cellular processes (22, 23). Protein–pseudoknot interactions have been proposed to modulate translational regulation or replication initiation, with the relevant protein–pseudoknot association having been shown directly in a few cases (24, 25).
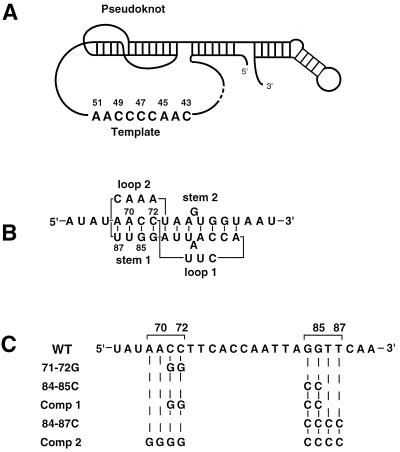
Figure 1.
Ciliate TER structure and pseudoknot sequence alterations. (A) The conserved core structure of ciliate TER determined by phylogenetic covariation and structural probing (4), highlighting the pseudoknot domain that forms into a quasicontinuous helix on stacking of the two stems, and the wild-type template sequence from T. thermophila. (B) Detail of the pseudoknot domain from T. thermophila with sequence, position numbers, and standard designations of stems 1 and 2 as well as loops 1 and 2. (C) Sequence alterations used within the pseudoknot domain. The top line shows the wild-type pseudoknot RNA sequence from T. thermophila. Specific changes are shown directly below the wild-type residues that have been changed. Comp 1 and Comp 2, compensatory mutations 1 and 2.
We report that the pseudoknot found in the TER from Tetrahymena is critical for the stable assembly in vivo of a catalytically active telomerase. Mutations in TER that disrupt base pairing of the pseudoknot prevent its stable assembly with the catalytic RT component of telomerase, p133 (20), but allow the stable assembly of RNP complexes containing the TER-associated proteins p80 and p95 (16).
MATERIALS AND METHODS
Strains and Transformation.
T. thermophila strains used for all experiments were CU427, ChxA/ChxA (cy-sens, VI) and CU428, Mpr/Mpr (6-mp sens, VII). They were kindly provided by P. J. Bruns, (Cornell University, Ithaca, NY). Electroporation was performed essentially as described by Gaertig and Gorovsky (26). The vector used for electroporation was prD4-1 (27).
Mutagenesis.
Mutagenesis was performed by overlap extension by using PCR as described (28), except for the following modifications. PCR consisted of 20 cycles of denaturation (30 s at 94°C), annealing (30 s at 50°C), and extension (30 s at 72°C) under conditions suggested by the manufacturer of the DNA Thermal Cycler (Perkin—Elmer/Cetus). Mutant TERs were inserted into the vector prD4-1 (27) at the XhoI site within the vector polylinker. Potential mutant genes were carefully sequenced to detect possible unwanted errors by Taq polymerase.
Telomerase Preparation.
Partially purified mutant and wild-type telomerase were prepared as described (29) with the following modifications. Populations of mutant and wild-type TER gene transformants were grown to late logarithmic phase in 50-ml cultures with shaking at 100 rpm at 30°C. Cells were washed once with Dryl’s solution (1.7 mM sodium citrate/2.4 mM sodium phosphate/2 mM CaCl2) and incubated in Dryl’s solution at 30°C with shaking at 100 rpm for ≈24 h. S-100 extract was loaded onto a DEAE–agarose (Bio-Rad) column and eluted with 300 mM sodium acetate after washing with six column volumes of 200 mM sodium acetate. Preparations from the previous column were adjusted to 500 mM sodium acetate and desalted by loading onto an octyl-Sepharose (Amersham Pharmacia) column and washing with six column volumes of TMG buffer [10 mM Tris⋅HCl, pH 8.0/1 mM MgCl2/10% (vol/vol) glycerol/0.1 mM phenylmethylsulfonyl fluoride]. Telomerase was eluted with TMG buffer containing 1% Triton X-100, and samples were kept frozen at −80°C until needed.
Telomerase Reactions.
Full-length primer was purified on 20% polyacrylamide/8 M urea gels and eluted passively with shaking at 100 rpm overnight at 37°C. Primer was then decontaminated by passage over Sep-Pak C-18 (Waters). Telomerase reactions were performed and prepared for electrophoresis essentially as described (29). Final reaction mixtures contained 50 mM Tris (pH 8.5 at 25°C), 1 mM spermidine, 1 mM dithiothreitol, 1 mM MgCl2, 1 μM primer, 1.5 μM [α-32P]dGTP, and 100 μM cold dTTP. Reactions were incubated at 30°C for 45 min unless otherwise specified. Elongation products from the in vitro reaction were run on 10% polyacrylamide/8 M urea gels. Gels were dried and exposed to x-ray films, generally for 1–2 days at −80°C before development.
Native Gel Electrophoresis.
Aliquots of S-100 preparations (≈20 μl) were separated on native gels composed of 3.5% polyacrylamide (60:1, acrylamide:bis-acrylamide), 0.45% agarose, and 50 mM Tris-acetate (pH 8.0) in running buffer of 50 mM Tris-acetate (pH 8.0). Gels were run at 200 V at 4°C. The gels were then placed in 50% (vol/vol) urea at room temperature for 30 min to dissociate the RNP complex. RNA was then transferred by electroblotting onto a nylon filter and then UV cross-linked to the nylon filter. Northern hybridization was performed as described above by using the Church and Gilbert method (30).
Western Analysis.
Telomerase RNP complexes were separated on native gels as described above and then transferred onto a nitrocellulose membrane by electroblotting. Western blot analysis was performed with an enhanced chemiluminescence detection kit (Amersham Pharmacia) according to the manufacturer’s instructions. Kathy Collins (University of California, Berkeley) kindly provided p80, p95, and p133 antibodies (20).
RESULTS AND DISCUSSION
To discover the role of the pseudoknot within TER from T. thermophila, we examined the consequences of altering the sequence within stem 1 (Fig. 1 B and C). Mutant TER genes were introduced into developing Tetrahymena cells on a high copy-number vector as described (13, 27). After early establishment of the introduced TER gene, telomerase was partially purified from logarithmically growing cells and studied in vitro to determine the effects of these alterations (13). In this overexpression system, mutations in the TER that allow accumulation of stable RNA and assembly of an active telomerase RNP complex quickly swamp out and replace the endogenous wild-type TER in the telomerase RNP. This replacement occurs because the copy number of the exogenous mutant gene is several hundred times greater than that of the endogenous wild-type gene. However, in this system, mutations in the introduced TER gene that destabilize TER or prevent RNP assembly allow the endogenous wild-type RNA to become the dominant species within the cell population, hence allowing endogenous wild-type telomerase to be active and telomeric DNA to be maintained.
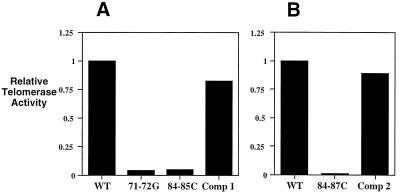
Disruption and compensatory alterations were made within stem 1 of the TER pseudoknot (Fig. 1B). Two separate dinucleotide substitutions on both sides of stem 1 of the pseudoknot, each disrupting the pseudoknot (71–72G and 84–85C; Fig. 1C), greatly reduced the total cellular telomerase activity levels by 20- to 25-fold (Fig. 2A). The compensatory alteration designated Comp 1 (Fig. 1C), which restored the base-pairing potential of stem 1, restored telomerase activity to near wild-type levels (Fig. 2A). A four-base disruption of the entire stem 1 (84–87C; see Fig. 1C) also greatly reduced the total telomerase activity level (Fig. 2B), which again was restored (Fig. 2B) by making the compensatory mutation (Comp 2; see Fig. 1C). Populations of cells transformed with the 71–72G, 84–85C, Comp 1, or Comp 2 TER alleles were not grossly different from wild-type cells in morphology, population doubling rate, or telomere length. However, in cell lines transformed with the pseudoknot disruption allele 84–87C, which completely eliminated stem 1, a cellular morphological phenotype with variable penetrance became apparent after about 10 fissions: some mutant cells were rounded and enlarged, although this morphological phenotype was usually distinct from the previously characterized “monster cell” phenotypes caused by other TER mutations (8). The morphologically altered 84–87C transformant cells failed to divide, as determined by single-cell isolation experiments (data not shown). Cells with wild-type appearance, the prevalent form in most 84–87C transformant lines, took over the culture after about 25 population doublings.
Figure 2.
Relative telomerase activity levels of disruptions and compensatory alterations within the pseudoknot domain compared with wild-type telomerase. Telomerase activity levels were normalized by using cell numbers and total protein content of partially purified preparations. The results represent an average of three separate experiments with a minimum of two lines for each sample. (A) Two-base pseudoknot disruptions, 71–72G and 84–85C, and the corresponding compensatory alteration, Comp 1, compared with activity levels for wild-type telomerase (WT). (B) Four-base pseudoknot disruption, 84–87C, and the corresponding compensatory alteration, Comp 2, compared with activity level for wild-type telomerase (WT).
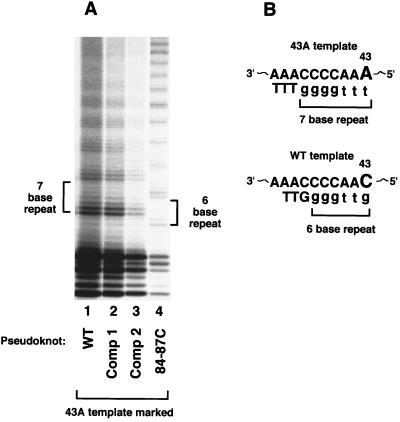
These results showed that disrupting the pseudoknot of TER interfered with total telomerase activity within cells. However, the restoration of telomerase activity levels to near wild-type levels by the compensatory mutations might theoretically have resulted from either restoration of a functional telomerase or, conversely, failure of the compensatory mutant TERs to compete effectively with wild-type endogenous TER for stable RNP assembly. Such a failure to compete would then have permitted telomerase containing the wild-type endogenous TER to predominate in these cells. The observed telomerase activity levels and cellular phenotypes of the different mutants did not distinguish between these two possibilities. Therefore, to test the effects of the pseudoknot mutations on telomerase activity directly, we marked the template of the pseudoknot TER molecules. We used a previously characterized C-to-A base substitution at position 43 (the 43A mutation), which is at the 5′ end of the template and is the last position copied (Fig. 3; ref. 13). Clonal lines transformed with the 43A mutant TER and containing essentially fully substituted 43A telomerase are viable, and their 43A telomerase activity is readily distinguished from wild-type telomerase activity (13). In standard telomerase assays, wild-type telomerase synthesizes products with a six-base repeat, G4T2, identified as a ladder of reaction products with a six-base periodicity when resolved by denaturing PAGE (Fig. 3, lane 4). The 43A mutant telomerase synthesizes clearly distinguishable seven-base G4T3 repeats (13). We used the 43A mutant to mark and monitor the activity of the pseudoknot mutant TERs. Using this approach, we found that the pseudoknot disruption 84–87C TER produced no detectable functional telomerase. Instead, the wild-type endogenous TER accounted for the functional telomerase in these cells, as shown by the six-base repeat pattern in Fig. 3, lane 4. In contrast, telomerase from cells transformed with wild–type, Comp 1, or Comp 2 TER genes marked with the 43A template each produced the seven-base repeat pattern characteristic of the 43A mutant telomerase (Fig. 3, lanes 1–3). This result directly showed that the compensatory pseudoknot TERs, Comp 1 and Comp 2, were functional. Therefore, restoring the pseudoknot structure, even though the entire original sequence of stem 1 and its G/C content were changed in Comp 2 mutants, allowed efficient formation of an active telomerase.
Figure 3.
Marking the template in combination with pseudoknot alterations. (A) Standard telomerase in vitro assay reactions separated on 10% PAGE. Cells were transformed with various pseudoknot wild-type or pseudoknot-altered TER genes that were marked at the template with the 43A template mutation. Lane 1, wild-type (WT) at pseudoknot; lane 2, Comp 1; lane 3, Comp 2; lane 4, 84–87C. (B) Schematic of 43A and wild-type templates. Telomerase with a 43A template change synthesizes a seven-base repeat consisting of G4T3. Wild-type telomerase synthesizes a six-base repeat consisting of G4T2. Note that cell numbers and protein content were not normalized between these different lines and that telomerase was harvested from cells in stationary phase culture.
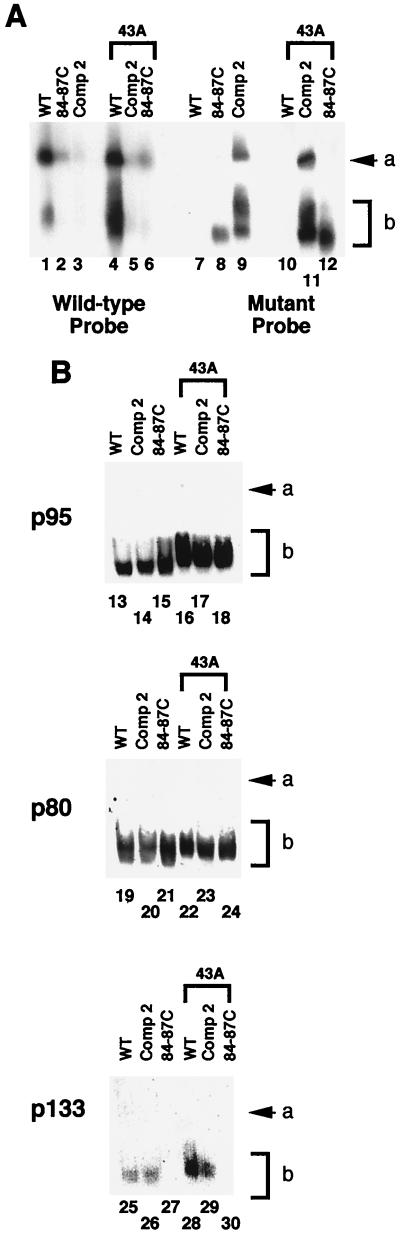
To characterize the effects of altering the pseudoknot on telomerase RNP assembly, RNP complexes containing TER were analyzed by native gel electrophoresis, blotting, and hybridization to TER sequence probes. In this fractionation, wild-type TER migrated in two locations: band a and a broad band b (Fig. 4A, Lane 1). The nucleic acid in both bands was RNA (as opposed to DNA) consisting of the sense strand of TER, as shown by strand-specific hybridization probes and nuclease-sensitivity tests (data not shown). Both bands migrated far behind deproteinized free TER (data not shown). No free TER was detected in the native preparations. Although cells transformed with the pseudoknot disruption 84–87C contained some endogenous wild-type TER, its levels were greatly reduced as shown by hybridization with a wild-type TER-specific probe (Fig. 4, lanes 2 and 6). This finding was consistent with the reduced level of telomerase activity recovered from cells transformed with the 84–87C disruption mutant gene (compare Fig. 2B with Fig. 4A, lanes 2 and 6). The same blot was reprobed with a mutant-specific probe that hybridizes to both 84–87C and Comp 2 TERs but not wild-type TER (Fig. 4A, lanes 7–12). The 84–87C TER reproducibly appeared as one band within the region of broad band b (Fig. 4A, lanes 8 and 12). Hence, even though relatively high levels of 84–87C TER were present within 84–87C transformants (Fig. 4A, lanes 8 and 12), the endogenous wild-type TER provided the active telomerase in these cells (Fig. 3A, lane 4), albeit at a reduced level (Fig. 2B). Notably, no 84–87C TER signal was detectable in band a (Fig. 4A, lanes 8 and 12).
Figure 4.
TER-associated protein complexes separated by native gel electrophoresis. (A) Northern analysis of native complexes. Northern blots probed with either a wild-type-specific end-labeled oligonucleotide (5′-ATTTGAACCTAATTGG-3′) that annealed only to wild-type TER (lanes 1–6) or a mutant-specific end-labeled oligonucleotide (5′-ATTTGGGGGTAATTGG-3′) that annealed only to 84–87C and Comp 2 TERs (lanes 7–12). These antisense oligonucleotides anneal to residues 92–77 of T. thermophila TER. Lane 1 and 7, wild-type; lane 2 and 8, 84–87C; lane 3 and 9, Comp 2; lane 4 and 10, wild-type with 43A template; lane 5 and 11, Comp 2 with 43A template; lane 6 and 12, 84–87C with 43A template. (B) Western analysis of native complexes. Western blots probed with antibody against p95 (lanes 13–18), p80 (lanes 19–24), or p133 (lanes 25–30). Lanes 13, 19, and 25, wild-type; lanes 14, 20, and 26, Comp 2; lanes 15, 21, and 27, 84–87C; lanes 16, 22, and 28, wild-type with 43A template; lanes 17, 23, and 29, Comp 2 with 43A template; lanes 18, 24, and 30, 84–87C.
Comp 2 RNA was present in both band a and broad band b (Fig. 4A, lanes 9 and 11) along with low levels of endogenous wild-type TER (Fig. 4A, lanes 3 and 5). The Comp 2-specific RNA signal in the region of broad band b frequently resolved into two partially separated bands (Fig. 4A, lanes 9 and 11). These Comp 2 broad band b species migrated slightly more slowly than the single, faster-migrating TER RNP species present in 84–87C preparations (Fig. 4A, compare lanes 8 and 12 with lanes 9 and 11). Although such splitting of broad band b was not always apparent, it was observed only with Comp 2 telomerase preparations and in no case with wild-type or 84–87C preparations analyzed on the same gel. Hence, broad band b might contain two or more biochemically or conformationally distinct telomerase complexes that are more readily resolved when the complexes contain Comp 2 RNA.
Western analysis was used to determine whether known TER-associated proteins comigrated with the TER RNAs in bands a and b. When the native gel was probed with antibodies specific for either p95 or p80, signal appeared only in the broad band b region and was nearly equal in all samples (Fig. 4B, lanes 13–24). In contrast, p133, the RT protein subunit of telomerase, was detected only in wild-type and Comp 2 lanes (Fig. 4B, lanes 25, 26, 28, and 29) and was absent from 84–87C lanes (Fig. 4B, lanes 27 and 30). On longer exposures, a weak signal was observed, which was presumably attributable to the low levels of endogenous wild-type TER complexed with p133. Thus, strikingly, p133 was unable to complex with 84–87C TER (Fig. 4, lanes 27 and 30), but association of TER with p133 was restored by restoration of the pseudoknot structure (Fig. 4B, lanes 26 and 29).
These results show that an intact TER pseudoknot topology is essential for in vivo RNP assembly with TERT. In contrast, both the pseudoknot and TERT are not required for TER to associate with proteins p80 and p95. Therefore, Fig. 4 indicates that at least three TER-containing complexes exist in the cell: the complex in band a, the slower-migrating complex of broad band b containing p133 and possibly other protein components, and the faster-migrating complex of broad band b containing at least p80 and p95. We conclude that, although the pseudoknot disruption 84–87C TER is present at relatively high steady-state levels and is associated with p95 and p80, such RNP complexes lack the catalytic component p133 and are not functional. Perhaps p80 and p95 function to chaperone TER to a specific cellular location for assembly and/or to form an RNP complex with TER to protect the RNA from degradation or to sequester the RNA in an inactive form to regulate telomerase activity. Using telomerase assays performed on in vitro reconstituted preparations, Autexier and Greider (31) found that the pseudoknot was not required for positive telomerase activity in their system. Therefore, the critical role of the pseudoknot is the in vivo stable assembly of an active telomerase. We propose that certain conditions can compensate for the lack of a pseudoknot in vitro.
In the Comp 2 TER, which formed a telomerase complex with apparently normal function, all the original bases within stem 1 were altered, but the same number of potential base pairs was restored. Thus, the topology of the pseudoknot is important, rather than the specific sequence of stem 1. The telomerase RT component p133 might interact directly with the pseudoknot or the pseudoknot might provide some conformation that allows p133 assembly. Other RNA pseudoknots are critical for specific protein binding. The gene 32 protein from T4 phage binds to a pseudoknot at the 5′ end of its own mRNA (24), and the ribosomal protein S15 also binds to a pseudoknot in its mRNA (25), leading to autoregulation in both cases. NMR studies show interaction between loops within the grooves of corresponding stems and specific structure at stem–stem junctions (32, 33). The NMR spectroscopic structure of the transfer RNA-like pseudoknot from turnip yellow mosaic virus suggested that this topological structure had “internal mobility” or structural flexibility at helical junctions that may be critical for regulating protein binding (33). Structural flexibility might also be important for the TER pseudoknot for proper telomerase function and regulation in vivo. Certain structural alterations of the TER pseudoknot, for example unwinding of stem 1 or other changes, might promote the dissociation of TERT and negatively regulate telomerase activity.
Previously, no function had been defined for any region of TER outside the template. Previously, several pseudoknots that specifically bound with high affinity to HIV type 1 RT were selected randomly in vitro (34). Here, we have reported a biological function for a natural, conserved pseudoknot structure in promoting complex formation with TERT.
Acknowledgments
We thank He Wang for valuable intellectual and technical assistance during this project, John Prescott, Chris Smith, and Yehuda Tzfati for critical reading of the manuscript and for helpful suggestions, and Kathy Collins for kindly providing antibody against p80, p95, and p133. This work was supported by National Institutes of Health Grant GM26259 to E.H.B.
ABBREVIATIONS
- RNP
ribonucleoprotein
- TER
telomerase RNA
- RT
reverse transcriptase
- TERT
protein RT subunit
- Comp n
compensatory mutation n
References
- 1.Blackburn E H. Annu Rev Biochem. 1992;61:113–129. doi: 10.1146/annurev.bi.61.070192.000553. [DOI] [PubMed] [Google Scholar]
- 2.Zakian A M, Prescott D M. Trends Cell Biol. 1996;6:29–33. doi: 10.1016/0962-8924(96)81035-x. [DOI] [PubMed] [Google Scholar]
- 3.Greider C W. Annu Rev Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- 4.Blackburn E H. In: Telomerase RNA Structure and Function. Simons R W, Grunberg-Manago M, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1998. pp. 669–693. [Google Scholar]
- 5.Shay J W, Werbin H, Wright W E. Ciba Found Symp. 1997;211:148–155. doi: 10.1002/9780470515433.ch10. [DOI] [PubMed] [Google Scholar]
- 6.Bodnar A G, Ouellette M, Frolkis M, Holt S E, Chiu C-P, Morin G B, Harley C B, Shay J W, Lichtsteiner S, Wright W E. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 7.Greider C W, Blackburn E H. Nature (London) 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 8.Yu G-L, Bradley J D, Attardi L D, Blackburn E H. Nature (London) 1990;344:126–132. doi: 10.1038/344126a0. [DOI] [PubMed] [Google Scholar]
- 9.Lingner J, Hughes T R, Shevchenko A, Mann M, Lundblad V, Cech T R. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 10.Nakamura T M, Morin G B, Chapman K B, Weinrich S L, Andrews W H, Lingner J, Harley C B, Cech T R. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 11.Martin-Rivera L, Herrera E, Albar J P, Blasco M A. Proc Natl Acad Sci USA. 1998;95:10471–10476. doi: 10.1073/pnas.95.18.10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakamura T M, Cech T R. Cell. 1998;92:587–590. doi: 10.1016/s0092-8674(00)81123-x. [DOI] [PubMed] [Google Scholar]
- 13.Gilley D, Lee M S, Blackburn E H. Genes Dev. 1995;15:2214–2226. doi: 10.1101/gad.9.18.2214. [DOI] [PubMed] [Google Scholar]
- 14.Prescott J, Blackburn E H. Genes Dev. 1997;15:528–540. doi: 10.1101/gad.11.4.528. [DOI] [PubMed] [Google Scholar]
- 15.Roy J, Fulton T B, Blackburn E H. Genes Dev. 1998;12:3286–3300. doi: 10.1101/gad.12.20.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collins K, Kobayashi R, Greider C W. Cell. 1995;2:677–686. doi: 10.1016/0092-8674(95)90529-4. [DOI] [PubMed] [Google Scholar]
- 17.Lingner J, Cech T R. Proc Natl Acad Sci USA. 1996;93:10712–10717. doi: 10.1073/pnas.93.20.10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrington L, McPhail T, Mar V, Zhou W, Oulton R, Bass M B, Arruda I, Robinson M O. Science. 1997;275:973–977. doi: 10.1126/science.275.5302.973. [DOI] [PubMed] [Google Scholar]
- 19.Nakayama J L. Cell. 1997;88:875–884. doi: 10.1016/s0092-8674(00)81933-9. [DOI] [PubMed] [Google Scholar]
- 20.Collins K, Gandhi L. Proc Natl Acad Sci USA. 1998;21:8485–8490. doi: 10.1073/pnas.95.15.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCormick-Graham M, Romero D P. Nucleic Acids Res. 1995;11:1091–1097. doi: 10.1093/nar/23.7.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pleij C W A. Curr Opin Struct Biol. 1994;4:337–344. [Google Scholar]
- 23.Tinoco I., Jr Nucleic Acids Symp Ser. 1997;36:49–51. [PubMed] [Google Scholar]
- 24.McPheeters D S, Stormo G D, Gold L. J Mol Biol. 1988;232:89–104. doi: 10.1016/0022-2836(88)90634-1. [DOI] [PubMed] [Google Scholar]
- 25.Benard L, Mathy N, Grunberg-Manago M, Ehresmann B, Ehresmann C, Portier C. Proc Natl Acad Sci USA. 1998;95:2564–2567. doi: 10.1073/pnas.95.5.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaertig J, Gorovsky M A. Proc Natl Acad Sci USA. 1992;89:9196–9200. doi: 10.1073/pnas.89.19.9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu G-L, Blackburn E H. Proc Natl Acad Sci USA. 1989;86:8487–8491. doi: 10.1073/pnas.86.21.8487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 29.Greider C W, Blackburn E H. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- 30.Church G M, Gilbert W. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Autexier C, Greider C W. Nucleic Acids Res. 1998;26:787–795. doi: 10.1093/nar/26.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen L X, Tinoco I., Jr J Mol Biol. 1995;247:963–978. doi: 10.1006/jmbi.1995.0193. [DOI] [PubMed] [Google Scholar]
- 33.Kolk M H, van der Graaf M, Wijmenga S S, Pleij C W A, Heus H A, Hilbers C W. Science. 1998;280:434–438. doi: 10.1126/science.280.5362.434. [DOI] [PubMed] [Google Scholar]
- 34.Tuerk C, MacDougal S, Gold L. Proc Natl Acad Sci USA. 1992;89:6988–6992. doi: 10.1073/pnas.89.15.6988. [DOI] [PMC free article] [PubMed] [Google Scholar]