State of the field
The complexity of biological membranes has motivated the development of a wide variety of simpler model systems whose size, geometry, and composition can be tailored with great precision. Approaches highlighted in this review are illustrated in Figure 1 including vesicles, supported bilayers, and hybrid membrane systems. These have been used to study problems ranging from phase behavior to membrane fusion. Experimental membrane models continue to advance in complexity with respect to architecture, size, and composition, as do computer simulations of their properties and dynamics. Analytical techniques such as imaging secondary ion mass spectrometry have also been developed and refined to give increasing spatial resolution and information content on membrane composition and dynamics.
Figure 1. Model membrane systems.
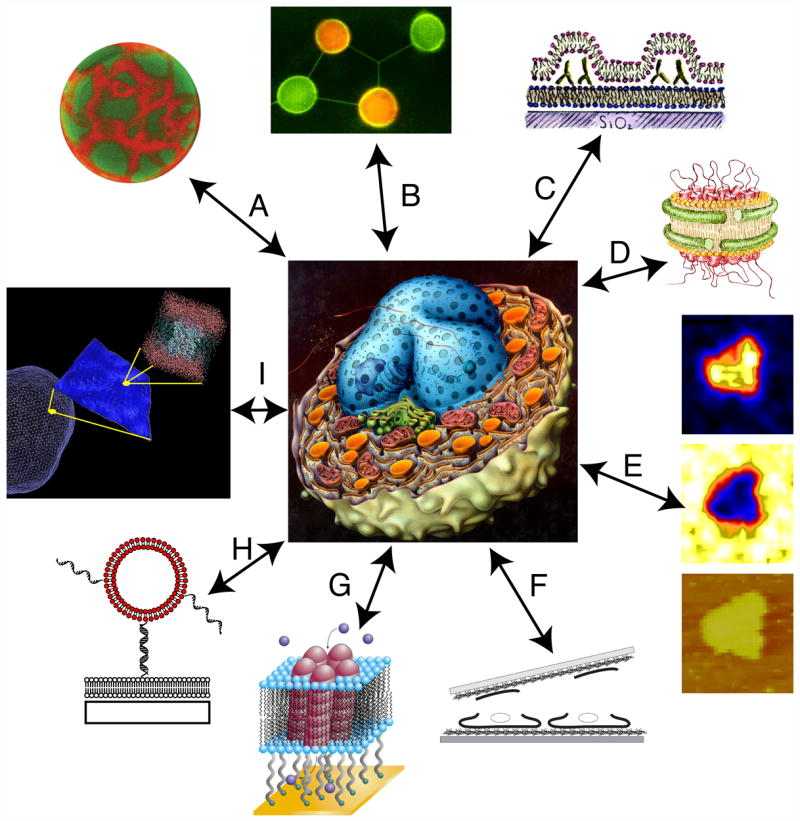
A collection of model lipid membrane systems discussed in this review and surrounding a schematic diagram of a cell, drawn by Tomo Narashima, that emphasizes the large number of different membrane surfaces. In clockwise order, beginning from the upper left: (A) Giant unilamellar vesicles and blebs; (B) Networks of giant vesicles connected by lipid microtubules; (C) Ruptured GUV’s on solid supported bilayers; (D) Membrane nanodiscs containing transmembrane proteins; (E) Supported lipid bilayers analyzed by NanoSIMS; (F) Ruptured cell membranes on solid supports; (G) Bilayers tethered to a solid support containing ion channels; (H) Vesicles tethered to a supported lipid bilayer by DNA; (I) Visual representation of multi-scale simulations.
Introduction
The most basic function of biological membranes is to define a boundary, whether between or within cells and organelles (Figure 1, center). Many cellular processes depend upon the membrane’s ability to separate different areas while allowing communication and tightly regulated transport within and across membranes to occur. All cell-cell communication and further assembly into tissues, organs and organisms are mediated by such interactions. Biological membranes vary tremendously in composition even within a eukaryotic cell, and their organization must be dynamic in order to mediate and modulate conformational changes, signaling, trafficking, and recognition. Because they play such a fundamental role and because natural membranes are so complex, many different model systems have been created that retain the essential lipid bilayer structure, but simplify the system so that the roles of individual components can be assessed and that their organization and dynamics can be visualized.
Model systems and approaches that have seen great progress in the last few years are shown schematically in Figure 1 in the order (clockwise) that we will discuss them. These include: bilayers in the form of vesicles ranging in size from tens of nanometers (small unilamellar vesicles, SUV’s) to tens of microns (giant unilamellar vesicles, GUV’s) that are either free-standing or tethered to supports; planar supported bilayers either interacting directly with a solid substrate or tethered to the substrate; bilayer islands wrapped by proteins; and fragments from natural cell membranes. In many cases, detailed reviews about these systems have been published in the last two years, and these are referenced along with specific advances that are highlighted. Nearly every imaging method has been applied to model and native membranes, especially in the planar format which matches well with surface-sensitive techniques originally developed for studies in materials science, electronic materials, geochemistry, and catalysis. Because even the simplest model systems involve the self assembly of many lipid molecules in water, they push the limits of atomistic simulations and have led to attempts to marry fully atomistic models with coarse grained or continuum approaches. A strong synergy between simulations on all length scales and experiments is a major driver of this field.
Free-standing vesicles, giant vesicles and discs (A–D)
Giant unilamellar vesicles (GUV’s) have diameters on the order of one to ten microns and have been instrumental in determining the phase behavior of binary and ternary lipid mixtures in bilayers. Phase separation (see Fig 1A) is often visualized by the partitioning of fluorescent probes into gel, liquid-ordered (lo), and liquid-disordered (ld) phases [1]. A comparison of 26 probe molecules reveals that their behavior is highly dependent on the identity of the fluorophore as well as of the membrane anchor [2•]. Furthermore, the composition of the phase domain can result in different dye-partitioning properties. For instance, DiI dyes with long, saturated tail groups partition into the lo phase of ternary mixtures of DSPC, DOPC, and cholesterol, but into the ld phase in mixtures of sphingomyelin, DOPC, and cholesterol. The balance among these factors is difficult to predict, which indicates that caution is necessary when conducting experiments and interpreting results obtained by fluorescence.
GUV’s are typically created using well-defined mixtures of pure lipids, but still unclear is the relationship between the behavior of these model systems and actual biological membranes which are much more complex mixtures of lipids, proteins, and saccharides. Reconstitution of proteins into giant vesicles is challenging due to a dehydration step that is common to most protocols to form GUV’s from lipid stocks. Methods for addressing this problem include only partially drying the lipid-protein mixture before electroformation [3] or adding sucrose to stabilize proteins against denaturation during the process [4]. Giant plasma membrane vesicles (GPMV’s), or blebs, are a more direct source of biological lipid material. In response to chemical stresses, the plasma membrane of cells protrudes and detaches to form these blebs, which resemble GUV’s and can be manipulated using many of the same techniques. The lipid composition of blebs is thought to resemble that of the parent cell; they also contain membrane proteins which diffuse into the detached area during bleb formation. Dye partitioning of incorporated fluorescently-labeled lipids indicates that GPMV’s derived from rat basal leukemia mast cells can separate into co-existing lo and ld phases, though at sub-physiological temperatures [5•]. The phase partitioning behaviors of fluorescently labeled membrane proteins such as and IgE receptor and Lyn-kinase were observed, sometimes disagreeing with the expected behavior based on detergent extraction studies.
GUV’s have also been used as the basis for novel lipid architectures. Orwar’s group has used micromanipulation to form networks of GUV’s connected by lipid tubules with diameters roughly 100–300 nm that can be used to study reaction dynamics in confined volumes [6•]. Two vesicles are connected by a single lipid tubule and contain two reactants which are mixed by allowing the connected GUV’s to merge. Because a connection has already been made via the lipid tubule, no further disruption of the lipid layers is necessary to achieve fusion and content is retained. These networks approximate the compartmentalization of cellular functions in separate organelles (they may be closely related to recently observed structures [7]), and can be made to be as complex as needed (see Fig 1B). Lipid tubules can also be pulled from GUV’s by motor proteins [8] or optical tweezers [9•], and the growth of these structures can be related to mechanical properties and phase behavior of the membrane source material. Networks can also be formed using material from blebs, allowing manipulation of complex lipid mixtures, as well as potentially cytoplasmic content [10].
When exposed to supported lipid bilayers (SLB’s), GUV’s can rupture to form unilamellar membrane flakes that interact transiently with the SLB [11]. These membrane flakes have been used to study the reorganization dynamics of proteins between the two bilayers, approximating inter-cellular junctions (see Fig 1C). As GUV’s were ruptured over fluorescently labeled proteins bound to a SLB, the protein distribution changed from even coverage to form separated aggregates on the surface [12]. The resulting pattern can be related to the fluctuation dynamics of the ruptured GUV and the protein mobility in the SLB. This work is highly relevant to questions of cell-cell communication, such as in the immunological synapse [13•] where ligand-receptor binding at the membrane results in reorganization of proteins and initiation of an immune response.
Free-standing membranes consisting of a circular area of lipid bilayer surrounded by a membrane scaffolding protein (MSP) derived from apolipoprotein A–I, known as nanodiscs, have been developed for isolating and displaying integral membrane proteins [14–16]. The diameters of nanodiscs are very uniform for any given preparation and range from 8–13 nm depending on the sequence of the recombinant MSP used (Fig 1D). By themselves, nanodiscs fail to define an internal and external volume and are thus unable to probe questions of transport across membranes; however, they provide a controlled and homogeneous environment that is well suited for measurements of ligand binding. Also, since a controlled number of membrane proteins can be captured (though not necessarily oriented in the same way), the possible consequences of protein associations can be probed, e.g. the importance (or lack thereof) of dimerization of G-protein coupled receptors (GPCRs). To date, bacteriorhodopsin, GPCRs, and ion channel monomers have been successfully inserted into these structures. Alami et al. use nanodiscs to study the binding of the membrane protein complex SecYEG to the cytosolic protein SecA, which is implicated in translocon transport [17•].
Membranes on solid supports (E–G)
The supported lipid bilayer (SLB) is typically formed by vesicle fusion or Langmuir transfer to a suitable surface. The mechanism of vesicle fusion continues to be studied with respect to the vesicle lipid composition and surface properties of the solid support [18, 19•]. SLB’s offer many advantages to an experimentalist, including ease of preparation, stability, patterning [20], and the availability of a wide variety of surface sensitive techniques for characterization. Our lab has recently applied a specialized type of imaging secondary ion mass spectrometry (SIMS) using the NanoSIMS to obtain high spatial resolution images of SLB’s with high sensitivity and composition information [21, 22••]. Because the Cs+-ion primary beam used to generate negative secondary ions fragments the target molecules into atomic or small molecular ions, the lipids or proteins in the SLB must be specifically isotopically labeled in order to assign a particular mass (i.e. 13CH− or C15N−) to its parent molecule. This method is able to distinguish co-existing gel and liquid phases at a resolution of roughly 100 nm as well as determine the mole fractions of the two lipid components within each phase (see Fig 1E). One advantage of these experiments is that fluorescent labels, which have been shown to influence phase behavior, are not needed to visualize the sample. The high sensitivity of the NanoSIMS instrument makes it possible to detect small amounts of membrane incorporated or associated proteins as well as lipids. Other groups have used time-of-flight (TOF)-SIMS measurements to study the organization of lipid components in supported monolayers [23]. TOF-SIMS offers the advantage that larger molecular fragments are obtained, obviating the need for isotopic labeling with the trade-off of lower sensitivity and spatial resolution compared to the NanoSIMS. In a remarkable paper, Ostrowski et al. were able to detect the accumulation of a particular lipid component at the junctions of mating protozoan cells [24••]. This example illustrates the great potential of SIMS to complement other imaging methods by providing a genuine chemical analysis of membranes with high spatial resolution.
To create supported membranes taken from biological sources, Vogel uses a coverslip coated with poly-L-lysine to rip off fragments of the upper cell membrane (see Fig 1F) [25, 26•]. If a substrate machined to have arrays of 500 nm holes is used, the technique creates sets of free-standing membranes which seal the holes. This allows independent access to either side of the cell membrane for measurements of transport across the bilayer or other directional reactions. A complementary method has also been developed to mount the basal cellular membrane by sonicating an adhered cell [27]. Using an appropriate support and biosynthetic incorporation of isotopic labels, cell membrane lipids and proteins could potentially be analyzed using SIMS to allow composition imaging.
A major shortcoming of solid supported bilayers is that incorporation of trans-membrane proteins often leads to loss of their lateral mobility and function. Several strategies have been developed to address this problem, including variations on the membranes over holes approach described above. One is to assemble bilayers on softer supports such as polymer cushions [28]. Ongoing work by the Tamm group uses Langmuir transfers and/or vesicle fusion to create polymer supported bilayers with different lipid mixtures in the two leaflets [29•]. They deposited such an asymmetric bilayer where the lipid composition, if used to make symmetric bilayers, is expected to lead to phase separation in the proximal leaflet but not the distal. Depending on the specific combinations used, domain formation in the former can induce domain formation in the latter, suggesting a mechanism for similar behavior in biological systems whose membranes are similarly asymmetric.
Another strategy for forming bilayers to preserve protein function is to use long-chain tethers to remove the membrane from the solid support (see Fig 1G). These tethered bilayers can be formed on a variety of surfaces (e.g. silica or gold) by modifying the anchoring molecule using surface coupling chemistry (silane or thiol, respectively) [30]. When using a metal support, electrical measurements can be performed to monitor the activity of ion channels incorporated into the bilayer. Vockenroth et al. have developed such a system to incorporate the M2 domain of the nicotinic acetylcholine receptor, pentamers of which form an ion channel which allow cations to traverse the membrane selectively based on their size [31•]. This work is directed towards making devices for electrical measurements of ion channels [32]. Free-standing planar bilayers such as black lipid membranes are an incredibly fruitful model system for studying membrane proteins. We focus here on progress towards forming such structures on scaffolds suitable for use as sensors or other devices. Although conventional free-standing membranes formed over solid substrates with holes are only moderately stable, hole sizes can be scaled down and the membrane surrounded by polymers such as an agarose matrix [33•]. This has recently been described as an approach to making electric sensors using single alpha hemolysin (αHL) pore proteins to act as voltage gates which can be modified to recognize specific analytes. Taken together, this collection of carefully designed and assembled interfaces suggests that the long-sought goal of constructing artificial devices based on membrane components and exploiting their exquisite sensitivity and selectivity may be realized.
Complex assemblies: tethered vesicles (H)
Several groups have tethered lipid vesicles to solid supported bilayers using DNA hybridization [34, 35•, 36] or biotin-streptavidin [37••, 38–40] recognition elements. These approaches combine many of the advantages of a supported bilayer, now in a supporting role, with free-standing systems. In particular, proteins incorporated into tethered vesicles are shielded from the solid support by the presence of the SLB. Our group and others have shown that DNA-tethered vesicles (see Fig 1H) are constrained to diffuse in two dimensions in a plane parallel to the surface with a diffusion constant roughly five times less than that of fluorescently labeled lipids [41]. Interactions between vesicles displaying reactive membrane components such as DNA or proteins can be observed as vesicles diffuse and collide with each other, and this begins to approximate the next level of reaction and organization (Y-HM Chan et al., unpublished). Complex behavior is observed when charged vesicles are tethered onto a patterned SLB containing charged lipids and then subjected to an electric field parallel to the bilayer surface [42•]. The competition between electroosmotic and electrophoretic forces on the vesicles, coupled to the rearrangement of the charged lipids in the SLB, can lead to a focusing effect where a vesicle’s position depends on its charge. This raises the potential to use these vesicles to detect binding of membrane receptors to ligands if the net charge on the vesicle changes.
Vesicles tethered with biotin-streptavidin coupling via biotinylated lipids are often observed not to diffuse, so this system is not useful for studying interactions between tethered vesicles; however, this can be an excellent system for encapsulating reactants in small volumes for fluorescence measurements down to the single-molecule level [38–40]. Also, these tethered vesicles can react with free vesicles flowed over the surface from bulk solution, and this was recently used to study lipid mixing between single pairs of vesicles mediated by yeast SNARE proteins [37••]. The stages of vesicle fusion were distinguished by the extent of Förster resonant energy transfer (FRET) due to mixing of fluorescently labeled lipids from the two vesicles, though content mixing was not demonstrated. Time traces indicate several pathways from docking to outer and inner leaflet mixing, with many displaying a hemifusion intermediate and/or kiss-and-run behavior.
In addition to acting as tethers, DNA oligomers can also act as recognition elements to mediate fusion between lipid vesicles. This is achieved when the orientations of the complementary DNA incorporated into separate SUV’s are such that hybridization brings the bilayer surfaces in close proximity (as is believed to occur with SNARE proteins). Stengel et al. show that cholesterol anchored DNA in small unilamellar vesicles can lead to lipid mixing as visualized by FRET donor dequenching, though content mixing was not reported [43•]. Independent experiments in our lab using the lipid anchor described in [35•] also lead to lipid and content mixing. The ability to change DNA sequence gives great control over the properties of hybridization reaction, potentially making it a powerful tool for studying membrane fusion.
Multi-scale simulations (I)
Computational modeling of lipid membranes ranges in complexity from continuum to full-atomistic descriptions, and the level of detail is typically chosen based on the size of the system being studied. Many problems in membrane biology, however, involve interactions and dynamics over a large range of length-scales. Multi-scale simulations (see Fig 1I) bridge this range by modeling different components of membranes at various levels of complexity [44, 45•, 46]. This approach has been used to model membrane proteins, with a full-atomistic model for the protein and a mesoscopic [45•] or coarse-grained [46] model for the lipid bilayer and solvent. This gives a balance between retaining enough detail to describe the salient features of the system while allowing larger scale simulations to run with limited computing power.
Atomistic [47•] and coarse-grained [48•] simulations have been performed on the fusion of vesicles roughly 15 nm in diameter. These suggest that upon stalk formation, lipid mixing proceeds through several hemifusion intermediates to achieve full fusion on the order of nano- to microseconds, which is substantially faster than is observed experimentally. Future improvements to these models could include achieving larger vesicle sizes and the incorporation of membrane proteins, and these papers highlight the demand for increasingly complex simulations in order to address this important problem.
Conclusions and Outlook
The model systems and analytical techniques presented in this review provide only a small sample of the diversity and creativity of research being performed on model membranes. Each system has individual advantages when probing questions of basic science or used for device applications. As the ability to manipulate membranes improves, continuing challenges include incorporating and observing complex membrane protein assemblies, device multiplexing and robustness, and connecting experimental and theoretical results. It is likely that the next generation of model systems will involve more complex compositions (lipids and proteins) and combinations of the assemblies illustrated in Figure 1, leading to insights into essential biological processes such as membrane fusion and trafficking, signaling, cell-cell recognition, and organization into tissue, as well as insight into the origins of life [49] and the creation of entirely artificial entities that may capture essential features of living systems.
Acknowledgments
We thank Gerald Feigenson, Owe Orwar, Jay Groves, Franck Duong, Horst Vogel, Ingo Köper, and Gregory Voth for permission to include figures from their publications and Tomo Narashima for the cell shown schematically at the center of Figure 1. This work was supported in part by grants from the NSF Biophysics Program, NIH GM069630 and by the MRSEC Program of the NSF under Award DMR-0213618 (CPIMA). The authors declare no conflicts of interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Feigenson GW. Phase boundaries and biological membranes. Annu Rev Biophys Biomol Struct. 2007;36:63–77. doi: 10.1146/annurev.biophys.36.040306.132721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.•.Baumgart T, Hunt G, Farkas ER, Webb WW, Feigenson GW. Fluorescence probe partitioning between Lo/Ld phases in lipid membranes. Biochim Biophys Acta. 2007 doi: 10.1016/j.bbamem.2007.05.012. The partitioning behavior of 26 fluorophores into lo and ld phases in GUV’s is reported. Most lipid-conjugated fluorophores partition into the ld phase, while DiI derivatives and polyaromatic hydrocarbons showed more mixed behavior. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Girard P, Pecreaux J, Lenoir G, Falson P, Rigaud J, Basserau P. A new method for the reconstitution of membrane proteins into giant unilamellar vesicles. Biophys J. 2004;87:419–429. doi: 10.1529/biophysj.104.040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doeven MK, Folgering JHA, Krasnikov V, Geertsma ER, van den Bogaart G, Poolman B. Distribution, lateral mobility and function of membrane proteins incorporated into giant unilamellar vesicles. Biophys J. 2005;88:1134–1142. doi: 10.1529/biophysj.104.053413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.•.Baumgart T, Hammond AT, Sengupta P, Hess ST, Holowka DA, Baird BA, Webb WW. Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proc Natl Acad Sci USA. 2007;104:3165–3170. doi: 10.1073/pnas.0611357104. Giant plasma membrane vesicles derived from mammalian cells are shown to exhibit phase separation at sub-physiological temperatures. The phase partitioning behavior of fluorescent lipids and labeled proteins are also described. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.•.Karlsson A, Sott K, Markström M, Davidson M, Konkoli Z, Orwar O. Controlled initiation of enzymatic reactions in micrometer-sized biomimetic compartments. J Phys Chem B. 2005;109:1609–1617. doi: 10.1021/jp0459716. Networks of GUV’s connected by lipid tubules are used to mix enzyme and substrate incorporated into the vesicle contents. This approaches the level of organization found in biological systems, and allows controlled sequences of reactions to be observed. [DOI] [PubMed] [Google Scholar]
- 7.Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 8.Leduc C, Campàs O, Zeldovich KB, Roux A, Jolimaitre P, Bourel-Bonnet L, Goud B, Joanny J-F, Bassereau P, Prost J. Cooperative extraction of membrane nanotubes by molecular motors. Proc Natl Acad Sci USA. 2004;101:17096–17101. doi: 10.1073/pnas.0406598101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.•.Roux A, Cuvelier D, Nassoy P, Prost J, Bassereau P, Goud B. Role of curvature and phase transition in lipid sorting and fission of membrane tubules. EMBO J. 2005;24:1537–1545. doi: 10.1038/sj.emboj.7600631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bauer B, Davidson M, Orwar O. Direct reconstitution of plasma membrane lipids and proteins in nanotube-vesicle networks. Langmuir. 2006;22:9329–9332. doi: 10.1021/la060828k. Lipid tubules pulled from GUV’s by optical tweezers show depletion lo partitioning lipids and cholesterol with respect to the parent vesicle. Fission of the tubes is observed at the boundaries between phase separated lo and ld domains. [DOI] [PubMed] [Google Scholar]
- 11.Groves JT. Bending mechanics and molecular organization in biological membranes. Annu Rev Phys Chem. 2007;58:697–717. doi: 10.1146/annurev.physchem.56.092503.141216. [DOI] [PubMed] [Google Scholar]
- 12.Parthasarathy R, Groves JT. Coupled membrane fluctuations and protein mobility in supported intermembrane junctions. J Phys Chem B. 2006;110:8513–8516. doi: 10.1021/jp055730d. [DOI] [PubMed] [Google Scholar]
- 13.••.Mossman KD, Campi G, Groves JT, Dustin ML. Altered TCR signaling from geometrically repatterned immunological synapses. Science. 2005;310:1191–1193. doi: 10.1126/science.1119238. Immunological synapses are formed between T-cells exposed to a patterned supported lipid bilayer displaying complementary protein ligands simulating an antigen presenting cell. The patterned surface constrains T-cell receptor reorganization, and a dependence of T-cell receptor signaling on position is observed. [DOI] [PubMed] [Google Scholar]
- 14.Nath A, Atkins WM, Sligar SG. Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry. 2007;46:2059–2069. doi: 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]
- 15.Boldog T, Li M, Hazelbauer GL. Using nanodiscs to create water-soluble transmembrane chemoreceptors inserted in lipid bilayers. Methods Enzymol. 2007;423:317–335. doi: 10.1016/S0076-6879(07)23014-9. [DOI] [PubMed] [Google Scholar]
- 16.Bhat S, Sorci-Thomas MG, Tuladhar R, Samuel MP, Thomas MJ. Conformational adaptation of apolipoprotein A–I to discretely sized phospholipid complexes. Biochemistry. 2007;46:7811–7821. doi: 10.1021/bi700384t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.•.Alami M, Dalal K, Lelj-Garolla B, Sligar SG, Duong F. Nanodiscs unravel the interaction between the SecYEG channel and its cytosolic partner Sec A. EMBO J. 2007;26:1995–2004. doi: 10.1038/sj.emboj.7601661. SecYEG complexes in lipid nanodiscs are found to bind only the monomeric form of their partner SecA. The binding is also found to be dependent on a key amino acid in the SecY protein as well as the lipid composition of the nanodisc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richter RP, Brisson AR. Following the formation of supported lipid bilayers on mica: A study combining AFM, QCM-D, and ellipsometry. Biophys J. 2005;88:3422–3433. doi: 10.1529/biophysj.104.053728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.•.Richter RP, Bérat R, Brisson AR. Formation of solid-supported lipid bilayers: An integrated view. Langmuir. 2006;22:3497–3505. doi: 10.1021/la052687c. This paper is an excellent synthesis of experiments elucidating the mechanism of the vesicle fusion reaction to form supported lipid bilayers and its dependence on factors including lipid composition, electrostatics, divalent cations, and surface properties. [DOI] [PubMed] [Google Scholar]
- 20.Yee CK, Amweg ML, Parikh AN. Membrane photolithography: Direct micropatterning and manipulation of fluid phospholipids membranes in the aqueous phase using deep-UV light. Adv Mater. 2004;16:1184–1189. [Google Scholar]
- 21.Marxer CG, Kraft ML, Weber PK, Hutcheon ID, Boxer SG. Supported membrane composition analysis by secondary ion mass spectrometry with high lateral resolution. Biophys J. 2005;88:2965–2975. doi: 10.1529/biophysj.104.057257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.••.Kraft ML, Weber PK, Longo ML, Hutcheon ID, Boxer SG. Phase separation of lipid membranes analyzed with high-resolution secondary ion mass spectrometry. Science. 2006;313:1948–1951. doi: 10.1126/science.1130279. NanoSIMS imaging was performed on two-component, phase-separated lipid bilayers. Isotopic labeling of the lipids allowed composition to be quantified based on the collection of molecular ions, leading to images with high spatial resolution and information content. [DOI] [PubMed] [Google Scholar]
- 23.McQuaw CM, Sostarecz AG, Zheng L, Ewing AG, Winograd N. Lateral heterogeneity of dipalmitoylphosphatidylethanolamine-cholesterol Langmuir-Blodgett films investigated with imaging time-of-flight secondary ion mass spectrometry and atomic force microscopy. Langmuir. 2005;21:807–813. doi: 10.1021/la0479455. [DOI] [PubMed] [Google Scholar]
- 24.••.Ostrowski SG, Van Bell CT, Winograd N, Ewing AG. Mass spectrometric imaging of highly curved membranes during Tetrahymena mating. Science. 2004;305:71–73. doi: 10.1126/science.1099791. TOF-SIMS composition imaging is performed on a complex biological sample, mating Tetrahymena cells. The junction between cells shows depleted levels of phosphocholine lipids and elevation of the cone-shaped 2-aminoethylphosphonolipid. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Danelon C, Perez J-B, Santschi C, Brugger J, Vogel H. Cell membranes suspended across nanoaperture arrays. Langmuir. 2006;22:22–25. doi: 10.1021/la052387v. [DOI] [PubMed] [Google Scholar]
- 26.•.Perez J-B, Martinez KL, Segura J-M, Vogel H. Supported cell-membrane sheets for functional fluorescence imaging of membrane proteins. Adv Funct Mater. 2006;16:306–312. Poly-L-lysine coated slides are placed on top of cultured cells then removed, creating a supported membrane taken directly from biological sources for further study. This is a general method for capturing complex membrane composition in a planar format. [Google Scholar]
- 27.Drees F, Reilein A, Nelson WJ. Cell-adhesion assays: Fabrication of an E-cadherin substratum and isolation of lateral and basal membrane patches. Methods Mol Biol. 2004;294:303–320. doi: 10.1385/1-59259-860-9:303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka M, Sackmann E. Polymer-supported membranes as models of the cell surface. Nature. 2005;437:656–663. doi: 10.1038/nature04164. [DOI] [PubMed] [Google Scholar]
- 29.•.Kiessling V, Crane JM, Tamm LK. Transbilayer effects of raft-like lipid domains in asymmetric planar bilayers measured by single molecule tracking. Biophys J. 2006;91:3313–3326. doi: 10.1529/biophysj.106.091421. Asymmetric bilayers are formed on a polymer cushion where the proximal leaflet is expected to phase separate but the distal leaflet is not. Induced domain formation in the distal leaflet was observed for certain compositions indicating coupling between the bilayer leaflets. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Atanasov V, Atansova PP, Vockenroth IK, Knorr N, Köper I. A molecular toolkit for highly insulating tethered bilayer lipid membranes on various substrates. Bioconjugate Chem. 2006;17:631–637. doi: 10.1021/bc050328n. [DOI] [PubMed] [Google Scholar]
- 31.•.Vockenroth IK, Atanasova PP, Long JR, Jenkins ATA, Knoll W, Köper I. Functional incorporation of the pore forming segment of AChR M2 into tethered bilayer lipid membranes. Biochim Biphys Acta. 2007;1768:1114–1120. doi: 10.1016/j.bbamem.2007.02.006. A defect free tethered bilayer incorporating the pore forming domain of an ion channel is assembled on a gold electrode. Ions of different size can be distinguished, and the tether identity also affects the performance of the device. [DOI] [PubMed] [Google Scholar]
- 32.Keizer HM, Dorvel BR, Andersson M, Fine D, Price RB, Long JR, Dodabalapur A, Köper I, Knoll W, Anderson PAV, Duran RS. Functional ion channels in tethered bilayer membranes—implications for biosensors. Chem Bio Chem. 2007;8:1246–1250. doi: 10.1002/cbic.200700094. [DOI] [PubMed] [Google Scholar]
- 33.•.Kang X-F, Cheley S, Rice-Ficht AC, Bayley H. A storable encapsulated bilayer chip containing a single protein nanopore. J Am Chem Soc. 2007;129:4701–4705. doi: 10.1021/ja068654g. The αHL protein membrane pore protein is assembled in a bilayer spanning a small Teflon hole on an electrode. This device is able to detect the presence of chemical analytes based on electrical current, and can be developed into robust sensors. [DOI] [PubMed] [Google Scholar]
- 34.Yoshina-Ishii C, Boxer SG. Arrays of mobile tethered vesicles on supported lipid bilayers. J Am Chem Soc. 2003;125:3696–3697. doi: 10.1021/ja029783+. [DOI] [PubMed] [Google Scholar]
- 35.•.Yoshina-Ishii C, Miller GP, Kraft ML, Kool ET, Boxer SG. General method for modification of liposomes for encoded assembly on supported bilayers. J Am Chem Soc. 2005;127:1356–1357. doi: 10.1021/ja043299k. Vesicles are tethered to a SLB using complementary DNA which has been covalently coupled a lipid-like tail group. The sequence specificity of DNA allows arrays of vesicles to be made using patterned bilayers and their lateral mobility can be exploited for studies of vesicle-vesicle interactions. [DOI] [PubMed] [Google Scholar]
- 36.Indriati P, Höök F. Bivalent cholesterol-based coupling of oligonucleotides to lipid membrane assemblies. J Am Chem Soc. 2004;126:10224–10225. doi: 10.1021/ja048514b. [DOI] [PubMed] [Google Scholar]
- 37.••.Yoon T-Y, Okumus B, Zhang F, Shin Y-K, Ha T. Multiple intermediates in SNARE-induced membrane fusion. Proc Natl Acad Sci USA. 2006;103:19731–19736. doi: 10.1073/pnas.0606032103. Vesicles presenting yeast SNARE proteins are tethered to an SLB and reacted with vesicles in solution presenting the cognate proteins. Several mechanisms for outer then inner leaflet mixing (many including hemifusion intermediates) are classified based on FRET analysis of lipid labels. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rhoades E, Gussakovsky E, Haran G. Watching proteins fold one molecule at a time. Proc Natl Acad Sci USA. 2003;100:3197–3202. doi: 10.1073/pnas.2628068100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stamou D, Duschl C, Delamarche E, Vogel H. Self-assembled microarrays of attoliter molecular vessels. Angew Chem Int Ed. 2003;42:5580–5583. doi: 10.1002/anie.200351866. [DOI] [PubMed] [Google Scholar]
- 40.Bolinger P-Y, Stamou D, Vogel H. Integrated nanoreactor systems: Triggering the release and mixing of compounds inside single vesicles. J Am Chem Soc. 2004;126:8594–8595. doi: 10.1021/ja049023u. [DOI] [PubMed] [Google Scholar]
- 41.Yoshina-Ishii C, Chan Y-HM, Johnson JM, Kung LA, Lenz P, Boxer SG. Diffusive dynamics of vesicles tethered to a fluid supported bilayer by single-particle tracking. Langmuir. 2006;22:5682–5689. doi: 10.1021/la0534219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.•.Yoshina-Ishii C, Boxer SG. Controlling two-dimensional tethered vesicle motion using an electric field: Interplay of electrophoresis and electro-osmosis. Langmuir. 2006;22:2384–2391. doi: 10.1021/la0526277. The dynamics of vesicles tethered on a patterned SLB and subjected to an external electric field were observed. A subtle balance between electrophoretic and electroosmotic forces, in conjunction with electrophoretic reorganization of the lipids in the SLB, can lead to focusing effects where the steady-state position of the vesicles is dependent on their charge. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.•.Stengel G, Zahn R, Höök F. DNA-induced programmable fusion of phospholipid vesicles. J Am Chem Soc. 2007;129:9584–9585. doi: 10.1021/ja073200k. Cholesterol-anchored DNA introduced into lipid vesicles can mediate inner and outer leaflet mixing as shown by FRET dequenching. This can lead to insights into the mechanisms for fusion. [DOI] [PubMed] [Google Scholar]
- 44.Ayton GS, Noid WG, Voth GA. Multiscale modeling of biomolecular systems: in serial and in parallel. Curr Opin Struct Biol. 2007;17:192–198. doi: 10.1016/j.sbi.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 45.•.Ayton GS, Voth GA. Multiscale simulation of transmembrane proteins. J Struct Biol. 2007;157:570–578. doi: 10.1016/j.jsb.2006.10.020. This simulation study uses a combined coarse-grained and continuum model to describe membrane proteins and the lipid bilayer, respectively. The protein shows a subtle conformational change when the mechanical properties of the bilayer are adjusted. [DOI] [PubMed] [Google Scholar]
- 46.Shi Q, Izvekov S, Voth GA. Mixed atomistic and coarse-grained molecular dynamics: Simulation of a membrane-bound ion channel. J Phys Chem B. 2006;110:15045–15048. doi: 10.1021/jp062700h. [DOI] [PubMed] [Google Scholar]
- 47.•.Knecht V, Marrink S-J. Molecular dynamics simulations of lipid vesicle fusion in atomic detail. Biophys J. 2007;92:4254–4261. doi: 10.1529/biophysj.106.103572. Atomistic simulations of vesicle fusion show progression from stalk formation to a stable hemifusion to the appearance fusion pore on the order of 10 ns. The density of lipids in the bilayer junction is reduced during hemifusion, but voids are not observed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.•.Kasson PM, Kelley NW, Singhal N, Vrljic M, Brunger AT, Pande VS. Ensemble molecular dynamics yields submillisecond kinetics and intermediates of membrane fusion. Proc Natl Acad Sci USA. 2006;103:11916–11921. doi: 10.1073/pnas.0601597103. A coarse grained model is used to simulated the fusion of lipid vesicles. Many mechanistic pathways are observed, and the timescale of fusion depends on the separation distance between the vesicles’ surfaces. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen IA, Salehi-Ashtiani K, Szostak JW. RNA catalysis in model protocell vesicles. J Am Chem Soc. 2005;127:13213–13219. doi: 10.1021/ja051784p. [DOI] [PMC free article] [PubMed] [Google Scholar]


