Abstract
Xenomonitoring (detection of filarial larvae or their DNA in mosquitoes) is a sensitive marker for assessing the endemicity of filariasis and a useful tool for evaluating elimination programs. To examine the fate of microfilariae (mf) and filarial DNA in vector competent and non-competent mosquito strains, we compared the detection of Brugia malayi parasites by dissection and by quantitative real-time polymerase chain reaction (PCR) in three different mosquito strains. We conclude that PCR is much more sensitive than dissection for detecting filarial larvae, especially their remnants in mosquitoes. However, parasite DNA can be detected in both vector and non-vector mosquitoes for two weeks or longer after they ingest mf-positive blood. Thus, although xenomonitoring with vector and non-vector mosquito species may be a sensitive method for indirectly detecting filarial parasites in human populations, positive test results for parasite DNA in mosquitoes do not necessarily prove that transmission is ongoing in the study area.
INTRODUCTION
Filarial nematodes infect more than 150 million people in many tropical and subtropical regions of the world. Lymphatic filariasis (caused by Wuchereria bancrofti, Brugia malayi, and B. timori) may lead to acute fever attacks and chronic hydrocele and lymphedema. This mosquito-borne disease has been targeted by the World Health Organization for elimination by the year 2020. The key strategy of the Global Program to Eliminate Lymphatic Filariasis (GPELF) is to eliminate the microfilariae (mf) from the human host through mass drug administration (MDA) of the population at risk of infection with the aim to interrupt transmission.1 Sensitive and specific diagnostic tools are required to monitor the success of MDA and to establish endpoints for intervention.
The examination of mosquito vectors for ingested MF, developing larvae, and infective third-stage larvae (L3) offers the opportunity to monitor the impact of control programs without disturbing the human population. Traditionally, mosquitoes have been examined by dissection and microscopic identification of filarial larvae. In areas with low infection rates, high-throughput screening is required and dissection of individual mosquitoes is not cost-effective; therefore, a number of polymerase chain reaction (PCR) assays have been developed for the screening of mosquito pools.2–4 These methods hold great promise to be widely used for monitoring effects of MDA or other interventions. During a five-year MDA campaign in Egypt, decreases in mosquito infection rates determined by PCR tests of filarial DNA from pools of blood-fed mosquitoes correlated well with decreases in other infection parameters in humans (mf, circulating filarial antigen, and antibody prevalence rates).5 However, the significance of mosquito pool screening PCR results for guiding lymphatic filariasis elimination programs is still not established.
Filarial vector capacity is limited by the number of mf ingested by a mosquito, and less than 100% of them develop into infective L3 within competent vectors.6–9 In contrast, no ingested MF develop into L3s in non-competent vectors. Dead filarial larvae or their remnants may remain free or encapsulated in the hemocoel or in different mosquito tissues (i.e., midgut or thorax). This can provide a source of worm DNA for detection by PCR. It has been suggested that recently blood-fed mosquitoes may have higher detection rates for filarial DNA.2,3 However, there is no experimental evidence to support this hypothesis and it is not known how long DNA from non-developing or dead filarial larvae remain detectable by PCR. Therefore, the aim of the present study was to determine the fate of mf and their DNA in vector competent and non-competent mosquito strains to better define the role of mosquito screening as a monitoring tool for lymphatic filariasis control. We infected three different strains of mosquitoes with B. malayi and examined them by dissection and quantitative real-time PCR for developing larvae and parasite DNA, respectively.
MATERIALS AND METHODS
Mosquito colonies
Aedes aegypti black-eyed Liverpool (AeL), Ae. aegypti Rockefeller (AeR), and Culex pipiens Iowa (Cu) used in this study were maintained at the University of Wisconsin-Madison, as previously described.10,11 These three mosquito strains were selected for this study based on their vector competence for B. malayi. AeL is a competent laboratory vector of B. malayi in which worms complete development to infective stage larvae, and AeR and Cu are both refractory mosquito strains. In AeR, mf penetrate the midgut and enter thoracic muscle cells where development ceases. In contrast, B. malayi mf do not penetrate the midgut in Cu mosquitoes but die in the midgut lumen (Christensen BM, unpublished data). Laboratory animals were handled according to guidelines approved by the Animal Care Committee at the University of Wisconsin-Madison.
Mosquito exposure to infective blood
Brugia malayi (TRS strain) infected cat blood was obtained from the National Institute of Allergy and Infectious Diseases/National Institute of Health Filariasis Research Reagent Repository Center (http://www.filariasiscenter.org) with mf densities of 28–54 mf/20 μL. Mosquitoes were fed using water-jacketed membrane feeders fitted with fresh chicken skin membranes.12 Control mosquitoes were allowed to feed on uninfected dog blood from similar membrane feeders. Fully engorged mosquitoes were separated, placed in 0.5-liter paper cartons, and provided 0.3 M sucrose solution ad libitum. Mosquitoes were sampled for dissection or DNA extraction on days 0 (two hours post-blood feeding), 1, 2, 3, 4, 7, 10, 14, and 21 post-ingestion of mf-positive blood (pi).
Mosquito dissection
Individual mosquitoes were dissected, and the head, thorax, midgut, and abdomen (with midgut removed) were examined separately for parasites. Each body part was placed in a separate drop of Aedes saline13 and teased apart with 0.15-mm insect pin probes. Mosquito tissues were examined for filarial worms with a compound microscope using phase contrast optics. Approximately 15 minutes were spent examining tissues from each dissected mosquito.
Extraction of DNA
Four mosquitoes were pooled (only three on day 0 pi) and stored at −20°C. For extraction of DNA, mosquitoes were homogenized in a parafilm-sealed, 2-mL, round-bottom reaction tube containing 180 μL of phosphate-buffered saline and one zinc-plated 4.5-mm steel BB (Daisy Outdoor Products, Rogers, AR) by vortexing for 20 minutes using a 24-sample vortex adapter (Laney S, unpublished data). Samples were briefly centrifuged and extraction was performed with the DNeasy kit (Qiagen, Valencia, CA) as described previously.3 DNA was eluted in 100 μL of AE buffer (Qiagen) (75 μL for samples from day 0 with a pool size of three mosquitoes) and 1 μL of template was used in a 25-μL real-time PCR.
Quantitative real-time PCR
Real-time PCR targeting the Hha I repeat of B. malayi was performed as described previously.14 Two different assays were used that amplify either the entire 320-basepair repeat (Taqman assay; Applied Bio-systems, Foster City, CA) or a 120-basepair fragment of the repeat (MGB Eclipse assay; Nanogen, Bothell, WA). All PCRs were carried out in duplicate. The samples were retested if only one of the duplicates was found positive or if the cycle threshold value (Ct) was ≥ 39. The sample Ct value was calculated as the arithmetic mean of duplicates. In all real-time PCRs, water was used as a no-template negative control and 100 pg of B. malayi DNA was used as a positive control.
Data analysis
Group differences were assessed with the chi-square test. The Poolscreen program version 2.0 was used to estimate mosquito infection rates.15
RESULTS
Detection of B. malayi larvae by dissection
Two independent blood-feeding experiments were performed. Fifty-two individual AeL mosquitoes were dissected at various time points post-infection (pi). Brugia malayi larvae were detected in 44 (85%) of the dissected mosquitoes (Figure 1A). Among 44 dissected AeR, B. malayi mf were only found in 10 mosquitoes (23%), with most being recovered during the first two days pi. A single, non-moving, and completely melanized mf was recovered from the thorax of one mosquito at day 4, and another single motile mf with little melanin deposition was recovered from the abdomen of another AeR mosquito on day 10 (Figure 1C). In 44 examined Cu mosquitoes, only a single non-moving mf was detected in the midgut two hours pi (Figure 1E).
Figure 1.
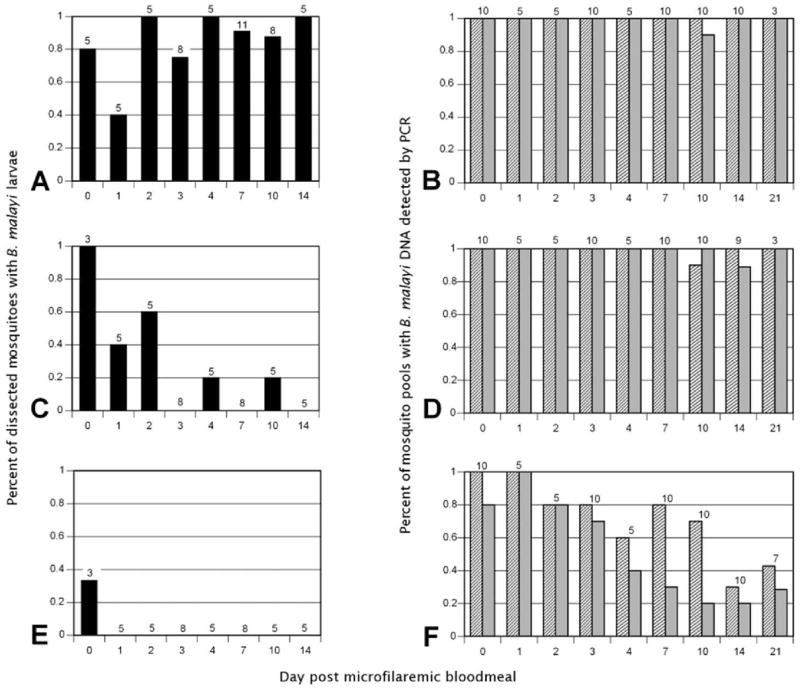
Detection of Brugia malayi in mosquito vectors. Membrane feeders containing microfilaremic cat blood were used to feed Aedes aegypti Liverpool (competent vector; A and B), Ae. aegypti Rockefeller (non-competent vector; C and D) and Culex pipiens (non-competent vector; E and F). Brugia malayi was detected by dissection of individual mosquitoes (black bars; A, C, and E) or by DNA detection using TaqMan real-time polymerase chain reaction (PCR) (gray bars) and MGB Eclipse real-time PCR (striped bars) with DNA isolated from mosquito pools (B, D, and F). Numbers above bars indicate individual mosquitoes dissected (A, C, and E) or mosquito pools tested (B, D, and F).
Detection of B. malayi DNA in pools of mosquitoes by real-time PCR
In two experiments, DNA was extracted from 272 AeL, 268 AeR, and 308 Cu mosquitoes in 68, 67, and 77 pools, respectively, of 4 mosquitoes each at various time points pi. In addition, DNA was prepared for each mosquito strain from 60 uninfected mosquitoes in 15 pools for each strain. Two different real-time PCR assays were used to detect parasite DNA. The Taqman assay has essentially the same sensitivity as conventional PCR and uses the entire Hha I repeat as a target, and the more sensitive MGB Eclipse assay uses a slightly different chemistry and detects a short fragment of the Hha I repeat. The Taqman assay showed that all pools of AeL were positive over the study period of 21 days pi, with the exception of 1 of 10 pools on day 10 pi (Figure 1B). Using the Poolscreen algorithm, this would correspond to an estimated B. malayi DNA detection rate in single mosquitoes of 65% (95% confidence interval [CI] = 50–85%). Examination of the same pools by the Eclipse MGB assay showed B. malayi DNA in all pools, and a DNA detection rate for mosquitoes could not be estimated by Poolscreen.
Screening of AeR pools showed similar results. All pools were positive for filarial DNA using the Taqman and the MGB assays with exception of a single pool for each method on day 14 pi and day 10 pi, respectively (Figure 1D). This corresponds to an estimated B. malayi DNA detection rate in single mosquitoes of 65% (95% CI = 48–85%).
Culex mosquito pools had somewhat different results. Eighty percent of the pools were Taqman positive for B. malayi DNA up to day 3 pi, although only 26% from later collection times were positive (Figure 1F). With the more sensitive MGB Eclipse assay, 82% of the pools through 10 days pi were positive, and on day 14 pi and 21 days pi only 35% were positive. Poolscreen estimates for DNA detection rates in Cu mosquitoes were 14% (95% CI = 10–20%) with the Taqman assay and 28% (95% CI = 21–36%) with the Eclipse MGB assay.
Quantitation of B. malayi DNA in pools of mosquitoes by real-time PCR
Cycle threshold (Ct) values (with fluorescence signals that exceed background noise levels to indicate a positive real-time PCR) are inversely correlated with the amount of target DNA present in the sample. In general, the Taqman assay had significantly higher Ct values compared with the MGB Eclipse assay (Figure 2; P < 0.05). Cycle threshold values for Cu mosquitoes on days 0 and 1 were equivalent to or lower than those for AeL and AeR. Therefore, the lower B. malayi DNA detection rates and DNA concentrations in Cu at later time points cannot be explained by a reduced uptake of mf during the blood meal.
Figure 2.
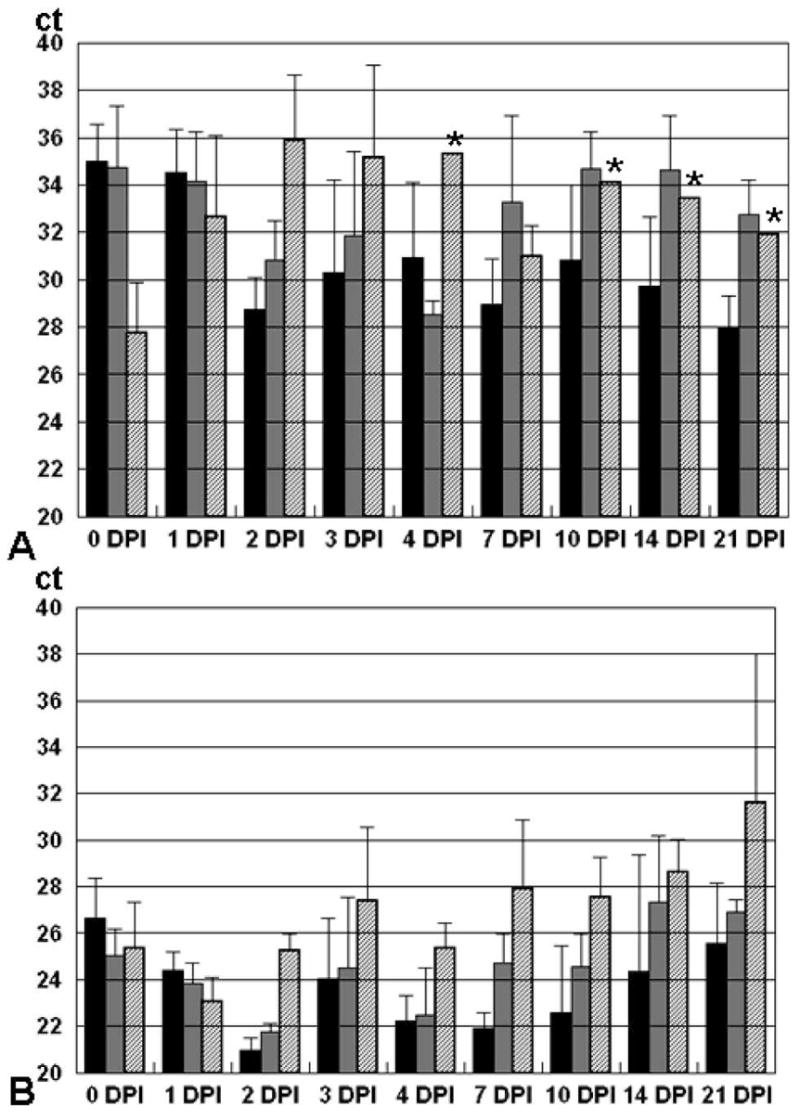
Quantitation of Brugia malayi DNA in mosquito pools by real-time polymerase chain reaction Cycle threshold (Ct) values for Taqman (A)– and MGB Eclipse (B)–positive mosquito pools at various time points after a microfilaria -positive blood meal. DPI = days post-infection. Solid bars = Aedes aegypti Liverpool; gray bars = Ae. aegypti Rockefeller; hatched bars = Culex pipiens. Error bars indicate standard deviation; stars indicate less than three positive pools.
Detection of parasite DNA in pools of mosquitoes after a second blood meal by real-time PCR
In the Aedes strains, mf penetrate the midgut wall within a few hours after an infective blood meal. In contrast, in Cx. pipiens, the mf do not penetrate the midgut wall and are trapped in the midgut. Therefore, we tested the hypothesis whether a second blood meal on an uninfected host would help to clear microfilarial DNA from the midgut. Culex mosquitoes were fed on mf-positive cat blood and reared in the insectary for 14 days. A group of mosquitoes were fed again on day 7 pi on a non-infected chicken, and another group acted as a control with no blood feeding. Mosquitoes were screened for filarial DNA on day 14 pi and 21 pi using Taqman and MGB Eclipse PCRs (Table 1). Although some mosquitoes cleared filarial DNA, there was no significant difference in DNA detection rates between the two groups of mosquito pools (P > 0.05, by chi-square test). Thus, filarial DNA was detectable in some Cu mosquitoes after a second blood meal taken one week after mf ingestion.
Table 1.
Detection rate of Brugia malayi DNA in pools of Culex pipiens that initially fed on microfilaremic blood on day 0 and on uninfected blood 7 days later*
|
B. malayi microfilaremic blood† |
||||||
|---|---|---|---|---|---|---|
| Uninfected blood
|
TaqM
|
MGB Eclipse
|
||||
| DPI | TaqM | MGB | Non–re-fed | Re-fed‡ | Non–re-fed | Re-fed‡ |
| 0 | 0 (1) | 0 (1) | 100 (5) | NA | 100 (5) | NA |
| 10 | 0 (1) | 0 (1) | 75 (4) | 50 (4) | 75 (4) | 50 (4) |
| 14 | 0 (1) | 0 (1) | 25 (4) | 33 (3) | 25 (4) | 67 (3) |
Parasite DNA was detected by TaqM or MGB Eclipse real-time polymerase chain reaction. Values are percentage of mosquito pools positive for B. malayi (no. of pools tested). DPI = days post-ingestion of B. malayi; NA = not available.
By membrane feeders containing microfilaremic cat blood with 54 MF/20 μl.
Oviposition substrate was provided from 4 to 7 DPI; mosquitoes were then allowed to feed on an uninfected chicken 7 DPI.
DISCUSSION
We studied the fate of mf and filarial DNA with three different mosquito strains by dissection and real-time PCR. These included a competent laboratory vector (AeL), a non-transmitting strain that supports migration of MF (AeR), and a non-transmitting species (Cu) in which mf do not penetrate the midgut. Filarial DNA was detected in almost all pools of AeL and AeR for up to three weeks pi. Filarial DNA was also detected in 80% of Cu pools in the first three days pi and in smaller proportions of pools for up to three weeks pi. In addition, a second blood meal on an uninfected host had no influence on DNA persistence in non-vector Cu mosquitoes.
The natural vectors of B. malayi are mainly Anopheles (nocturnally periodic strains) and Mansonia (subperiodic strains) species.16 AeL and Ae. togoi are efficient laboratory vectors whereas Culex species are not considered to be vectors for B. malayi either in nature or in the laboratory. Dissection of mosquitoes showed that in AeL filarial larvae of different developmental stages were detectable in 85% of the mosquitoes. In contrast, filarial larvae were found mainly during the first two days pi in AeR and only two partly melanized mf were detected at later time points in this strain. A pronounced defense mechanism, such as complete melanization and encapsulation of filarial larvae, does not occur in this strain of Ae. aegypti. This is in agreement with earlier studies of AeR infected with the closely related B. pahangi.17 Few filarial larvae were detected in Cu, and mf were not found outside the midgut. This confirmed earlier observations that mf are unable to penetrate the midgut wall of this mosquito strain (Christensen BM, unpublished data). Staining of filarial larvae using hemalum may enhance the sensitivity of mosquito dissection to detect larvae or parasite fragments. However, it is unlikely that a significant number of larvae were overlooked by our careful dissection procedure because larvae were unambiguously identified by dissection in most of the AeL mosquitoes. Therefore, we postulate that remnants of filarial larvae remain free or encapsulated in the refractory mosquito strains AeR and Cu and that parasite DNA in these remnants is detected by PCR.
We used a relatively small pool size of only four mosquitoes to study the fate of B. malayi DNA in different mosquito strains. With the exception of the negative controls, all mosquitoes were allowed to feed on mf-positive blood. This experimental design ensured that immediately after blood feeding, all pools would contain at least one mf-positive mosquito that could be detected by real-time PCR. Previous evaluation of these real-time PCR assays indicated their high sensitivity.14 Because of the high copy number of the Hha I repeat and the short target sequence of the MGB Eclipse assay, we have estimated that a single cell of B. malayi can be detected in 100 μL of blood. We have no evidence that this assay is less sensitive for the detection of B. malayi DNA in pooled mosquitoes. It can be assumed that all positive pools contain at least one B. malayi cell (or an equivalent amount of DNA). Increasing the pool size to 20 mosquitoes using uninfected mosquitoes should not influence the sensitivity. Therefore, we conclude that the MGB Eclipse assay should be capable of detecting a single B. malayi cell in a pool of up to 20 mosquitoes.
In eastern Indonesia, Anopheles barbirostris is the vector for Brugia timori, a sibling species of B. malayi that can be detected by identical PCR assays.3 In the same area, Culex spp. (especially Cx. tritaeniorhynchus) are the predominant human-biting mosquitoes, but they are never found to be infected with Brugia larvae. However, we have sometimes detected Brugia DNA in Culex pools, and we assumed that this was due to mf ingested by these mosquitoes with blood meals. The results of the present study prove that Brugia DNA can persist in non-vector Culex species for up to three weeks, which is essentially the entire life span of the of the mosquito in nature. These results are consistent with results from blood meal identification studies in mosquitoes and other blood-sucking arthropods, which show that host DNA can be detected for 2–3 days or significantly longer after a blood meal, depending on the species, the molecular marker, and the DNA detection method used.18 Therefore, our laboratory results explain why filarial DNA can be detected in non-vector mosquito species in filariasis-endemic areas.
The main strategy used in the GPELF is MDA, which attempts to interrupt transmission by reducing the source of mf available to vector mosquitoes. Monitoring of vectors for filarial larvae and their DNA is an important diagnostic tool in intervention programs.5,19–21 There are essentially two monitoring strategies: xenomonitoring and transmission monitoring (Table 2). Our study has clearly shown that vector and non-vector mosquitoes may have filarial DNA detectable by PCR for weeks after they have fed on a microfilaremic host. The results indicate that mosquito collection for xenomonitoring using DNA detection assays should be performed after mf in the human population have decreased and at least two weeks after intervention when new generations of mosquitoes are present. Although high DNA detection rates in pools of a particular mosquito species may suggest this species is a competent vector, it does not provide proof of its ability to support parasite development and transmission. Our studies show that a non-vector mosquito species might have a high filarial DNA prevalence rate (similar to AeR in our experimental setting), and the actual vector is another species that may have a wider host range and less frequent contact with MF-positive blood meals. The L3s have to be detected to assess infectivity. Theoretically, this could be done by the detection of stage-specific transcripts using reverse transcriptase–PCR. However, RNA detection is technically more demanding than DNA detection, and it is difficult to identify candidate genes that are not expressed in other larval stages in mosquitoes. Heads and bodies were screened separately for the large-scale assessment of black flies for transmission of the filarial parasite Onchocerca volvulus by poolscreen PCR.22 Infective-stage larvae were assumed to be present only in the heads, and body pools may be judged only as indicators of vector-parasite contact. This strategy is less suitable for lymphatic dwelling filarial worms because infective-stage larvae are often not limited to the head of the mosquito.
Table 2.
Comparison of mosquito screening for xenomonitoring and transmission monitoring of lymphatic filariasis*
| Xenomonitoring | Transmission monitoring | |
|---|---|---|
| Mosquito samples | Any human-biting (anthropophilic) mosquitoes, no detailed species identification required | Only vector competent species (cytospecies), proper identification required |
| Sampling method | Human bait or indoor resting. Identification of anthropophilic species required when light or gravid traps are used. | Human bait or indoor resting. Identification of vector species (cytospecies) required when light or gravid traps are used. |
| Collection of large numbers of samples | Easy | Easy, if the vector is the predominant mosquito species |
| Screening method | Dissection of individual mosquitoes (cumbersome), pool screening for filarial DNA by conventional or real-time PCR (more sensitive) | Dissection of individual mosquitoes (cumbersome), mass dissection, pool screening for L3-specific filarial mRNA (experimental, not yet validated, requires special techniques) |
| Identification of vectors | Requires microscopy (detection of L3), high DNA detection rates in a mosquito may indicate vector competence, but confirmation required | Yes, by microscopy or detection of L3-specific mRNA (identification of 100% L3-specific transcripts difficult) |
| Indicator of prevalence in humans | Yes, indirectly | Yes, indirectly |
| Indicator for transmission | No. Probably, if DNA is detected in vector species | Yes, directly |
PCR = polymerase chain reaction; L3 = third-stage larvae.
Our study underlines the potential value of vector pool screening PCR, but has shown that such data have to be interpreted very carefully. Human-biting mosquitoes can be screened to detect mosquito-mf contacts, regardless of their vector competence. Dissection is far less sensitive than poolscreen PCR for that purpose. However, poolscreen PCR provides no direct indicator for the presence of developing or infective-stage larvae. For advanced elimination programs, xenomonitoring of vector and non-vector mosquitoes using poolscreen PCR is a sensitive method for indirectly detecting filarial parasites in human populations and the potential for transmission. However, positive parasite DNA test results in mosquitoes do not necessarily prove that significant transmission is ongoing in the study area.
Acknowledgments
We thank Dr. John McCall (University of Georgia, Athens, GA) and the National Institute of Allergy and Infectious Diseases/National Institutes of Health Filariasis Research Reagent Repository Center for providing microfilaremic cat blood.
Financial support: This work was supported in part by the National Institutes of Health grant AI-35855 (Gary J. Weil) and AI-19769 (Bruce M. Christensen).
References
- 1.Ottesen EA. Lymphatic filariasis: treatment, control and elimination. Adv Parasitol. 2006;61:395–441. doi: 10.1016/S0065-308X(05)61010-X. [DOI] [PubMed] [Google Scholar]
- 2.Bockarie MJ, Fischer P, Williams SA, Zimmerman PA, Griffin L, Alpers MP, Kazura JW. Application of a polymerase chain reaction-ELISA to detect Wuchereria bancrofti in pools of wild-caught Anopheles punctulatus in a filariasis control area in Papua New Guinea. Am J Trop Med Hyg. 2000;62:363–367. doi: 10.4269/ajtmh.2000.62.363. [DOI] [PubMed] [Google Scholar]
- 3.Fischer P, Wibowo H, Pischke S, Ruckert P, Liebau E, Ismid IS, Supali T. PCR-based detection and identification of the filarial parasite Brugia timori from Alor Island, Indonesia. Ann Trop Med Parasitol. 2002;96:809–821. doi: 10.1179/000349802125002239. [DOI] [PubMed] [Google Scholar]
- 4.Rao RU, Atkinson LJ, Ramzy RM, Helmy H, Farid HA, Bockarie MJ, Susapu M, Laney SJ, Williams SA, Weil GJ. A real-time PCR-based assay for detection of Wuchereria bancrofti DNA in blood and mosquitoes. Am J Trop Med Hyg. 2006;74:826–832. [PMC free article] [PubMed] [Google Scholar]
- 5.Ramzy RM, El Setouhy M, Helmy H, Ahmed ES, Abd Elaziz KM, Farid HA, Shannon WD, Weil GJ. Effect of yearly mass drug administration with diethylcarbamazine and albendazole on bancroftian filariasis in Egypt: a comprehensive assessment. Lancet. 2006;367:992–999. doi: 10.1016/S0140-6736(06)68426-2. [DOI] [PubMed] [Google Scholar]
- 6.Albuquerque CM, Cavalcanti VM, Melo MA, Vercosa P, Regis LN, Hurd H. Bloodmeal microfilariae density and the uptake and establishment of Wuchereria bancrofti infections in Culex quinquefasciatus and Aedes aegypti. Mem Inst Oswaldo Cruz. 1999;94:591–596. doi: 10.1590/s0074-02761999000500005. [DOI] [PubMed] [Google Scholar]
- 7.Farid HA, Hammad RE, Hassan MM, Ramzy RM, El Setouhy M, Weil GJ. Effects of combined diethylcarbamazine and albendazole treatment of bancroftian filariasis on parasite uptake and development in Culex pipiens L. Am J Trop Med Hyg. 2005;73:108–114. [PubMed] [Google Scholar]
- 8.Farid HA, Hammad RE, Soliman DA, El Setouhy MA, Ramzy RM, Weil GJ. Relationships between Wuchereria bancrofti microfilaria counts in human blood and parasite uptake and maturation in Culex pipiens, with observations on the effects of diethylcarbamazine treatment on these parameters. Am J Trop Med Hyg. 2003;68:286–293. [PubMed] [Google Scholar]
- 9.Gad AM, Hammad RE, Farid HA. Uptake and development of Wuchereria bancrofti in Culex pipiens L. and Aedes caspius Pallas. J Egypt Soc Parasitol. 1996;26:305–314. [PubMed] [Google Scholar]
- 10.Christensen BM, Sutherland DR. Brugia pahangi: exsheathment and midgut penetration in Aedes aegypti. Trans Am Microsc Soc. 1984;103:423–433. [Google Scholar]
- 11.Bartholomay LC, Farid HA, Ramzy RM, Christensen BM. Culex pipiens pipiens: characterization of immune peptides and the influence of immune activation on development of Wuchereria bancrofti. Mol Biochem Parasitol. 2003;130:43–50. doi: 10.1016/s0166-6851(03)00143-9. [DOI] [PubMed] [Google Scholar]
- 12.Rutledge LC, Ward RA, Gould DJ. Studies on the feeding response of mosquitoes to nutritive solutions in a new membrane feeder. Mosq News. 1964;24:407–419. [Google Scholar]
- 13.Hayes RO. Determination of a physiological solution for Aedes aegypti (L.) J Econ Entomol. 1953;46:624–627. [Google Scholar]
- 14.Rao RU, Weil GJ, Fischer K, Supali T, Fischer P. Detection of Brugia parasite DNA in human blood samples by real-time PCR. J Clin Microbiol. 2006;44:3887–3893. doi: 10.1128/JCM.00969-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katholi CR, Unnasch TR. Important experimental parameters for determining infection rates in arthropod vectors using pool screening approaches. Am J Trop Med Hyg. 2006;74:779–785. [PubMed] [Google Scholar]
- 16.World Health Organization. Lymphatic filariasis: the disease and its control. Fifth report of the WHO Expert Committee on Filariasis. World Health Organ Tech Rep Ser. 1992;821:1–71. [PubMed] [Google Scholar]
- 17.Christensen BM, Sutherland DR, Gleason LN. Defense reactions of mosquitoes to filarial worms: comparative studies on the response of three different mosquitoes to inoculated Brugia pahangi and Dirofilaria immitis microfilariae. J Invertebr Pathol. 1984;44:267–274. doi: 10.1016/0022-2011(84)90024-7. [DOI] [PubMed] [Google Scholar]
- 18.Mukabana WR, Takken W, Knols BG. Analysis of arthropod bloodmeals using molecular genetic markers. Trends Parasitol. 2002;18:505–509. doi: 10.1016/s1471-4922(02)02364-4. [DOI] [PubMed] [Google Scholar]
- 19.Goodman DS, Orelus JN, Roberts JM, Lammie PJ, Streit TG. PCR and mosquito dissection as tools to monitor filarial infection levels following mass treatment. Filaria J. 2003;2:11. doi: 10.1186/1475-2883-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plichart C, Sechan Y, Davies N, Legrand AM. PCR and dissection as tools to monitor filarial infection of Aedes polynesiensis mosquitoes in French Polynesia. Filaria J. 2006;5:2. doi: 10.1186/1475-2883-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams SA, Laney SJ, Bierwert LA, Saunders LJ, Boakye DA, Fischer P, Goodman D, Helmy H, Hoti SL, Vasuki V, Lammie PJ, Plichart C, Ramzy RM, Ottesen EA. Development and standardization of a rapid, PCR-based method for the detection of Wuchereria bancrofti in mosquitoes, for xenomonitoring the human prevalence of bancroftian filariasis. Ann Trop Med Parasitol. 2002;96(Suppl 2):S41–S46. doi: 10.1179/000349802125002356. [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez-Perez MA, Katholi CR, Hassan HK, Unnasch TR. Large-scale entomologic assessment of Onchocerca volvulus transmission by poolscreen PCR in Mexico. Am J Trop Med Hyg. 2006;74:1026–1033. [PubMed] [Google Scholar]


