Abstract
At tight junctions (TJs), claudins with four transmembrane domains are incorporated into TJ strands. Junctional adhesion molecule (JAM), which belongs to the immunoglobulin superfamily, is also localized at TJs, but it remains unclear how JAM is integrated into TJs. Immunoreplica electron microscopy revealed that JAM showed an intimate spatial relationship with TJ strands in epithelial cells. In L fibroblasts expressing exogenous JAM, JAM was concentrated at cell–cell adhesion sites, where there were no strand-like structures, but rather characteristic membrane domains free of intramembranous particles were detected. These domains were specifically labeled with anti-JAM polyclonal antibody, suggesting that JAM forms planar aggregates through their lateral self-association. Immunofluorescence microscopy and in vitro binding assays revealed that ZO-1 directly binds to the COOH termini of claudins and JAM at its PDZ1 and PDZ3 domains, respectively. Furthermore, another PDZ-containing polarity-related protein, PAR-3, was directly bound to the COOH terminus of JAM, but not to that of claudins. These findings led to a molecular architectural model for TJs: small aggregates of JAM are tethered to claudin-based strands through ZO-1, and these JAM aggregates recruit PAR-3 to TJs. We also discuss the importance of this model from the perspective of the general molecular mechanisms behind the recruitment of PAR proteins to plasma membranes.
Keywords: JAM; PAR-3; claudin; ZO-1; tight junction
Introduction
Tight junctions (TJs)* are located at the most apical part of lateral membranes of epithelial and endothelial cells and are implicated in multiple functions such as the barrier, fence, and signaling functions (Anderson and van Itallie, 1995; Tsukita et al., 1999, 2001). On freeze fracture electron microscopy, TJs appear as a set of continuous anastomosing intramembranous strands or fibrils (TJ strands) within plasma membranes (Staehelin, 1974). TJ strands have been thought to represent units of integral membrane proteins polymerized linearly within lipid bilayers, but until recently, such proteins have not been identified.
Occludin and claudins (claudin-1–24) are now known as constituents of TJ strands (Furuse et al., 1993, 1998a). Both occludin and claudins bear four transmembrane domains, but did not show any sequence similarity with each other. When claudins were overexpressed in mouse L fibroblasts, claudin molecules were polymerized within plasma membranes to reconstitute TJ strands (Furuse et al., 1998b). Another type of integral membrane protein, the junctional adhesion molecule (JAM), was also reported to be localized at TJs (Martin-Padura et al., 1998). JAM belongs to the immunoglobulin superfamily: it has a single transmembrane domain, and its extracellular portion is thought to be folded into two immunoglobulin-like domains. JAM was shown to be involved in cell–cell adhesion/junctional assembly of epithelial/endothelial cells (Martin-Padura et al., 1998; Bazzoni et al., 2000a; Liu et al., 2000; Palmeri et al., 2000), as well as in the extravasation of monocytes through endothelial cells (Martin-Padura et al., 1998), but our knowledge on its localization and function at TJs is still fragmentary.
Most claudin species, as well as JAM, end in Val at their COOH termini, suggesting that these COOH termini directly bind to PDZ domains. Indeed, three related PDZ-containing proteins, ZO-1, ZO-2, and ZO-3, are known to be concentrated at TJs. ZO-1 (∼220 kD) was first identified as an antigen for a mAb raised against the junction-enriched fraction from the liver (Stevenson et al., 1986). Then, ZO-2 (∼160 kD) was identified as a protein that was coimmunoprecipitated with ZO-1 (Gumbiner et al., 1991). A phosphorylated 130-kD protein was also found in the ZO-1 immunoprecipitate (Balda et al., 1993) and is now called ZO-3. Cloning and sequencing cDNAs encoding these molecules showed that all have three PDZ domains (PDZ1–3), one SH3 domain, and one GUK domain, in this order from their NH2 termini (Itoh et al., 1993; Willott et al., 1993; Jesaitis and Goodenough, 1994; Haskins et al., 1998). Among these three PDZ domains, PDZ1 domain was recently shown to bind directly to the COOH termini of claudins (Itoh et al., 1999).
Recently, another intriguing PDZ-containing protein, a mammalian homologue of PAR-3, was reported to be concentrated at TJs (Izumi et al., 1998). PAR-3, which contains three PDZ domains, was initially identified in C. elegans as a product of one of six partitioning-defective genes (par-1–6) that are essential for the first asymmetric divisions of early embryos (Kemphues et al., 1988; Guo and Kemphues, 1996). A mammalian homologue of PAR-3 was identified as a binding partner for atypical PKCs (ASIP) in epithelial cells (Izumi et al., 1998). As TJs are involved in the establishment of epithelial polarity, the molecular mechanism behind the recruitment of PAR-3 to TJs, as well as its physiological function at TJs, now attracts increasing interest.
Thus, for a better understanding of the molecular architecture of TJs, the most pressing questions concern the molecular mechanisms underlying the recruitment of JAM and PAR-3 (and their binding proteins) to TJs. In this study, we examined the detailed localization of JAM at TJs and the interactions between JAM and underlying PDZ-containing proteins including PAR-3. The results obtained led us to propose a new molecular architectural model for TJs that could explain how JAM and PAR-3 are recruited to TJs.
Results and discussion
Intimate spatial relationship between JAM and TJ strands in epithelial cells
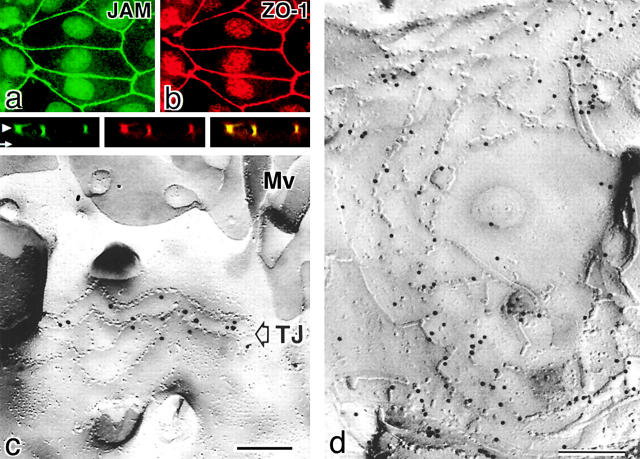
To date, JAM has been shown to be concentrated at TJs in epithelial cells at both the immunofluorescence and immunoelectron microscopic levels (Martin-Padura et al., 1998), but the spatial relationship between JAM and TJ strands remains unclear. As shown in Fig. 1 , a and b, polyclonal antibodies (pAbs) specific for the cytoplasmic domain of human JAM exclusively stained the ZO-1–positive intercellular junctional regions of cultured MDCK cells (Martin-Padura et al., 1998). Using these pAbs, we performed immunoreplica analyses developed by Fujimoto (1995): when freeze fracture replicas obtained from MDCK cells were incubated with anti-JAM pAb, the TJ region was specifically labeled (Fig. 1 c), and most of the immunogold particles were distributed on and around TJ strands, showing an intimate spatial relationship between JAM and TJ strands (Fig. 1 d). Taking the spatial resolution of this labeling technique into consideration (Fujimoto, 1995), however, it remains difficult to distinguish between the following two possibilities: (a) JAM is directly incorporated into TJ strands or (b) JAM laterally associates with TJ strands.
Figure 1.
JAM in TJs of epithelial cells. (a and b) Double immunofluorescence staining of cultured MDCK cells with anti-JAM pAb (C4) (a) and anti–ZO-1 mAb (b). JAM was precisely colocalized with ZO-1 at cell–cell adhesion sites. (Bottom) Show vertical-sectional views and their merged view generated from confocal images. Arrowhead, apical level; arrow, basal level. (c and d) Freeze fracture replicas obtained from cultured MDCK cells were immunolabeled with anti-JAM pAb (C-tail). The TJ region was specifically labeled with immunogold particles (c), and most immunogold particles showed an intimate spatial relationship with TJ strands (d). (c and d) The TJ region on the E-face and P-face, respectively, was labeled. The E-face–associated labeling is not easily explained according to Fujimoto's original paper (1995), but similar labeling was observed with anticlaudin and antioccludin pAbs, suggesting that this type of labeling was characteristic to TJ constituents, i.e., that JAM molecules are tightly associated with TJ strands. Mv, microvilli. Bars, 100 nm.
No strand formation in L transfectants expressing JAM
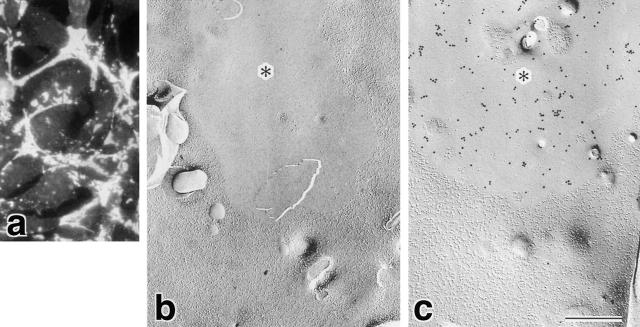
As previously shown, when claudin-1 was expressed in L fibroblasts, at cell–cell adhesion sites of these transfectants (C1L cells) claudin-1 molecules were polymerized into TJ strand-like structures (Furuse et al., 1998b). To evaluate the above two possibilities, it was examined whether JAM has an ability to reconstitute TJ strand-like structures when expressed in L fibroblasts. We then transfected L fibroblasts with JAM cDNA and immunofluorescently stained stable transfectants (JL cells) with anti-JAM pAb. Similar to CHO cells transfected with JAM (Martin-Padura et al., 1998; Bazzoni et al., 2000b), in JL cells, JAM was concentrated at cell–cell borders not as lines, but as planes (Fig. 2 a). When these JL cells were intensively examined by freeze fracture replica electron microscopy, no strand-like structures were observed. Instead, the very characteristic membrane domains free of intramembranous particles were frequently detected on the P-face of JL cells (Fig. 2 b, *). As integral membrane proteins with a single membrane-spanning domain such as JAM is not visualized as intramembranous particles by freeze fracture replica electron microscopy, we supposed that the particle-free fracture domain would be occupied by laterally aggregated JAM molecules. Supporting this notion, when these freeze fracture replicas were labeled with anti-JAM pAb, the particle-free fracture domain was exclusively labeled with gold particles (Fig. 2 c).
Figure 2.
Lateral aggregation of JAM in L transfectants. (a) Immunofluorescence microscopy of JL cells with anti-JAM pAb (C4). JAM was concentrated at cell–cell adhesion sites as planes. (b) Freeze fracture replica image of cell–cell adhesion sites of JL cells. Characteristic intramembranous particle-free domains (*) were frequently observed. (c) The particle-free domains of JL cells were specifically labeled with pAb-specific for the cytoplasmic domain of JAM (anti-JAM pAb, C-tail). Bar, 200 nm.
JAM was reported to be expressed at high levels in cells lacking TJs such as platelets (Williams et al., 1999), suggesting that JAM is not directly involved in the formation of TJ strands. Therefore, at TJs in epithelial cells, JAM molecules may aggregate laterally, and these small aggregates are tightly and laterally associated with TJ strands (i.e., claudin-based linear polymers). Consistent with this conclusion, recent biochemical analyses reported that the soluble form of the extracellular domain of JAM forms dimers in solution (Bazzoni et al., 2000a). The next question then is how these two distinct types of polymers are linked at TJs in molecular terms.
Direct association of JAM with PDZ3 domain of ZO-1
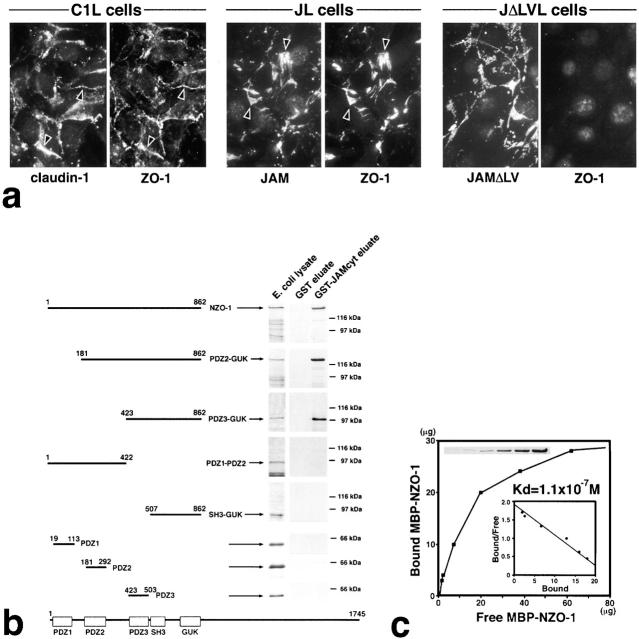
Recently, COOH termini of claudins were shown to directly bind to PDZ1 domain of ZO-1 (and also ZO-2/ZO-3) (Itoh et al., 1999). Considering that ZO-1 is a multidomain protein, it is tempting to speculate that JAM is associated with claudins through ZO-1. First, we examined the ability of JAM to recruit endogenous ZO-1 in L transfectants, JL cells (Fig. 3 a). As previously reported (Itoh et al., 1999), in C1L cells expressing claudin-1, endogenous ZO-1 was recruited to the claudin-based cell–cell adhesion sites. Similarly, in JL cells, endogenous ZO-1 was concentrated precisely at JAM-based cell–cell adhesion sites. In contrast, when the JAM mutant lacking its COOH-terminal −LV (JAMΔLV) was expressed in L fibroblasts (JΔLVL cells), these JAM mutants were concentrated at cell–cell borders, but ZO-1 was not recruited. These findings are consistent with a recent report on the association of JAM with ZO-1 (Bazzoni et al., 2000b) and suggest that some of the PDZ domains of ZO-1 interact with the COOH-terminal end of JAM.
Figure 3.
Interaction between JAM and ZO-1. (a) Recruitment of endogenous ZO-1 to cell–cell adhesion sites in L transfectants. C1L cells, JL cells, or JAM lacking its COOH-terminal –LV (JΔLVL cells) were double stained. Claudin-1, JAM, and JAMΔLV were all concentrated at cell–cell adhesion sites. Claudin-1 and JAM, but not JAMΔLV, recruited ZO-1 to cell–cell contact sites (arrowheads). (b) Eight distinct portions of ZO-1 were produced as recombinant fusion proteins with maltose-binding protein (MBP) in E. coli. Their crude lysates containing recombinant proteins (E. coli lysate) were mixed with beads conjugated with GST or GST fusion protein with the cytoplasmic domain of JAM (GST-JAMcyt). Bound proteins were then eluted from GST-conjugated beads (GST eluate) or GST-JAMcyt–conjugated beads (GST-JAMcyt eluate), and each eluate was subjected to SDS-PAGE followed by Coomassie brilliant blue staining. Among eight types of MBP fusion proteins, only MBP–NZO-1, MBP-PDZ2-GUK, and MBP-PDZ3-GUK were bound to GST-JAMcyt. (c) Quantitative analysis of the binding between MBP-NZO-1 and GST-JAMcyt. Glutathione–Sepharose bead slurry containing GST-JAMcyt was incubated with E. coli lysate containing various amounts of MBP–NZO-1. The amounts of MBP–NZO-1 in the E. coli lysate and each eluate (inset) were estimated by comparing the Coomassie Brilliant blue staining intensity of bands. The binding was saturable, and Scatchard analysis (inset) indicated that the K d value was 1.1 × 10−7 M.
We performed in vitro binding assays to examine whether JAM binds to ZO-1 directly and, if so, which PDZ domain of ZO-1 is responsible for this binding. First, we produced maltose-binding protein (MBP) fusion protein with NH2-terminal half of ZO-1 in Escherichia coli (MBP-NZO-1 containing three PDZs, SH3, and GUK), and their crude lysate containing recombinant MBP-NZO-1 was mixed with beads conjugated with the GST fusion protein with the cytoplasmic domain of JAM (GST-JAMcyt). Bound proteins were then eluted from beads, and each eluate was subjected to SDS-PAGE followed by Coomassie brilliant blue staining. As shown in Fig. 3 b, MBP-NZO-1 was specifically associated with GST-JAMcyt, suggesting the direct interaction of PDZ domains of ZO-1 with JAM. However, unexpectedly, recombinant MBP-PDZ1, MBP-PDZ2, and MBP-PDZ3 produced in E. coli showed no binding affinity to GST-JAMcyt, probably due to some conformational problems in these recombinant proteins. We performed further binding analyses with various deletion mutants of MBP-NZO-1 and found that the PDZ1 domain, which specifically binds to claudins (Itoh et al., 1999), is not required for ZO-1-JAM binding and that at least the PDZ3 domain is required (Fig. 3 b). We then estimated the dissociation constant between GST-JAMcyt and MBP-NZO-1 as previously described (Itoh et al., 1999). The binding was saturable, and Scatchard analysis revealed a single class of affinity-binding sites with a Kd of 1.1 × 10−7 M (Fig. 3 c).
Taking into consideration that the COOH termini of claudins bind to PDZ1 of ZO-1 at a Kd of 1.3 × 10−7 M (Itoh et al., 1999), it can be speculated that ZO-1 tethers JAM to claudin-based strands at TJs in epithelial cells. As PDZ1 domains of ZO-2 and ZO-3 show affinity to claudins (Itoh et al., 1999), ZO-2 and ZO-3 would also be involved in the recruitment of JAM to TJ strands.
Recruitment of PAR-3 to JAM, not to claudin-1
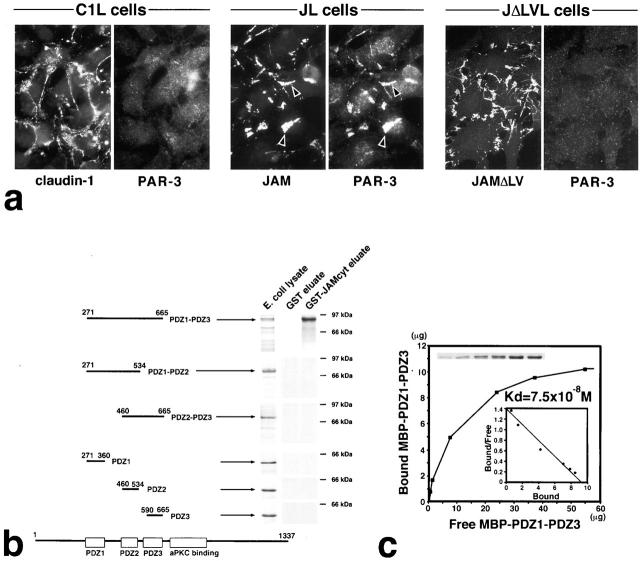
During the course of this study, we noticed that, in JL cells, endogenous PAR-3 was also recruited to JAM-based cell–cell adhesion sites: when JL cells were double stained with anti-JAM mAb and PAR-3/ASIP pAb, JAM and PAR-3 showed precise colocalization at cell–cell adhesion sites (Fig. 4 a). JAMΔLV did not recruit PAR-3 to cell–cell adhesion sites (Fig. 4 a). These findings suggested that, at least in the L fibroblast transfection system, JAM recruits PAR-3 to the plasma membrane at cell–cell adhesion sites through the interaction between the COOH terminus of JAM and PDZ domains of PAR-3. In marked contrast, in C1L cells endogenous PAR-3 was not recruited to the claudin-1–based cell–cell adhesion sites (Fig. 4 a).
Figure 4.
Interaction between JAM and PAR-3. (a) Recruitment of endogenous PAR-3 to cell–cell adhesion sites in L transfectants. C1L cells, JL cells, or JΔLVL cells were double stained. Claudin-1, JAM, and JAMΔLV were all concentrated at cell–cell adhesion sites. JAM, but not claudin-1–JAMΔLV, recruited PAR-3 to cell–cell contact sites (arrowheads). (b) Six distinct portions of PAR-3 were produced as recombinant fusion proteins with MBP in E. coli, and then the same in vitro binding analysis as described in the legend to Fig. 3 b was performed. Among six types of MBP fusion proteins, only MBP–PDZ1-PDZ3 was bound to GST-JAMcyt. (c) Quantitative analysis of the binding between MBP–PDZ1-PDZ3 of PAR-3 and GST-JAMcyt. K d value was determined to be 7.5 × 10−8 M.
In vitro binding analyses were performed between PAR-3 and GST-JAMcyt. As the recombinant MBP fusion protein with full-length PAR-3 was not produced in E. coli in sufficient amounts for in vitro binding, we first produced and used the deletion mutant of MBP–PAR-3 lacking most of the non-PDZ region (MBP-PDZ1/2/3–PAR-3; PDZ1-PDZ3 in Fig. 4 b) with the expectation that PDZ domains are involved in the PAR-3–JAM interaction. As shown in Fig. 4, b and c, MBP-PDZ1/2/3–PAR-3 specifically bound to JAM with a Kd of 7.5 × 10−8 M. We attempted to determine which PDZ domain of PAR-3 is responsible for JAM binding, but again the results obtained were complex (Fig. 4 b). The recombinant PDZ1, PDZ2, or PDZ3 domain of PAR-3 produced in E. coli showed no binding affinity to GST-JAMcyt. Furthermore, when the PDZ1 or PDZ3 domain was deleted from MBP-PDZ1/2/3–PAR-3, its binding ability to JAM was abolished. Therefore, the PDZ domain responsible for JAM binding has not been assigned in PAR-3 by in vitro binding assay, but it is safe to say that PDZ domains of PAR-3 directly bind to the COOH terminus of JAM.
A molecular architectural model of TJs
We were then led to the speculative model of the molecular architecture of TJs shown in Fig. 5 a, which could explain the recruitment of JAM as well as PAR-3 to TJs: it is expected that the cytoplasmic surface of individual TJ strands appears like a toothbrush consisting of densely packed numerous short COOH-terminal cytoplasmic tails of claudins. The cytoplasmic surface of strands would then strongly attract the PDZ1 domain of ZO-1 (and also ZO-2/ZO-3). JAM would be recruited and tethered to TJ strands through the direct binding of its COOH terminus to the PDZ3 domain of ZO-1. JAM molecules laterally aggregate to form oligomers, which would allow the recruitment of additional JAM molecules around TJ strands. Since these JAM molecules would be free of ZO-1, they could recruit PAR-3 and then its binding proteins, such as atypical PKC, PAR-6, and Cdc42, to TJs (Joberty et al., 2000; Lin et al., 2000; Qiu et al., 2000; Suzuki et al., 2001).
Figure 5.
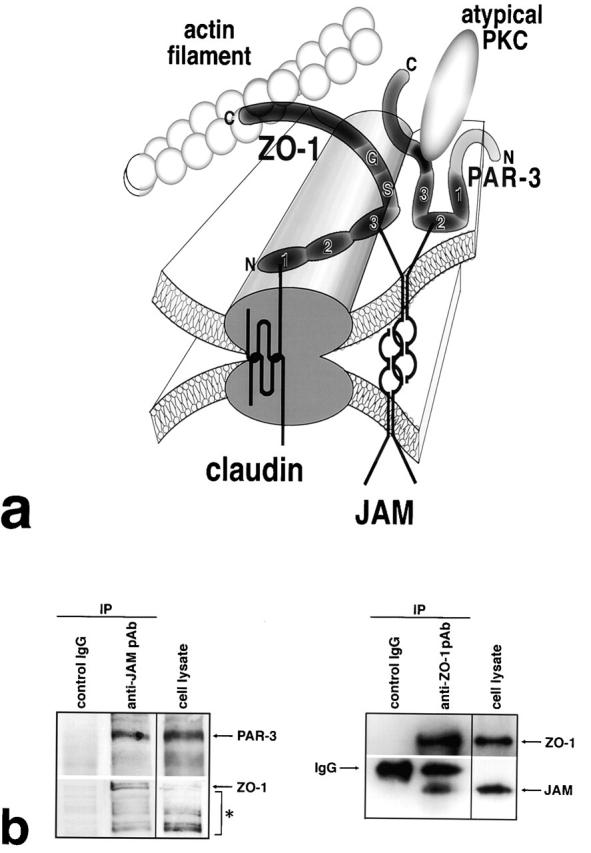
Molecular interactions within TJs in epithelial cells. (a) A model for the molecular architecture of TJs. See details in the text. COOH-terminal domain of ZO-1 is associated with actin filaments. ZO-2 and ZO-3 may also function as cross-linkers like ZO-1 (Itoh et al., 1999), and occludin in TJ strands may also be cross-linked to JAM by ZO-1, although these were not depicted in this model. (b) Immunoprecipitation. As mAb specific for the extracellular domain of JAM recognizes human JAM, but not mouse JAM, human T84 cells were used. (Left) JAM was immunoprecipitated from T84 cell lysate with anti-JAM mAb. Immunoblotting with anti–PAR-3/ASIP pAb or anti–ZO-1 mAb detected PAR-3 or ZO-1, respectively, in the JAM immunoprecipitate. *Degradation products of ZO-1. (Right) ZO-1 was immunoprecipitated with anti–ZO-1 pAb. Immunoblotting with anti-JAM pAb detected JAM in the ZO-1 immunoprecipitate. PAR-3 was undetectable under the condition used in this experiment, but this would be reasonable since PAR-3 is not directly associated with ZO-1.
Presently, this model is still speculative; it is technically difficult to experimentally demonstrate the molecular interactions depicted in this model in epithelial cells, since all these molecules are organized into very insoluble structures in TJs. Exceptionally, the association of PAR-3 and ZO-1 with JAM was detected in epithelial cells by immunoprecipitation experiments: cultured epithelial cells (T84) were solubilized, and JAM was immunoprecipitated with anti-JAM mAb specific for the extracellular portion of JAM. As shown in Fig. 5 b, immunoblotting with anti–PAR-3/ASIP pAb or anti–ZO-1 pAb clearly detected PAR-3 or ZO-1 in the JAM immunoprecipitates, respectively. The association between JAM and ZO-1 was also confirmed by the immunoprecipitation with anti–ZO-1 pAb (Fig. 5 b).
In C. elegans germline cells, most PAR proteins were enriched at the cell periphery and localized to one or the other pole of cells undergoing asymmetric cell divisions (Guo and Kemphues, 1996; Kemphues, 2000). Detailed analyses of PAR mutants revealed that the asymmetric distribution of PAR proteins is important for their function as determinants of cell polarity in these cells. Therefore, there is a search for membrane proteins that recruit some PAR proteins to the plasma membrane and allow their asymmetric distribution. In this sense, JAM is the first protein shown to recruit PAR proteins to certain specified membrane domains. We showed here that JAM recruits PAR-3 to the cell–cell adhesion sites in L transfectants and TJs in epithelial cells. This finding provides an important clue as to how PAR signaling determines cell polarity in general.
Materials and methods
Antibodies and cells
Rat anti–mouse claudin-1 pAb, mouse anti–mouse ZO-1 mAb, rabbit anti–human JAM cytoplasmic domain pAb (C-tail)/mAb (2H11), and anti–human JAM extracellular domain mAb (3D8) were raised and characterized previously (Itoh et al., 1999; Ozaki et al., 1999). Another anti–human JAM cytoplasmic domain pAb (C4) was raised in rabbits. Rabbit anti–mouse PAR-3/ASIP pAb (Izumi et al., 1998) was a gift from Dr. S. Ohno (Yokohama City University, Yokohama, Japan). Anti–ZO-1 pAb was purchased from Zymed Laboratories.
cDNA transfection
The cDNA fragments encoding full-length human JAM and a JAM mutant lacking its COOH-terminal residues –LV were produced by PCR using human JAM cDNA (Ozaki et al., 1999) in the pcDNA vector as a template, and these fragments were subcloned into the mammalian expression vector pME18S. Mouse L cells were cotransfected with 2 μg of expression vector and 0.1 μg of pGKpuro and selected basically as described previously (Itoh et al., 1999).
Recombinant proteins and in vitro binding assay
The cDNA fragment encoding the cytoplasmic domain of JAM was produced by PCR, and the fragment was subcloned into the pGEX vector (Amersham Pharmacia Biotech). For production of maltose-binding protein fusion proteins with various ZO-1 mutants or with various PAR-3 mutants, the cDNAs were amplified by PCR and subcloned into the pMAL vector (New England Biolabs, Inc.) (Figs. 3 b and 4 b). These recombinant proteins were expressed in E. coli.
In vitro binding assay was performed basically as described previously (Itoh et al., 1999).
Immunofluorescence microscopy and immunoreplica electron microscopy
For immunofluorescence microscopy, cells plated on glass coverslips were rinsed in PBS, fixed with 1% formaldehyde in PBS for 15 min, and processed as described previously (Itoh et al., 1999). For immunoelectron microscopy for examining freeze fracture replicas, MDCK cells and L transfectants were fixed with 1% paraformaldehyde in 0.1 M phosphate buffer (pH 7.3) for 5 min and then processed as described by Fujimoto (1995).
Immunoprecipitation
T84 cells cultured on 9-cm dishes were washed twice with PBS and lysed in 1-ml extraction buffer (0.1% nonidet P-40, 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 10% glycerol, 5 mM MgCl2, 5 mM CaCl2) followed by sonication (5 times for 15 s). Cell lysates were clarified by centrifugation at 15,000 rpm for 20 min and incubated with 50 μl of protein G–Sepharose bead slurry (Zymed Laboratories) coupled with anti-JAM mAb (3D8), anti–ZO-1 pAb, or respective control IgG for 3 h. After washing five times with the extraction buffer, immunoprecipitates were eluted from beads with the SDS-PAGE sample buffer.
Acknowledgments
We thank Dr. S. Ohno for donating anti-ASIP pAb. We also thank all the members of our laboratory (Department of Cell Biology, Faculty of Medicine, Kyoto University) for helpful discussions. Our thanks are also due to Ms. K. Kamikubo-Tsuchihashi for excellent technical assistance.
This study was supported in part by a Grant-in-Aid for Cancer Research and a Grant-in-Aid for Scientific Research (A) from the Ministry of Education, Science and Culture of Japan to S. Tsukita, and by JSPS Research for the Future Program to M. Furuse.
Footnotes
Abbreviations used in this paper: C1L cell, L transfectant expressing claudin-1; JAM, junctional adhesion molecule; JL cell, L transfectant expressing JAM; MBP, maltose-binding protein; pAb, polyclonal antibody; TJ, tight junction.
References
- Anderson, J.M., and C.M. van Itallie. 1995. Tight junctions and the molecular basis for regulation of paracellular permeability. Am. J. Physiol. 269:G467–G475. [DOI] [PubMed] [Google Scholar]
- Balda, M.S., L. González-Mariscal, K. Matter, M. Cereijido, and J.M. Anderson. 1993. Assembly of the tight junction: the role of diacylglycerol. J. Cell Biol. 123:293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzoni, G., O.M. Martinez-Estrada, F. Mueller, P. Nelboeck, G. Schmid, T. Bartfai, E. Dejana, and M. Brockhaus. 2000. a. Homophilic interaction of junctional adhesion molecule. J. Biol. Chem. 275:30970–30976. [DOI] [PubMed] [Google Scholar]
- Bazzoni, G., O.M. Martinez-Estrada, F. Orsenigo, M. Cordenonsi, S. Citi, and E. Dejana. 2000. b. Interaction of junctional adhesion molecule with the tight junction components ZO-1, cingulin, and occludin. J. Biol. Chem. 275:20520–20526. [DOI] [PubMed] [Google Scholar]
- Fujimoto, K. 1995. Freeze-fracture replica electron microscopy combined with SDS digestion for cytochemical labeling of integral membrane proteins. Application to the immunogold labeling of intercellular junctional complexes. J. Cell Sci. 108:3443–3449. [DOI] [PubMed] [Google Scholar]
- Furuse, M., T. Hirase, M. Itoh, A. Nagafuchi, S. Yonemura, S. Tsukita, and S. Tsukita. 1993. Occludin: a novel integral membrane protein localizing at tight junctions. J. Cell Biol. 123:1777–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse, M., K. Fujita, T. Hiiragi, K. Fujimoto, and S. Tsukita. 1998. a. Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J. Cell Biol. 141:1539–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse, M., H. Sasaki, K. Fujimoto, and S. Tsukita. 1998. b. A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J. Cell Biol. 143:391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner, B., T. Lowenkopf, and D. Apatira. 1991. Identification of a 160-kDa polypeptide that binds to the tight junction protein ZO-1. Proc. Natl. Acad. Sci. USA. 88:3460–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, S., and K.J. Kemphues. 1996. Molecular genetics of asymmetric cleavage in the early Caenorhabditis elegans embryo. Curr. Opin. Genet. Dev. 6:408–415. [DOI] [PubMed] [Google Scholar]
- Haskins, J., L. Gu, E.S. Wittchen, J. Hibbard, and B.R. Stevenson. 1998. ZO-3, a novel member of the MAGUK protein family found at the tight junction, interacts with ZO-1 and occludin. J. Cell Biol. 141:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, M., A. Nagafuchi, S. Yonemura, T. Kitani-Yasuda, S. Tsukita, and S. Tsukita. 1993. The 220-kD protein colocalizing with cadherins in non-epithelial cells is identical to ZO-1, a tight junction-associated protein in epithelial cells: cDNA cloning and immunoelectron microscopy. J. Cell Biol. 121:491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, M., M. Furuse, K. Morita, K. Kubota, M. Saitou, and S. Tsukita. 1999. Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins. J. Cell Biol. 147:1351–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi, Y., T. Hirose, Y. Tamai, S. Hirai, Y. Nagashima, T. Fujimoto, Y. Tabuse, K.J. Kemphues, and S. Ohno. 1998. An atypical PKC directly associates and colocalizes at the epithelial tight junction with ASIP, a mammalian homologue of Caenorhabditis elegans polarity protein PAR-3. J. Cell Biol. 143:95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesaitis, L.A., and D.A. Goodenough. 1994. Molecular characterization and tissue distribution of ZO-2, a tight junction protein homologous to ZO-1 and the Drosophila discs-large tumor suppresser protein. J. Cell Biol. 124:949–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joberty, G., C. Petersen, L. Gao, and I.G. Macara. 2000. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat. Cell Biol. 2:531–539. [DOI] [PubMed] [Google Scholar]
- Kemphues, K. 2000. PARsing embryonic polarity. Cell. 101:345–348. [DOI] [PubMed] [Google Scholar]
- Kemphues, K.J., J.R. Priess, D.G. Morton, and N. Cheng. 1988. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell. 52:311–320. [DOI] [PubMed] [Google Scholar]
- Lin, D., A.S. Edwards, J.P. Fawcett, G. Mbamalu, J.D. Scott, and T. Pawson. 2000. A mammalian PAR-3-PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nat. Cell Biol. 2:540–547. [DOI] [PubMed] [Google Scholar]
- Liu, Y., A. Nustrat, F.J. Schnell, T.A. Reaves, S. Walsh, M. Pochet, and C.A. Parkos. 2000. Human junction adhesion molecule regulates tight junction resealing in epithelia. J. Cell Sci. 113:2363–2374. [DOI] [PubMed] [Google Scholar]
- Martin-Padura, I., S. Lostaglio, M. Schneemann, L. Williams, M. Romano, P. Fruscella, C. Panzeri, A. Stoppacciaro, L. Ruco, A. Villa, D. Simmons, and E. Dejana. 1998. Junctional adhesion molecule, a novel member of the immunoglobulin superfamily that distributes at intercellular junctions and modulates monocyte transmigration. J. Cell Biol. 142:117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki, H., K. Ishii, H. Horiuchi, H. Arai, T. Kawamoto, K. Okawa, A. Iwamatsu, and T. Kita. 1999. Cutting edge: combined treatment of TNF-α and IFN-γ causes redistribution of junctional adhesion molecule in human endothelial cells. J. Immunol. 163:553–557. [PubMed] [Google Scholar]
- Palmeri, D., A. van Zante, C.C. Huang, S. Hemmerich, and S.D. Rosen. 2000. Vascular endothelial junction-associated molecule, a novel member of the immunoglobulin superfamily, is localized to intercellular boundaries of endothelial cells. J. Biol Chem. 275:19139–19145. [DOI] [PubMed] [Google Scholar]
- Qiu, R.G., A. Abo, and G.S. Martin. 2000. A human homolog of the C. elegans polarity determinant par-6 links rac and cdc42 to PKCzeta signaling and cell transformation. Curr. Biol. 10:697–707. [DOI] [PubMed] [Google Scholar]
- Staehelin, L.A. 1974. Structure and function of intercellular junctions. Int. Rev. Cytol. 39:191–283. [DOI] [PubMed] [Google Scholar]
- Stevenson, B.R., J.D. Siliciano, M.S. Mooseker, and D.A. Goodenough. 1986. Identification of ZO-1: a high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J. Cell Biol. 103:755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, A., T. Yamanaka, T. Hirose, N. Manabe, K. Mizuno, M. Shimizu, K. Akimoto, Y. Izumi, T. Ohnishi, and S. Ohno. 2001. Atypical protein kinase C is involved in the evolutionarily conserved par protein complex and plays a critical role in establishing epithelia-specific junctional structures. J. Cell Biol. 152:1183–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita, S., M. Itoh, and M. Furuse. 1999. Structural and signaling molecules come together at tight junctions. Curr. Opin. Cell Biol. 11:628–633. [DOI] [PubMed] [Google Scholar]
- Tsukita, S., M. Furuse, and M. Itoh. 2001. Multifunctonal strands in tight junctions. Nat. Rev. Mol. Cell Biol. 2:285–293. [DOI] [PubMed] [Google Scholar]
- Williams, L.A., I. Martin-Padura, E. Dejana, N. Hogg, and D.L. Simmons. 1999. Identification and characterization of human junctional adhesion molecule (JAM). Mol. Immunol. 36:1175–1188. [DOI] [PubMed] [Google Scholar]
- Willott, E., M.S. Balda, A.S. Fanning, B. Jameson, C. Van Itallie, and J.M. Anderson. 1993. The tight junction protein ZO-1 is homologous to the Drosophila discs-large tumor suppresser protein of septate junctions. Proc. Natl. Acad. Sci. USA. 90:7834–7831. [DOI] [PMC free article] [PubMed] [Google Scholar]