Abstract
Cdc34/Ubc3 is a ubiquitin-conjugating enzyme that functions in targeting proteins for proteasome-mediated degradation at the G1 to S cell cycle transition. Elevation of Cdc34 protein levels by microinjection of bacterially expressed Cdc34 into mammalian cells at prophase inhibited chromosome congression to the metaphase plate with many chromosomes remaining near the spindle poles. Chromosome condensation and nuclear envelope breakdown occurred normally, and chromosomes showed oscillatory movements along mitotic spindle microtubules. Most injected cells arrested in a prometaphase-like state. Kinetochores, even those of chromosomes that failed to congress, possessed the normal trilaminar plate ultrastructure. The elevation of Cdc34 protein levels in early mitosis selectively blocked centromere protein E (CENP-E), a mitotic kinesin, from associating with kinetochores. Other proteins, including two CENP-E–associated proteins, BubR1 and phospho-p42/p44 mitogen-activated protein kinase, and mitotic centromere-associated kinesin, cytoplasmic dynein, Cdc20, and Mad2, all exhibited normal localization to kinetochores. Proteasome inhibitors did not affect the prometaphase arrest induced by Cdc34 injection. These studies suggest that CENP-E targeting to kinetochores is regulated by ubiquitylation not involving proteasome-mediated degradation.
Keywords: centromere; cell cycle; kinesin; microtubules; proteasome
Introduction
The expanding list of proteins to which ubiquitin is ligated includes transcription factors, tumor suppressors, protooncogenes, signal transduction receptors, and proteins involved in development, apoptosis, and DNA repair (Jentsch, 1992; Hochstrasser, 1996; Hershko and Ciechanover, 1998). Ubiquitin is first activated by a ubiquitin-activating enzyme (E1), transferred to a ubiquitin-conjugating enzyme (E2), and then covalently attached to a target protein, which can occur directly or may require a ubiquitin-protein ligase (E3). Subsequent ubiquitin units are conjugated to the initial molecule, forming a multiubiquitin chain on the target, which is recognized by the 26S proteasome. The target is then degraded, releasing free ubiquitin to be recycled in the process (for review see Hershko and Ciechanover, 1998). Although proteasome-mediated degradation is the most thoroughly studied outcome of multiubiquitin addition to proteins, several examples of reversible ubiquitin conjugation as a posttranslational regulatory modification are known. These include the attachment ubiquitin to cell surface membrane receptors (for review see Hicke, 2001), histone H2B (Robzyk et al., 2000), the large ribosomal subunit L28 (Spence et al., 2000), and the transcription factor Met4 (Kaiser et al., 2000). In addition, the conjugation of ubiquitin-like proteins such as SUMO/SMT3 can serve as modifiers in protein localization (Matunis et al., 1998), centromere function (Jiang and Koltin, 1996), or disjunction of homologues in meiosis (Apionishev et al., 2001).
The E2 Cdc34 (Ubc3) was identified by Goebl et al. (1988) as a ubiquitin-conjugating enzyme in budding yeast. The human homologue of Cdc34 was discovered by its ability to suppress budding yeast mec1, a DNA damage checkpoint mutation, and human Cdc34 is able to complement a cdc34-2 temperature-sensitive strain (Plon et al., 1993). All E2 enzymes, including Cdc34, contain a ubiquitin-conjugating domain of ∼16 kD, which includes the essential cysteine residue for thiolester formation with ubiquitin. Mutation of the catalytic cysteine to alanine destroys any ability of Cdc34 to form a bond with ubiquitin (Sung et al., 1990; Banerjee et al., 1995).
In yeast, Xenopus, and mammals, Cdc34 participates in a ubiquitylation pathway that involves an E3 termed the Skp1-Cdc53/cullin–F-box protein (SCF)* complex (for review see Deshaies, 1999). In budding yeast, one target of Cdc34/SCF activity is Sic1p, an inhibitor of Clb/Cdc28 that must be degraded for entry into S phase (Schwob et al., 1994; Feldman et al., 1997). Cdc34 also participates in the G1/S transition of animal cells, targeting the mammalian cyclin-dependent kinase inhibitor p27 for proteolysis (Pagano et al., 1995; Carrano et al., 1999; Sutterluty et al., 1999) and the Xenopus homologue Xic1 (Yew and Kirschner, 1997).
Many other proteins are degraded in a Cdc34-dependent manner, including Far1 (Henchoz et al., 1997), CDC6 (Drury et al., 1997), Gcn4 (Kornitzer et al., 1994), Gic2 (Jaquenoud et al., 1998), G1 cyclins (Deshaies et al., 1995; Yaglom et al., 1995; Willems et al., 1996), and HO endonuclease (Kaplun et al., 2000) in budding yeast, and inducible cAMP early repressor (ICERIIγ), activating transcription factor 5 (Pati et al., 1999), transcription factors Myo D (Song et al., 1998) and E2F-1 (Marti et al., 1999), and the transcriptional regulator B-Myb (Charrasse et al., 2000) in mammals.
Evidence also links Cdc34 to the G2/M phase of the cell cycle. Cdc34 is involved in the degradation of the budding yeast Cdk inhibitory kinase Swe1 (Kaiser et al., 1998) and the Xenopus homologue Wee1 (Michael and Newport, 1998). Both act to inhibit entry into mitosis (Mueller et al., 1995; Murakami and Vande Woude, 1998). Previous studies suggest that Cdc34 also plays an important role in the function of the budding yeast kinetochore complex called CBF3. One component of the complex, Ctf13p, is degraded through the Cdc34 pathway (Kaplan et al., 1997). In addition, overexpression of Cdc34 suppresses the growth defect in one mutant allele of another component called Ndc10p (Yoon and Carbon, 1995). In mammalian cells, Cdc34 was reported to associate with the mitotic spindle in anaphase, suggesting that it may play a role in events of late mitosis (Reymond et al., 2000).
In higher eukaryotes, the mitotic spindle microtubules attach to the kinetochores after nuclear envelope breakdown, and each chromosome moves individually to align at the metaphase plate. The mechanism regulating this alignment is unknown. We found that microinjection of recombinant human Cdc34 into cells inhibits chromosome movement to the metaphase plate (Bastians et al., 1999). Here, we examine this effect in more detail in rat kangaroo Ptk1 and porcine LLC-Pk cells. Microinjection of wild-type Cdc34 but not an inactive Cdc34 mutant into mammalian cells in early mitosis caused an arrest at prometaphase. Typical bipolar spindles formed in arrested cells. The ultrastructure of kinetochores and attachment of microtubules to kinetochores appeared normal. However, localization of the kinesin motor, centromere protein E (CENP-E), to mitotic kinetochores was inhibited in cells injected with wild-type Cdc34. The localization of other kinetochore proteins, including other motor proteins, was unaffected. Our results indicate that overexpression of Cdc34 specifically blocks CENP-E association with kinetochores and disrupts events of early chromosome movement in mitosis.
Results
Microinjection of wild-type Cdc34 protein arrests Ptk1 cells in prometaphase
In a previous study, we found unexpectedly that microinjection of Cdc34 into mammalian cells caused inhibition or delay of chromosome alignment at the metaphase plate (Bastians et al., 1999). Although initially reported to be a consequence of injection with the cys93→ser93–leu97→ser97 mutant, resequencing of the constructs from which the bacterially expressed proteins were prepared revealed that these original results were obtained after injection of wild-type Cdc34. Subsequently, we reanalyzed the effect in mitosis and compared chromosome behavior in Ptk1 cells injected with wild-type Cdc34 or inactive mutant Cdc34 protein in which the catalytic cysteine was replaced with alanine, Cdc34CA. Budding yeast Cdc34 protein harboring this same mutation is devoid of any ubiquitin-conjugating activity (Banerjee et al., 1995).
Mitotic progression of injected cells is shown in Fig. 1 and Table I. Cells injected during prophase with buffer required on average ∼30 min to reach metaphase after chromosomes achieved bipolar attachment. Initiation of anaphase began ∼14 min after metaphase, and cytokinesis was completed ∼20 min later. Cells injected in the cytoplasm at prophase with the inactive Cdc34CA protein progressed through mitosis at rates similar to buffer-injected cells (Fig. 1 B). In contrast, cells injected in the cytoplasm during prophase with wild-type Cdc34 arrested at prometaphase (Fig. 1, C and D). The level of Cdc34 protein expression does not appear to vary with cell cycle (Goebl et al., 1988; Reymond et al., 2000). By quantifying immunoblots and comparing a known amount of recombinant Cdc34 with extracts prepared from a known number of cells, we estimate that an individual Ptk1 cell contains ∼0.2 picograms of Cdc34 protein. Thus, the minimum amount of injected Cdc34 necessary to observe effects on chromosome movement was approximately five times the endogenous level. In cells injected with Cdc34, the chromosomes attached to microtubules of the mitotic spindle and oriented toward the poles after nuclear envelope breakdown. In most cells observed after injection of Cdc34, some chromosomes showed typical prometaphase chromosome oscillations (Fig. 2) . However, other chromosomes showed little movement, and many never aligned at the metaphase plate during the observation period. A few cells that were injected with wild-type Cdc34 in the early stages of mitosis did successfully align their chromosomes at the metaphase plate and arrested at metaphase (Table I). 12 of the cells injected at prophase with Cdc34 (> 3 mg/mL) included in Table I were followed for 2–6 h beyond nuclear envelope breakdown. 11 remained arrested in a prometaphase-like state, and 1 arrested at metaphase. Injection during late prometaphase usually delayed the onset of anaphase (Table I and Fig. 1 F). Injection of Cdc34 protein after anaphase onset did not impair completion of mitosis or cytokinesis (Table I). The prometaphase arrest was also observed when LLC-Pk cells in prophase were injected with Cdc34 (unpublished data).
Figure 1.
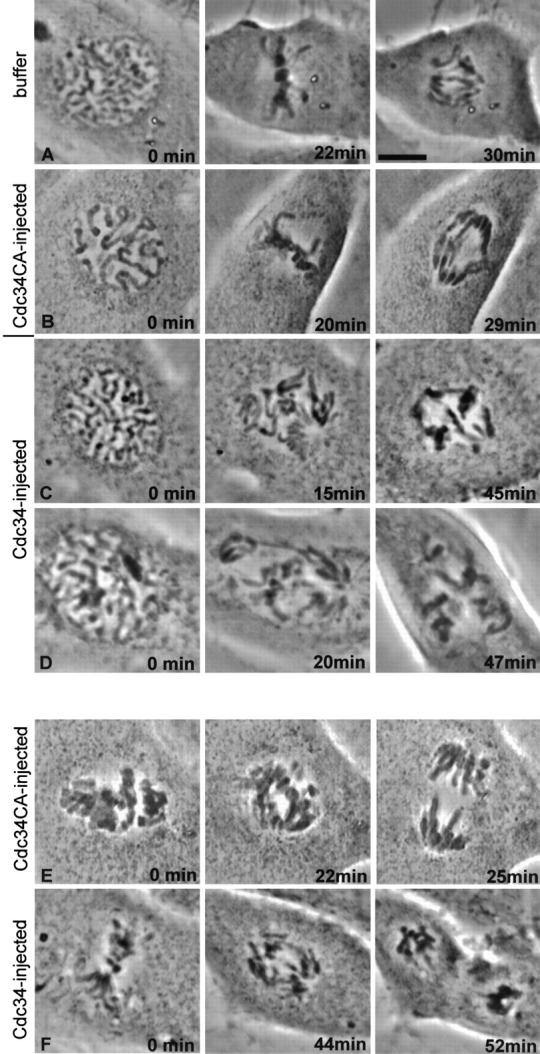
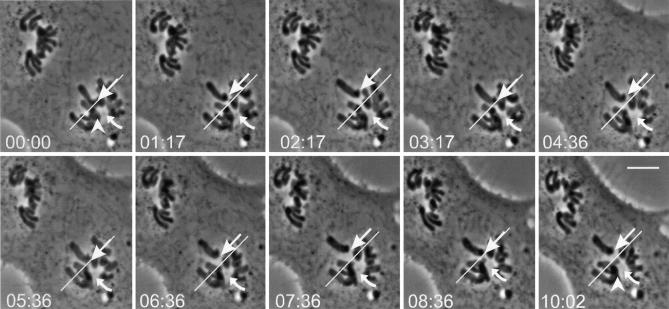
Injection of Cdc34 protein into prophase cells causes prometaphase arrest, and injection into prometaphase cells causes a delay at metaphase. Phase–contrast images of injected Ptk1 cells. (A) Buffer was injected into the cytoplasm of this cell during prophase. The nuclear envelope broke down within 2 min of injection, and chromosomes aligned ∼20 min later. The cell spent ∼8 min at metaphase and is shown ∼2 min after anaphase onset. (B) Recombinant Cdc34CA (a catalytically inactive form of Cdc34 protein) was injected into the cytoplasm of this cell during prophase. Progression through mitosis occurred with timing similar to the cell in A. (C and D) Injection of wild-type Cdc34 into cells at prophase. Both cells were injected in the cytoplasm just before capture of the first images. Nuclear envelope breakdown occurred normally in each cell. Many of the chromosomes of cells shown in B and C remained monooriented and never aligned at the metaphase plate during the 1-h observation period. (E) The prometaphase cell was injected with Cdc34CA protein immediately before the 0-min image was acquired. Chromosomes aligned, and the cell spent ∼10 min at metaphase then initiated anaphase. (F) The prometaphase cell was injected immediately before the first image was taken. After ∼40 min at metaphase, anaphase began. The cell is shown ∼3 min after anaphase onset. Bar, 5 μm.
Table I. Progression of microinfected cells through mitosis.
| Injection | Cell phase at injection |
Number of cells injected |
Number of cells arrested at prometaphase |
Number of cells arrested at metaphase |
Number of cells that completed mitosis |
|---|---|---|---|---|---|
| Buffer | Prophase | 8 | 8 | ||
| Prometaphase | 2 | 2 | |||
| Cdc34CA (≥3 mg/mL) | Prophase | 11 | 11 | ||
| Prometaphase | 4 | 4 | |||
| Cdc34CA (≥3 mg/mL) | Prophase | 104 | 89 | 5 | 10 |
| Prometaphase | 24 | 18 | 5 | 1 | |
| Late prometaphase | 11 | 4 | 7* | ||
| Anaphase | 6 | 6 | |||
| Cdc34 (3 mg/mL) denatured | Prophase | 6 | 6 | ||
| Prometaphase | 3 | 3 | |||
| Cdc34 (1.5 mg/mL) | Prophase | 11 | 7 | 4 | |
| Cdc34 (3 mg/mL) with 10 μm MG132 or β-lactone |
Prophase | 15 | 15 |
These cells spent on average 32.5 min at metaphase, which is significantly different from uninjected cells (P < 0.05).
Figure 2.
Chromosome oscillation during Cdc34 induced prometaphase arrest. (A) Ptk1 cell injected with Cdc34 protein at prophase. The spindle poles were widely separated at the time of nuclear envelope breakdown, and all of the chromosomes assumed an initial monopolar orientation. A curved arrow indicates the lower pole. The straight white line is drawn an equal distance from the pole in all panels to serve as a position reference. The chromosome indicated by the white arrow begins at the white line and then moves repeatedly away from and toward the lower pole during the observation period. In contrast, the chromosomes indicated by the white arrowhead in the first and last images undergo very little motion relative to the spindle pole. Time is indicated in minutes and seconds. A Quicktime video of the time-lapse phase image referring to Fig. 2 (Video 1) and additional examples of chromosome behavior in cells injected with Cdc34 protein (videos 2 and 3) are available at http://www.jcb.org/content/vol154/issue4/707. Bar, 10 μm.
Normal prometaphase characteristics are observed in the presence of high levels of Cdc34
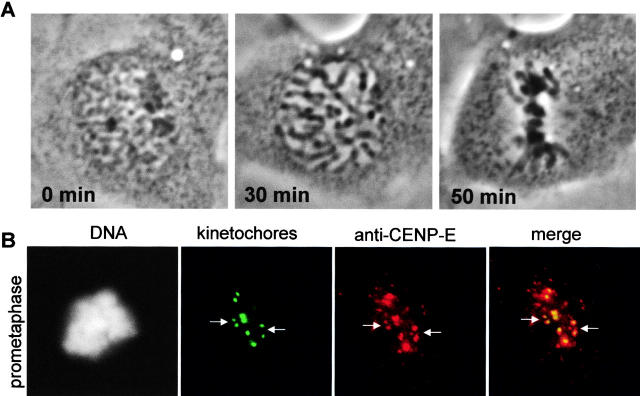
Spindles that formed in the Cdc34-injected cells were bipolar and contained astral, kinetochore, and nonkinetochore spindle microtubules (Fig. 3 A). This morphology resembled that of uninjected cells fixed in prometaphase. Ultrastructurally, kinetochores in cells injected with Cdc34 appeared identical to kinetochores of uninjected cells (Fig. 3 B). In injected cells, some chromosomes were found situated unusually close to the centriole at the spindle pole (Fig. 3 B, c, arrowhead). However, sister kinetochores (Fig. 3 B, c, white arrows) of chromosomes situated between the spindle poles (Fig. 3 B, c, black arrows) were widely separated, indicating the kinetochore area was under bipolar tension. We quantified the distances between sister kinetochores of chromosomes at the midplane and near the spindle poles in Cdc34-injected and uninjected cells (Table II). For both bioriented or monooriented chromosomes, we found no significant differences in the interkinetochore distances, comparing injected and uninjected cells. Nocodazole treatment reduced the interkinetochore distances of both injected and noninjected cells similarly (Table II.). These results suggest that the excess Cdc34 protein did not interfere with kinetochore attachment to microtubules or spindle assembly or cause structural alterations in the centromeric chromatin between sister kinetochores. Cells injected with Cdc34 protein were stably arrested in M phase as determined by several markers of M phase arrest including expression of phosphorylated histone H3 (Hendzel et al., 1997) and dispersed distribution of nuclear pore antigens (Snow et al., 1987) (Fig. S1 available at http://www.jcb.org/content/vol154/issue4/707).
Figure 3.
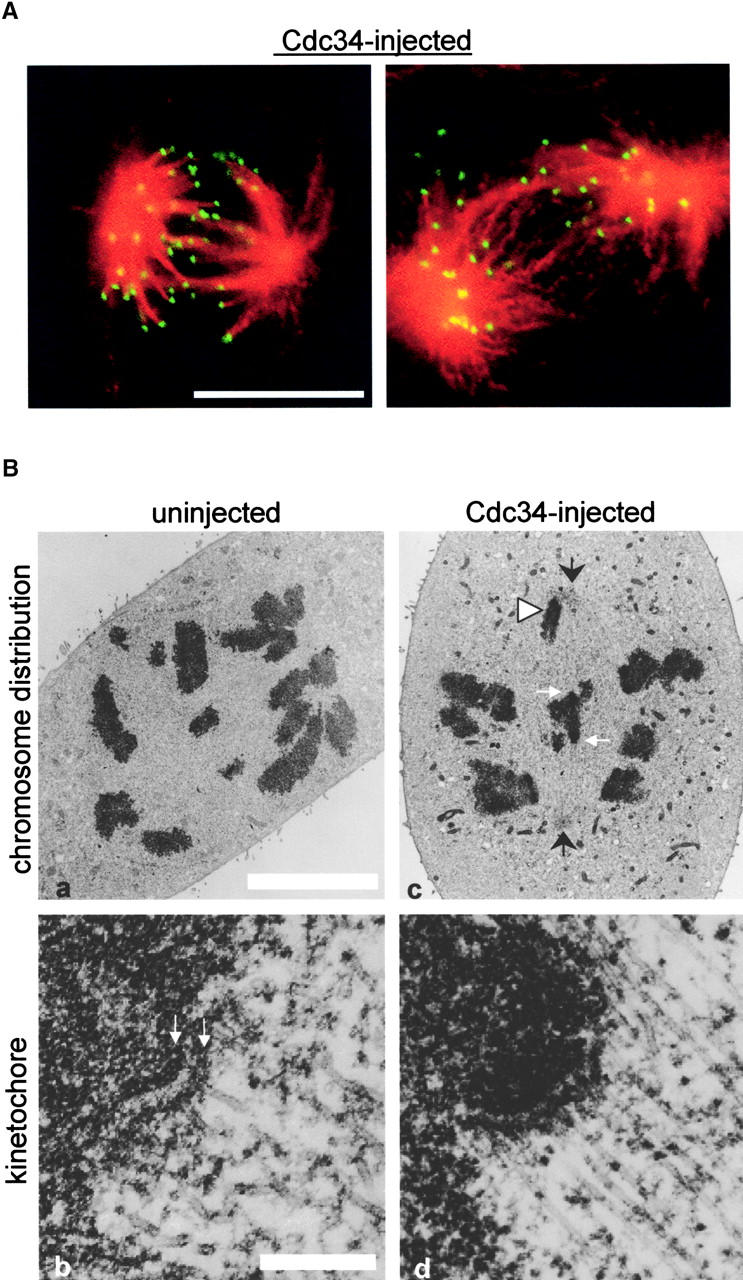
Bipolar spindles and normal kinetochores form in cells injected with Cdc34. (A) Cells were injected with Cdc34 protein, observed to undergo nuclear envelope breakdown, and were further observed for ∼45 min before fixation. Both fixed cells were labeled with antitubulin (red) and anticentromere (green) antibodies. (B) Ultrastructure of kinetochores in injected cells. The chromosome distribution of an uninjected prometaphase cell is shown (a) with an enlargement of a kinetochore from that cell (b). The inner and outer plates of the trilaminar structure are denoted (b, arrows). The chromosome distribution of a cell injected with Cdc34 (c) appears similar to the pattern in the uninjected cell. However, as often seen in Cdc34-injected cells a chromosome is situated very close to the spindle pole (arrowhead). Kinetochores of the injected cell (d) show a clear trilaminar structure with microtubules embedded in the outer plate. Bars: (A and B, a) 10 μm; (B, b) 2 μm.
Table II. Interkinetochore distances.
| Uninjected | Cdc34-injected | |
|---|---|---|
| Chromosomes at midplane | ||
| Average μm ± SD | 1.73 ± 0.38 | 1.68 ± 0.28 |
| Range | 1.11–2.59 | 1.36–2.33 |
| Sister kinetochores/cells | 24/10 | 21/12 |
| Chromosomes at poles | ||
| Average μm ± SD | 1.22 ± 0.19 | 1.35 ± 0.28 |
| Range | 0.9–1.59 | 0.7–1.86 |
| Sister kinetochores/cells | 15/8 | 15/7 |
| Nocodazole-treated | ||
| Average μm ± SD | 1.08 ± 0.21* | 1.23 ± 0.17* |
| Range | 0.66–1.35 | 0.85–1.59 |
| Sister kinetochores/cells | 18/4 | 26/4 |
Values are significantly different from the average interkinetochore distance of sister kinetochores at midplane and near the spindle poles of cells that were not exposed to nocodazole (P < 0.05).
Injection of Cdc34 causes mislocalization of CENP-E in mitotic cells
Inhibition of CENP-E function in mammalian cells by antibody microinjection or introduction of antisense constructs prevents congression of chromosomes to the metaphase plate (Schaar et al., 1997; Yao et al., 2000). Similar results are obtained by immunodepletion of CENP-E from Xenopus egg extracts (Wood et al., 1997). Because the chromosomes in cells injected with wild-type Cdc34 protein also fail to achieve metaphase alignment, we assessed the distribution of CENP-E in injected cells. In Fig. 4, C and D , injection of Cdc34 into Ptk1 cells inhibited CENP-E localization at kinetochores and caused CENP-E protein (red) to accumulate in irregular aggregates in the cytoplasm that did not colocalize with the kinetochores (green). Injection of the Cdc34CA enzymatically inactive mutant did not affect localization of CENP-E to kinetochores (Fig. 4 B). Mislocalization of CENP-E was not a property unique to Ptk1 cells, since injected LLC-Pk cells show similar displacement of CENP-E from the kinetochores into cytoplasmic aggregates (Fig. 4 F). Displacement of CENP-E by Cdc34 was specific for CENP-E. Mitotic centromere-associated kinesin (MCAK), another member of the kinesin superfamily (Wordeman and Mitchison, 1995; Walczak et al., 1996; Maney et al., 1998), and dynein, a minus end–directed motor protein that also localizes to kinetochores (Pfarr et al., 1990; Steuer et al., 1990) were not altered by microinjection of Cdc34 (Fig. 5, B and D) .
Figure 4.
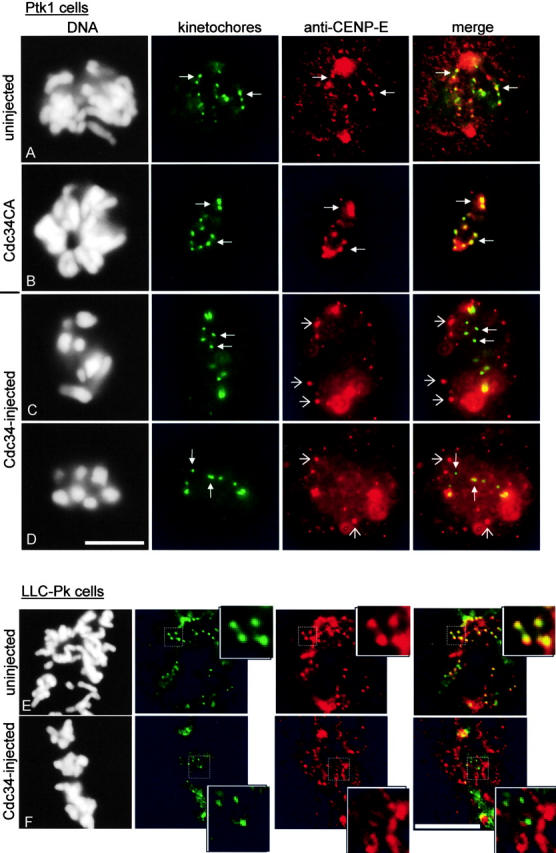
CENP-E is mislocalized in Cdc34-injected cells. (A) The association of CENP-E (red) with kinetochores of an uninjected cell. The kinetochores (green, closed arrows) appear yellow as a result of colocalization of anti-CENP-E and antikinetochore antibodies. (B) Injection of Cdc34CA did not affect CENP-E localization to sister kinetochores (B, closed arrows). (C and D) Cells were injected in prophase with Cdc34 protein. Both were incubated for ∼45 min after nuclear envelope breakdown was detected. Immunolabeling reveals that CENP-E is localized to distinct cytoplasmic aggregates (open arrows) but not on kinetochores (closed arrows). In some instances, yellow kinetochores in the merged image result from the aggregates of CENP-E being in close proximity to kinetochores rather than a true colocalization of anti–CENP-E and antikinetochore antibodies. (E and F) Insets show enlargements of the boxed regions of each cell and illustrate that CENP-E (red) is found on kinetochores (green) of an uninjected prometaphase LLC-Pk cell (E). CENP-E does not concentrate at kinetochores in the Cdc34-injected LLC-Pk cell (F). Each image of a cell was taken at the same focal plane. Bars, 10 μm.
Figure 5.
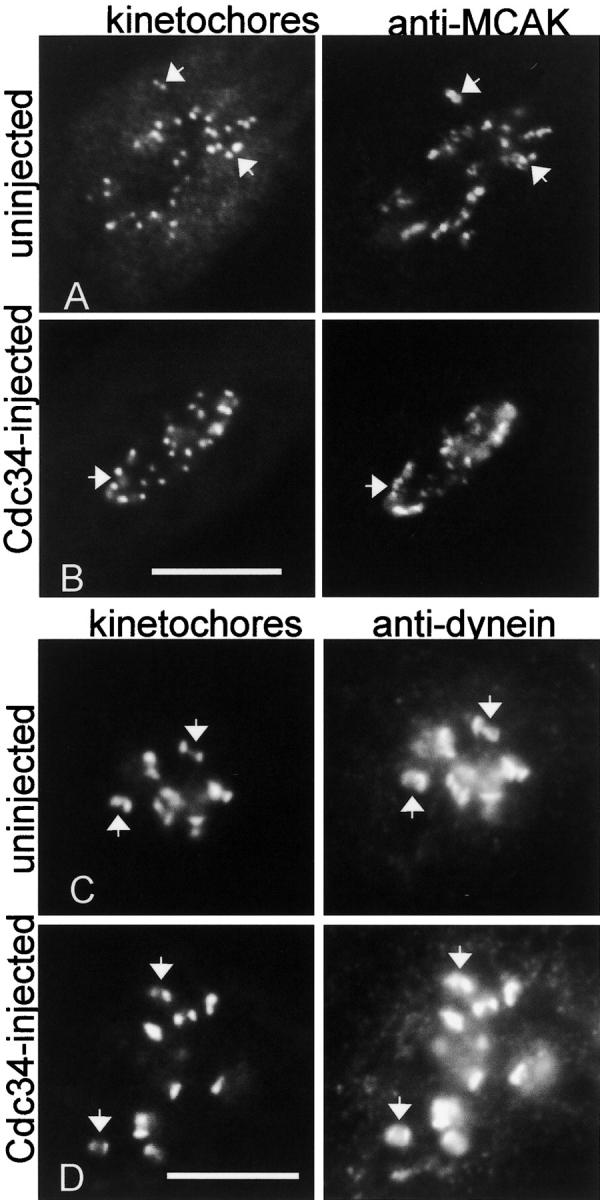
Cdc34 does not affect localization of MCAK and dynein to kinetochores. (A and B) The cell in A arrested at prometaphase after injection with Cdc34. Pairs of kinetochores in the Cdc34- injected and control cell and the corresponding MCAK localization (arrows) are indicated. (C and D) The cells were exposed to nocodazole for 30 min. Cdc34 protein was injected into the cell in C and was incubated for ∼45 min after nuclear envelope breakdown in the presence of the drug. Dynein association with kinetochore pairs (arrows) appears similar in the Cdc34-injected cell (C) and a neighboring uninjected cell (D). The two images for each cell were taken in the same focal plane. Bars, 10 μm.
Mitotic spindle localization of proteins other than CENP-E are not affected by injection of Cdc34
Certain kinases, namely the mitotic checkpoint kinase BubR1 and active Erk1/2 (extracellular signal regulated kinases 1 and 2 also called p42/p44 mitogen-activated protein kinase) localize to kinetochores during prometaphase and have been associated with CENP-E (Cahill et al., 1998; Zecevic et al., 1998; Chan et al., 1999). Other mitotic checkpoint proteins including Cdc20/p55CDC and Mad 2 that localize to kinetochores during M phase (Li and Benezra, 1996; Kallio et al., 1998) were unaffected by injection of Cdc34 (Fig. S2 available at http://www.jcb.org/content/vol154/issue4/707). In budding yeast, Cul1/Cdc53 associates with Cdc34 in all phases of the cell cycle (Mathias et al., 1998). In Ptk1 cells, we found that Cul1 is concentrated at centrosomes during mitosis. Injection of Cdc34 protein did not appear to affect the subcellular localization of any of these proteins in mitotic cells (Figs. S1 and S2 available at http://www.jcb.org/content/vol154/issue4/707). Finally, one trivial reason Cdc34 might affect cellular processes is simply by sequestering free ubiquitin. However, coinjection of a large molar excess of ubiquitin did not reverse the Cdc34-induced prometaphase arrest (unpublished data).
Proteasome inhibitors do not affect prometaphase chromosome movements or CENP-E localization to kinetochores; the effect of Cdc34 on chromosome movement is independent of proteasome activity
The injection of Cdc34 protein might affect the degradation of proteins during prometaphase, either causing an unscheduled degradation or inhibiting a required degradation. Most previous studies with rodent and HeLa cells have shown that proteasome inhibition blocks anaphase onset but does not affect chromosome alignment at the metaphase plate (Sherwood et al., 1993; Wojcik et al., 1996). Similarly, we found that treatment of Ptk1 and LLC-Pk cells with proteasome inhibitors MG132 or β-lactone–induced metaphase arrest without impairment of chromosome movement in prometaphase (Fig. 6 A).
Figure 6.
Proteasome inhibition does not prevent chromosome congression or CENP-E association with kinetochores but arrests cells at metaphase. (A) Live images of a Ptk1 cell incubated in MG132. The cell began chromosome condensation (0 min) after 30-min incubation in MG132. Nuclear envelope breakdown and alignment of chromosomes at the metaphase plate occurred normally. The cell arrested at metaphase as shown in the 50-min image. (B) Cell was detergent extracted and fixed after incubation in MG132 for ∼1 h. Immunolabeling with anti-kinetochore antibody (green) is followed by anti–CENP-E labeling (red) taken at the same focal plane. The yellow kinetochores represent colocalization of the two antibodies (closed arrows).
If the Cdc34 injection displaces CENP-E simply because it interferes with a required degradation, then loss of CENP-E at kinetochores and formation of cytoplasmic aggregates of CENP-E should also occur in cells exposed to proteasome inhibitors. However, we found that proteasome inhibition with MG132 (Fig. 6 A) or β-lactone (unpublished data) did not affect CENP-E localization to kinetochores (Fig. 6 B, yellow kinetochores), and thus the effect of Cdc34 on CENP-E in prometaphase is not likely mediated by inhibited proteolysis.
To determine whether Cdc34 induced unscheduled destruction of a normally stable protein in prometaphase, cells were treated with proteasome inhibitors 20–40 min before injection of Cdc34 protein. Prophase cells injected with Cdc34 protein while in medium containing MG132 or β-lactone completed nuclear envelope breakdown and arrested in prometaphase (Table I), similar to injected cells that were not incubated with proteasome inhibitor. Thus, inhibition of proteasome activity failed to rescue the injected cells from the prometaphase arrest induced by Cdc34.
Discussion
Cdc34 activity and CENP-E
Here, we show that injection of excess Cdc34 into mammalian cells selectively blocks the normal association of CENP-E with kinetochores and inhibits chromosome alignment at metaphase. We found that some chromosomes continued to undergo oscillatory movements in cells injected with Cdc34. However, monooriented chromosomes situated very near the poles underwent fewer apparent movements. In cells injected with Cdc34, many chromosomes near the spindle midplane exhibited normal bipolar connections. However, we detected monooriented chromosomes abnormally close to the spindle poles. We speculate that the physiological role of Cdc34 and/or CENP-E may be to rescue mono-oriented chromosomes near the poles by facilitating bipolar attachment of the distal kinetochores of monooriented chromosomes or by providing antipoleward directed forces.
CENP-E was identified as a kinetochore-associated protein by Yen et al. (1991) and as a plus end–directed motor by Wood et al. (1997). Direct interference with CENP-E function in Xenopus extracts or in mammalian cells inhibits chromosome alignment at metaphase (Schaar et al., 1997; Wood et al., 1997). Antisense inhibition of CENP-E levels in mammalian cells also results in a prometaphase arrest (Yao et al., 2000). The antisense treatment also induced a loss of tension between sister kinetochores, which did not occur upon injection of Cdc34 protein (Table II). Possibly antisense techniques may deplete CENP-E earlier or more thoroughly than does injection of Cdc34 protein.
As yet, we cannot determine whether the effect of microinjected Cdc34 on CENP-E localization is direct or indirect. Microinjection of a commercial anti-Cdc34 antibody had no apparent effects, but we do not know if this antibody inhibits Cdc34 activity in vivo (unpublished data). We have not detected coimmunoprecipitation of Cdc34 and CENP-E in cellular extracts. Also, immunolabeling of Cdc34-injected cells with anti-Cdc34 antibody did not reveal aggregates of Cdc34 protein in the cytoplasm as we had observed for CENP-E. Injected Cdc34 protein was distributed throughout the entire cell in a speckle pattern that is similar to previous findings (Reymond et al., 2000). In late mitosis, near the transition to G1, CENP-E levels fall, and proteasome inhibitors can block this decrease (Brown et al., 1994). If injection of Cdc34 resulted in premature degradation of CENP-E, we would expect to find low levels of CENP-E in the injected arrested cells. This did not appear to be the case. The idea that degradation is not involved in the mislocalization of CENP-E is supported by the observation that chemical inhibitors of proteasome function did not affect CENP-E association with kinetochores (Fig. 6). Although den Elzen and Pines (2001) reported recently that proteasome inhibitors block prometaphase chromosome congression, we (Fig. 6 A) and others (Sherwood et al., 1993; Wojcik et al., 1996) have found that mammalian cells treated with proteasome inhibitors arrest at metaphase with congressed chromosomes. Reversible addition of ubiquitin to proteins without proteasome degradation plays an important role as a regulatory posttranslational modification (Kaiser et al., 2000; Spence et al., 2000). We speculate that ligation or removal of ubiquitin on CENP-E or on associated proteins may serve to regulate chromosome congression.
Regulation of Cdc34 and the SCF during the cell cycle
SCF targeting of certain proteins occurs in all phases of the cell cycle (Galan and Peter, 1999). Cdc34 also associates with the cullin component of the SCF, Cdc53/Cul1, during the entire cell cycle (Mathias et al., 1998). In vertebrate cells, Cdc34 and the SCF components are required for centrosome duplication and localize to centrosomes (Freed et al., 1999). Injection of excess Cdc34 protein did not interfere with localization of the SCF component Cul1 to mitotic centrosomes (Fig. S1 available at http://www.jcb.org/content/ vol154/issue4/707).
The intracellular level of Cdc34 in yeast and in mammalian cells does not vary during the cell cycle (Goebl et al., 1988; Reymond et al., 2000). Although expressed at all stages, Cdc34 activity may be restricted by inhibitory mechanisms in early M phase. Possibly microinjection of excess Cdc34 overwhelms this regulation, leading to the untimely ubiquitylation of some target protein(s). An important next step will be identification of the mitotic target(s) of Cdc34 activity. Recently, Reimann et al. (2001) identified a vertebrate Skp1-interacting protein called early mitotic inhibitor (Emi)-1. Emi1 functions as an inhibitor of the anaphase-promoting complex/cyclosome, and overexpression of Emi1 induces a prometaphase arrest. Degradation of Emi1 occurs early in mitosis but is independent of anaphase-promoting complex/cyclosome activity. Degradation of Emi1 may require SCF activity.
Our search into the mitotic function of ubiquitin-conjugating enzymes revealed an unexpected effect of microinjecting excess wild-type Cdc34 in early mitosis: impairment of chromosome movement and arrest at prometaphase by the displacement of CENP-E from its normal association with kinetochores. This evidence suggests that Cdc34 may play a role in chromosome congression. Alternatively, it is possible that the introduction of excess Cdc34 protein into mitotic cells evokes a pathway normally suppressed during M phase. These experiments point to new roles for protein ubiquitylation in regulating chromosome movement in early mitosis.
Materials and methods
Cell culture
Ptk1 cells were cultured in MEM (Life Technologies) supplemented with 10% FBS, 20 mM Hepes buffer, 1× nonessential amino acids, 1 mM sodium pyruvate, 60 μg/ml penicillin, and 100 μg/ml streptomycin at 37°C under 5% CO2. LLC-Pk cells were cultured under identical conditions with the same supplements in DME (Life Technologies). Proteasome inhibitors MG132 and clasto-lactacystin-β-lactone (Calbiochem) were used in culture medium at 10 μM.
Gel electrophoresis and immunoblotting
Recombinant wild-type and mutant Cdc34 proteins were expressed in bacteria and purified as reported previously in Bastians et al. (1999). Purified recombinant proteins and Ptk1 cell extract proteins were separated by SDS-PAGE. Proteins were transferred to Immobilon-P membrane (Millipore). The membrane was blocked in 5% milk TBST (10 mM Tris, 150 mM NaCl, 0.059, Tween-20) for 30 min followed by incubation in anti-Cdc34 monoclonal antibody (Transduction Laboratories) at a dilution of 1:1,000 into TBST followed by anti-mouse HRP-conjugated (Jackson ImmunoResearch Laboratories) and visualized using chemiluminescence (Dupont-NEN Life Science Products). Characterization of the protein is shown in Fig. S3 (available at http://www.jcb.org/content/vol154/issue4/707).
Microinjection
Microinjection chambers were prepared by cutting an 18-mm hole into the bottom of a 60-mm plastic tissue culture dish. A 22-mm coverslip was sealed to the inside of the dish with silicone grease. Chambers were sterilized by inversion on a UV transilluminator (UVP Inc.) for 15 min. Ptk1 cells were cultured on inscribed glass coverslips or Bellco gridded coverslips for 2–3 d to 60% confluency, and transferred to a sterile chamber containing 4 mL of medium. The medium in the chamber was overlaid with light mineral oil before microinjection. The chamber was placed on a prewarmed temperature-controlled stage (Fryer Co. Inc.) at 34–36°C on a Diaphot microscope (Nikon). Microinjections were performed as described by Gorbsky et al. (1998). Cells were injected with wild-type Cdc34 (1.5–7 mg/ml), mutant Cdc34 (3–6 mg/ml), or buffer (50 mM Hepes-NaOH, ph 7.4, 5 mM KCl, 1.5 mM MgCl2, 1 mM DTT) to no more than 5% of their total volume. Images were collected with a digital cooled CCD camera (Photometrics) controlled with Metamorph software (Universal Imaging Corp.). Prophase cells were injected in the cytoplasm, whereas prometaphase or anaphase cells were injected away from the chromosomes to avoid any damage to DNA.
Electron microscopy
Cells grown on gridded coverslips were injected and followed in vivo, then carefully removed from the microinjection chamber, rinsed twice with PBS warmed to 37°C, and fixed 40 min with 3% glutaraldehyde (Electron Microscopy Sciences) in 0.1 M PO4 buffer, pH 6.9, that was also warmed to 37°C. Coverslips were postfixed in 2% osmium tetroxide and dehydrated. Injected cells were located and embedded in Epon 812 by inverting a Beem capsule over the appropriate area. After polymerization of the epon, the capsule was separated from the coverslip by alternate immersion into hot water and liquid nitrogen. Injected cells were relocated and the block trimmed to ensure that the only mitotic cells remaining on the block face had been injected. Each injected cell was sectioned and stained with uranyl acetate and lead citrate. Sections were examined in a Jeol 100CX electron microscope at 80 kV. Micrographs were scanned into Adobe Photoshop® 5.5, and final images were annotated in Microsoft PowerPoint 2000.
Immunofluorescence
Each coverslip was rinsed at room temperature with 60 mM Pipes, 25 mM Hepes, pH 6.9, 10 mM EGTA, 4 mM MgSO4 (PHEM). For CENP-E, dynein, and Cul1 immunolabeling, cells were lysed with 0.5% CHAPS detergent or 0.5% Triton X-100 in PHEM for 3 min and fixed for 15 min at room temperature with 1% freshly prepared formaldehyde in PHEM. Cells labeled with MCAK antibody were fixed for 20 min in 4% formaldehyde and then extracted in 0.5% CHAPS in PHEM. Cells labeled with antiphospho histone H3 antibody were fixed in 4% paraformaldehyde and extracted with 0.5% Triton X-100 prepared in PBS. For labeling of nuclear pore proteins with the RL-1 antibody, cells were fixed in 1% paraformaldehyde and extracted in 0.5% CHAPS in PHEM buffer. For tubulin labeling, cells were fixed in methanol at −20°C for 10 min. Cells were then rinsed three times with 10 mM MOPS, pH 7.2, 150 mM NaCl containing 0.05% Tween 20 (MBST) and blocked in 20% boiled normal goat serum in MBST. Cells were labeled with primary antibody diluted in 5% boiled normal goat serum for 45 min at room temperature. The following antibodies were used at the indicated dilutions: antitubulin ascites fluid (Dr. T. Frankfurter, University of Virginia) at 1:100, affinity purified rabbit anti–CENP-E antibody and anti-BubR1 (Dr. T. Yen, Fox Chase Cancer Center, Philadelphia, PA) at 1:500, purified MCAK polyclonal antibody (Dr. L. Wordeman, University of Washington School of Medicine, Seattle, WA) at 1:1,000, affinity purified antidynein antibody (Dr. K. Pfister, University of Virginia) at 1:500, antiserum to phosphorylated histone H3 (Dr. D. Allis, University of Virginia) at 1:5,000, RL-1 antinucleoporin monoclonal antibody (Dr. B. Paschal, University of Virginia) at 1:100, affinity purified anti-Cul1 antibody (Labvision/Neomarkers) at 1:200, affinity purified antiphospho p42/p44 mitogen-activated protein kinase antibody (Dr. M. Weber, University of Virginia) at 8.5 μg/ml, anti-p55CDC antibody (Santa Cruz Biotechnology) at 1:200, and rabbit anti-Mad2 antibody (Dr. E. Salmon, University of North Carolina, Chapel Hill, NC) at 4 μg/ml. In some cases, kinetochores of cells were colabeled with human nuclear (ANA) centromere serum (Cortex Biochem, Inc.) at a 1:500 dilution. Cells were washed for 15 min in three changes of MBST. Fluorescent secondary antibodies (Jackson ImmunoResearch Laboratories) were diluted in 5% boiled normal goat serum and applied at 1:400–1:2,000 for 45 min at room temperature. Coverslips were washed and then rinsed in DAPI at 0.1 μg/ml for 1 min. After rinsing twice in distilled water, coverslips were mounted in 10 μl Prolong (Molecular Probes).
Digital images of cells were obtained with either the Diaphot microscope or with a ZEISS Axioplan II equipped with a digital CCD Orca camera (C4742-45; Hamamatsu) controlled by a computer using Openlab software (Improvision, Inc.). Interkinetochore distances were measured from the center of one sister kinetochore to the center of the other sister using the measure distance functions in either Openlab or Metamorph software packages.
Online supplemental material
Supplemental Figs. 1–3 and videos 1–3, complementing the images presented in the print version of this article, are available online at http://www.jcb.org/content/vol154/issue4/707.
Supplemental Material
Acknowledgments
We thank Drs. David Castle, Kevin Pfister, and Rick Horwitz at the University of Virginia, Drs. Kerry Bloom and Ted Salmon at the University of North Carolina, and the members of their laboratories for providing space, equipment, and reagents to enable completion of this work. We thank Drs. Michele Pagano, Tim Yen, Linda Wordeman, Bryce Paschal, Tony Frankfurter, Mike Weber, and Dave Allis for supplying reagents. We also thank Dr. Jan Redick and Ms. Bonnie Sheppard for expert electron microscopy. We thank Dr. Penny Tavormina and Dr. Marie-George Come for critical reading of the manuscript and are grateful to Mr. John Daum for valuable technical assistance.
This work was supported by a grant from the National Institute of General Medical Sciences to G.J. Gorbsky. L.M. Toppper was supported by a National Cancer Institute predoctoral training award to the University of Virginia.
The online version of this article contains supplemental material.
L.M. Topper's present address is Department of Biology, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599.
Footnotes
Abbreviations used in this paper: CENP-E, centromere protein E; Emi, early mitotic inhibitor; MCAK, mitotic centromere-associated kinesin; SCF, skp1-Cdc53/cullin–F-box protein.
References
- Apionishev, S., D. Malhotra, S. Raghavachari, S. Tanda, and R.S. Rasooly. 2001. The Drosophila UBC9 homologue lesswright mediates the disjunction of homologues in meiosis I. Genes Cells. 6:215–224. [DOI] [PubMed] [Google Scholar]
- Banerjee, A., R.J. Deshaies, and V. Chau. 1995. Characterization of a dominant negative mutant of the cell cycle ubiquitin-conjugating enzyme Cdc34. J. Biol. Chem. 270:26209–26215. [DOI] [PubMed] [Google Scholar]
- Bastians, H., L.M. Topper, G.J. Gorbsky, and J.V. Ruderman. 1999. Cell cycle-regulated proteolysis of mitotic target proteins. Mol. Biol. Cell. 10:3927–3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, K.D., R.M.R. Coulson, T.J. Yen, and D.W. Cleveland. 1994. Cyclin-like accumulation and loss of the putative kinetochore motor CENP-E results from coupling continuous synthesis with specific degradation at the end of mitosis. J. Cell Biol. 125:1303–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill, D.P., C. Lengauer, J. Yu, G.J. Riggins, J.K.V. Willson, S.D. Markowitz, K.W. Kinzler, and B. Vogelstein. 1998. Mutations of mitotic checkpoint genes in human cancers. Nature. 392:300–303. [DOI] [PubMed] [Google Scholar]
- Carrano, A.C., E. Eytan, A. Hershko, and M. Pagano. 1999. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat. Cell Biol. 1:193–199. [DOI] [PubMed] [Google Scholar]
- Chan, G.K.T., S.A. Jablonski, V. Sudakin, J.C. Hittle, and T.J. Yen. 1999. Human BubR1 is a mitotic checkpoint kinase that monitors CENP-E functions at kinetochores and binds the cyclosome/APC. J. Cell Biol. 146:941–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrasse, S., I. Carena, V. Brondani, K.-H. Klempnauer, and S. Ferrari. 2000. Degradation of B-Myb by ubiquitin-mediated proteolysis: involvement of the Cdc34-SCFp45Skp2 pathway. Oncogene. 19:2986–2995. [DOI] [PubMed] [Google Scholar]
- Deshaies, R.J. 1999. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15:435–467. [DOI] [PubMed] [Google Scholar]
- Deshaies, R.J., V. Chau, and M. Kirschner. 1995. Ubiquitination of the G1 cyclin Cln2p by a Cdc34p-dependent pathway. EMBO J. 14:303–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Elzen, N., and J. Pines. 2001. Cyclin A is destroyed in prometaphase and can delay chromosome alignment and anaphase. J. Cell Biol. 153:121–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury, L.S., G. Perkins, and J.F. Diffley. 1997. The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 16:5966–5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman, R.M., C.C. Correll, K.B. Kaplan, and R.J. Deshaies. 1997. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 91:221–230. [DOI] [PubMed] [Google Scholar]
- Freed, E., K.R. Lacey, P. Huie, S.A. Lyapina, R.J. Deshaies, T. Stearns, and P.K. Jackson. 1999. Components of an SCF ubiquitin ligase localize to the centrosome and regulate the centrosome duplication cycle. Genes Dev. 13:2242–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan, J.-M., and M. Peter. 1999. Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc. Natl. Acad. Sci. USA. 96:9124–9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebl, M.G., J. Yochem, S. Jentsch, J.P. McGrath, A. Varshavsky, and B. Byers. 1988. The yeast cell cycle gene CDC34 encodes a ubiquitin-conjugating enzyme. Science. 41:1331–1335. [DOI] [PubMed] [Google Scholar]
- Gorbsky, G.J., R.H. Chen, and A.W. Murray. 1998. Microinjection of antibody to Mad2 protein into mammalian cells in mitosis induces premature anaphase. J. Cell Biol. 141:1193–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henchoz, S., Y. Chi, B. Catarin, I. Herskowitz, R.J. Deshaies, and M. Peter. 1997. Phosphorylation- and ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor Far1p in budding yeast. Genes Dev. 11:3046–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendzel, M.J., Y. Wei, M.A. Mancini, A. Van Hooser, T. Ranalli, B.R. Brinkley, D.P. Bazett-Jones, and C.D. Allis. 1997. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 106:348–360. [DOI] [PubMed] [Google Scholar]
- Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425–479. [DOI] [PubMed] [Google Scholar]
- Hicke, L. 2001. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2:195–201. [DOI] [PubMed] [Google Scholar]
- Hochstrasser, M. 1996. Protein degradation or regulation: Ub the judge. Cell. 84:813–815. [DOI] [PubMed] [Google Scholar]
- Jaquenoud, M., M.P. Gulli, K. Peter, and M. Peter. 1998. The Cdc42p effector Gic2p is targeted for ubiquitin-dependent degradation by the SCFGrr1 complex. EMBO J. 17:5360–5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch, S. 1992. The ubiquitin-conjugation system. Annu. Rev. Gen. 26:179–207. [DOI] [PubMed] [Google Scholar]
- Jiang, W., and Y. Koltin. 1996. Two-hybrid interaction of a human UBC9 homolog with centromere proteins of Saccharomyces cerevisiae. Mol. Gen. Genet. 251:153–160. [DOI] [PubMed] [Google Scholar]
- Kaiser, P., R.A. Sia, E.G. Bardes, D.J. Lew, and S.I. Reed. 1998. Cdc34 and the F-box protein Met30 are required for degradation of the Cdk-inhibitory kinase Swe1. Genes Dev. 12:2587–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser, P., K. Flick, C. Wittenberg, and S.I. Reed. 2000. Regulation of transcription by ubiquitination without proteolysis: Cdc34/SCFMet30–mediated inactivation of the transcription factor Met4. Cell. 102:303–314. [DOI] [PubMed] [Google Scholar]
- Kallio, M., J. Weinstein, J.R. Daum, D.J. Burke, and G.J. Gorbsky. 1998. Mammalian p55CDC mediates association of the spindle checkpoint protein Mad2 with the cyclosome/anaphase-promoting complex, and is involved in regulating anaphase onset and late mitotic events. J. Cell Biol. 141:1393–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, K.B., A.A. Hyman, and P.K. Sorger. 1997. Regulating the yeast kinetochore by ubiquitin-dependent degradation and skp1p-mediated phosphorylation. Cell. 91:491–500. [DOI] [PubMed] [Google Scholar]
- Kaplun, L., Y. Ivantsiv, D. Kornitzer, and D. Raveh. 2000. Functions of the DNA damage response pathway target Ho endonuclease of yeast for degradation via the ubiquitin-26S proteasome system. Proc. Natl. Acad. Sci. USA. 97:10077–10082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornitzer, D., B. Raboy, R.G. Kulka, and G.R. Fink. 1994. Regulated degradation of the transcription factor Gcn4. EMBO J. 13:6021–6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., and R. Benezra. 1996. Identification of a human mitotic checkpoint gene: hsMAD2. Science. 274:246–248. [DOI] [PubMed] [Google Scholar]
- Maney, T., A.W. Hunter, M. Wagenbach, and L. Wordeman. 1998. Mitotic centromere-associated kinesin is important for anaphase chromosome segregation. J. Cell Biol. 142:787–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti, A., C. Wirbelauer, M. Scheffner, and W. Krek. 1999. SCFSKP2 ubiquitin ligase/E2F-1 transcription factor complex formation underlies regulation of E2F-1 protein stability and S phase promoting function. Nat. Cell Biol. 1:14–19. [DOI] [PubMed] [Google Scholar]
- Mathias, N., C.N. Steussy, and M.G. Goebl. 1998. An essential domain within Cdc34p is required for binding to a complex containing Cdc4p and Cdc53p in Saccharomyces cerevisiae. J. Biol. Chem. 273:4040–4045. [DOI] [PubMed] [Google Scholar]
- Matunis, M.J., J. Wu, and G. Blobel. 1998. SUMO-1 modification and its role in targeting the RanGTPase-activating protein RanGAP1, to the nuclear pore complex. J. Cell Biol. 140:499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael, W.M., and J. Newport. 1998. Coupling of mitosis to the completion of S phase through Cdc34-mediated degradation of Wee1. Science. 282:1886–1889. [DOI] [PubMed] [Google Scholar]
- Mueller, P.R., T.R. Coleman, and W.G. Dunphy. 1995. Cell cycle regulation of a Xenopus Wee1-like kinase. Mol. Biol. Cell. 6:119–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami, M.S., and G.F. Vande Woude. 1998. Analysis of the early embryonic cell cycles of Xenopus; regulation of cell cycle length by Xe-wee1 and Mos. Development. 125:237–248. [DOI] [PubMed] [Google Scholar]
- Pagano, M., S.W. Tam, A.M. Theodoras, P. Beer-Romero, G. Del Sal, V. Chau, P.R. Yew, G.F. Draetta, and M. Rolfe. 1995. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science. 269:682–685. [DOI] [PubMed] [Google Scholar]
- Pati, D., M.L. Meistrich, and S.E. Plon. 1999. Human Cdc34 and Rad6B ubiquitin-conjugating enzymes target repressors of cyclic AMP-induced transcription for proteolysis. Mol. Cell. Biol. 19:5001–5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfarr, C.M., M. Coue, P.M. Grissom, T.S. Hays, M.E. Porter, and J.R. McIntosh. 1990. Cytoplasmic dynein is localized to kinetochores during mitosis. Nature. 345:263–265. [DOI] [PubMed] [Google Scholar]
- Plon, S.E., K.A. Leppig, H.N. Do, and M. Groudine. 1993. Cloning of the human homolog of the CDC34 cell cycle gene by complementation in yeast. Proc. Natl. Acad. Sci. USA. 90:10484–10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimann, J.D.R., E. Freed, J.Y. Hsu, E.R. Kramer, J.-M. Peters, and P.K. Jackson. 2001. Emi1 is a newly identified mitotic regulator that binds Cdc20 and inhibits the anaphase promoting complex. Cell. 105:645–655. [DOI] [PubMed] [Google Scholar]
- Reymond, F., C. Wirbelauer, and W. Krek. 2000. Association of human ubiquitin-conjugating enzyme CDC34 with the mitotic spindle in anaphase. J. Cell. Sci. 113:1687–1694. [DOI] [PubMed] [Google Scholar]
- Robzyk, K., J. Recht, and M.A. Osley. 2000. Rad6 dependent ubiquitination of histone H2B in yeast. Science. 287:501–504. [DOI] [PubMed] [Google Scholar]
- Schaar, B.T., G.T. Chan, P. Maddox, E.D. Salmon, and T.J. Yen. 1997. CENP-E function at kinetochores is essential for chromosome alignment. J. Cell Biol. 139:1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob, E., T. Bohm, M.D. Mendenhall, and K. Nasmyth. 1994. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 79:233–244. [DOI] [PubMed] [Google Scholar]
- Sherwood, S.W., A.L. Kung, J. Roitelman, R.D. Simoni, and R.T. Schimke. 1993. In vivo inhibition of cyclin B degradation and induction of cell-cycle arrest in mammalian cells by the neutral cysteine protease inhibitor N-acetylleucylleucylnorleucinal. Proc. Natl. Acad. Sci. USA. 90:3353–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow, C.M., A. Senior, and L. Gerace. 1987. Monoclonal antibodies identify a group of nuclear pore complex glycoproteins. J. Cell Biol. 104:1143–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, A., Q. Wang, M.G. Goebl, and M.A. Harrington. 1998. Phosphorylation of nuclear Myo D is required for its rapid destruction. Mol. Cell. Biol. 186:4994–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence, J., R.R. Gali, G. Dittmar, F. Sherman, M. Karin, and D. Finley. 2000. Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell. 102:67–76. [DOI] [PubMed] [Google Scholar]
- Steuer, E.R., L. Wordeman, T.E. Schroer, and M.P. Sheetz. 1990. Localization of cytoplasmic dynein to mitotic spindles and kinetochores. Nature. 345:266–268. [DOI] [PubMed] [Google Scholar]
- Sung, P., S. Prakash, and L. Prakash. 1990. Mutation of cysteine-88 RAD6 protein abolishes its ubiquitin-conjugating activity and various biological functions. Proc. Natl. Acad. Sci. USA. 87:2695–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterluty, H., E. Chatelain, A. Marti, C. Wirbelauer, M. Senften, U. Muller, and W. Krek. 1999. p45Skp2 promotes p27Kip1 degradation and induces S phase in quiescent cells. Nat. Cell Biol. 1:207–214. [DOI] [PubMed] [Google Scholar]
- Walczak, C.E., T.J. Mitchison, and A. Desai. 1996. XKCM1: a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 84:37–47. [DOI] [PubMed] [Google Scholar]
- Willems, A.R., S. Lanker, E.E. Patton, K.L. Craig, T.F. Nason, N. Mathias, R. Kobayashi, C. Wittenberg, and M. Tyers. 1996. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell. 86:453–463. [DOI] [PubMed] [Google Scholar]
- Wojcik, C., D. Schroeter, M. Stoehr, S. Wilk, and N. Paweletz. 1996. An inhibitor of the chymotrypsin-like activity of the multicatalytic proteinase complex (20S proteasome) induces arrest in G2-phase and metaphase in HeLa cells. Eur. J. Cell Biol. 70:172–178. [PubMed] [Google Scholar]
- Wood, K.W., R. Sakowicz, L.B. Goldstein, and D.W. Cleveland. 1997. Cenp-E is a plus end-directed kinetochore motor required for metaphase chromosome alignment. Cell. 91:357–366. [DOI] [PubMed] [Google Scholar]
- Wordeman, L., and T.J. Mitchison. 1995. Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin-related protein that associates with centromeres during mitosis. J. Cell Biol. 128:95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaglom, J., M.H. Linskens, S. Sadis, D.M. Rubin, B. Futcher, and D. Finley. 1995. p34Cdc28-mediated control of Cln3 cyclin degradation. Mol. Cell. Biol. 15:731–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, X., A. Abrieu, Y. Zheng., K.F. Sullivan, D.W. Cleveland. 2000. CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat. Cell Biol. 2:484–491. [DOI] [PubMed] [Google Scholar]
- Yen, T.J., D.A. Compton, D. Wise, R.P. Zinkowski, B.R Brinkley, W.C. Earnshaw, and D.W. Cleveland. 1991. CENP-E, a novel human centromere-associated protein required for progression from metaphase to anaphase. EMBO J. 10:1245–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yew, P.R., and M.W. Kirschner. 1997. Proteolysis and DNA replication: the CDC34 requirement in the Xenopus egg cell cycle. Science. 277:1672–1676. [DOI] [PubMed] [Google Scholar]
- Yoon, H.J., and J. Carbon. 1995. Genetic and biochemical interactions between an essential kinetochore protein, Cbf2p/Ndc10p, and the CDC34 ubiquitin-conjugating enzyme. Mol. Cell. Biol. 15:4835–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecevic, M., A.D. Catling, S.T. Eblen, L. Renzi, J.C. Hittle, T.J. Yen, G.J. Gorbsky, and M.J. Weber. 1998. Active MAP kinase in mitosis: localization at kinetochores and association with the motor protein CENP-E. J. Cell Biol. 142:1547–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.