Abstract
A considerable body of evidence points to a role for both cyclin E/cyclin-dependent kinase (cdk)2 activity and E2F transcription activity in the induction of S phase. We show that overexpression of cyclin E/cdk2 in quiescent cells induces S phase, that this coincides with an induction of E2F activity, and that coexpression of E2F enhances the cyclin E/cdk2-mediated induction of S phase. Likewise, E2F overexpression can induce S phase and does so in the apparent absence of cyclin E/cdk2 activity. In addition, although the inhibition of cyclin E/cdk2 activity blocks the induction of S phase after growth stimulation of normal mouse embryo fibroblasts, inhibition of cyclin E/cdk2 does not block S phase induction in Rb−/− cells where E2F activity is deregulated. These results point to the important roles for E2F and cyclin E/cdk2 in the induction of S phase. Moreover, the nature of the E2F targets and the suspected targets for cyclin E/cdk2 suggests a potential molecular mechanism for the collaborative action of cyclin E/cdk2 and E2F in the induction of S phase.
A variety of experiments have now led to the description of a complex regulatory pathway controlling the progression of mammalian cells out of a quiescent state, through G1, and into S phase. It is clear that G1 cyclin-dependent kinase (cdk) activity plays a critical role in this pathway. In particular, the inhibition of either D type cdk activity or cyclin E-dependent kinase activity prevents induction of S phase (1–3). At least part of the action of these kinases, particularly the D type cdks, involves the phosphorylation of Rb because the absence of Rb removes the requirement for D type cyclins in G1 progression (4–7). The role of Rb and Rb family members in cell growth control may largely be in the regulation of the E2F transcription factor. Substantial evidence now points to the critical role for E2F transcription activity in allowing cell-cycle progression through the G1/S restriction point. Inhibition of mammalian E2F activity, either by using a DP1 dominant-negative mutant (8) or an RNA ligand that selectively inhibits E2F activity (9), prevents S phase. Moreover, we have recently shown that E2F3 activity is required for G1/S progression in cycling fibroblast cells (10). In addition, E2F overexpression in mammalian cell cultures (11–14) or in Drosophila imaginal discs (15, 16) is sufficient to induce DNA replication. These studies thus suggest that the accumulation of E2F transcriptional activity is rate-limiting for transition from G1 into S phase; indeed, overexpression of E2F1, E2F2, or E2F3 in quiescent cells reduces the time spent in G1 following a serum stimulation (unpublished data).
Although we have shown that E2F accumulation can bypass a requirement for G1 cdk activity (17), placing E2F downstream of this regulatory step, it is also true that other experiments have suggested an importance for cdk-mediated phosphorylation in the initiation of replication independent of transcription (18–23). This latter finding suggests additional roles for cdk activity, besides the activation of E2F, in the induction of S phase that might be bypassed by the overproduction of E2F. To better define the relationship between the action of G1 cdk activity and E2F transcriptional activity in allowing cells to enter S phase, we have further explored the roles of these activities in S phase induction.
MATERIALS AND METHODS
Cells and Viruses.
Viral stocks were created as described (24), and the virus was purified by using CsCl density gradient centrifugation as described (25). Viral titers were determined by using an indirect immunofluorescent assay specific for the viral 72-kDa E2 gene product as described (26) and defined as focus-forming units (ffu) per ml. The construction of the recombinant viruses Ad-E2F1, Ad-E2F1/VP16, Ad-E2F2, and Ad-Con (a control virus, previously termed AdMb or Ad-CMV, lacking a cDNA insert) have been described (17, 24, 27). The Ad-p21 virus was similarly constructed by ligation of the BglII–BamHI fragment containing human p21 from CMV-p21 (17) into the BamHI-digested recombination plasmid pGEM-CMV. Ad-Cdk2 was constructed by ligation of the BamHI fragment from CMV-cdk2HA wt (28) into the BamHI-digested pGEM-CMV. Ad-CycE was constructed by ligation of the KpnI–XbaI fragment from pRc/CMV cycE (29) into the KpnI–XbaI digested pGEM-CMV.
REF52 cells were grown in DMEM containing 10% serum (5% fetal bovine serum and 5% calf serum). To bring cells to quiescence, REF52 cells were plated at ≈3,500 cells per cm2 and incubated overnight. The next day, the cells were washed once with DMEM, the culture medium was replaced with DMEM containing 0.25% serum, and cells were further incubated for 48 hr before virus infection. Cells on plates were infected in DMEM with 20 mM Hepes (pH 7.2) for 75 min at 37°C at a cell to volume ratio of 0.5 × 106 cells per ml (0.5 ml for a 35-mm plate, 2 ml for a 100-mm plate, or 5 ml for a 150-mm plate). After infection, four volumes of 0.25% serum/DMEM was added to each plate, and the cells were incubated at 37°C. Where indicated, the cells were subsequently serum-stimulated by replacement with culture media containing 10% serum.
For the p21 bypass experiments, density-arrested REF52 cells were trypsinized, resuspended in DMEM containing 10% serum, centrifuged, and resuspended in DMEM with 20 mM Hepes (pH 7.2) at 1 × 107 cells per ml. The higher cell to volume ratio in these infections, as compared with infection of a monolayer, resulted in similar levels of expression using a lower multiplicity of infection (moi). Virus was then added to the indicated moi, and the cells were infected in suspension for 20 min with constant, gentle swirling. After infection, the cells were replated at subconfluent densities (2 × 106 cells per 150-mm plate or 1 × 105 cells per 35-mm plate) in 10% serum in DMEM.
Northern Analysis.
Northern analysis was performed as described (26) except total RNA was isolated by using the Trizol method (GIBCO/BRL). The cDNAs used as probes for Northern analysis have been described (26, 31).
Kinase Assays.
Cells in 150-mm plates were harvested and immunoprecipitated, and associated kinase activity was determined as described for Rb kinase assays (17), except that where indicated, 5 μg of histone H1 was used as the substrate in place of glutathione S-transferase–Rb. Affinity-purified rabbit polyclonal antibodies, anti-cyclin E (SC-481), α-cyclin D1 (SC-450), α-cdk2 (SC-163), and α-cyclin B1 (SC-245) were purchased from Santa Cruz Biotechnology. A rabbit polyclonal antibody against cyclin A was also used (17).
Flow Cytometry and BrdUrd Incorporation.
REF52 cells on 100-mm dishes were processed for flow cytometry as described (14), with the following modifications: before resuspension in propidium iodide, cells were incubated in 1 ml of 2 M HCL containing 0.2 mg/ml pepsin for 30 min at room temperature; 3 ml of 0.1 M sodium tetraborate (pH 8.5) was then added, and the cells were pelleted for 5 min at 1,500 × g, washed once with PBS/1% BSA, and the cells were pelleted again. Cells were processed for BrdUrd incorporation as described (17).
Western Immunoblotting.
Rb protein was detected by Western blotting using the anti-Rb monoclonal IF8 from Santa Cruz Biotechnology as described (32). Endogenous Mcm2 protein was detected by using rabbit polyclonal anti-Mcm2 (generous gift of H. Kimura, University of Oxford, U.K.). Mcm3 protein was detected by using affinity-purified rabbit polyclonal antibody raised against a keyhole limpet hemocyanin-conjugated peptide derived from mouse Mcm3 sequence (VVCIDEFDKMSDMDRTA).
RESULTS
E2F and Cyclin E/cdk2 Collaborate for Efficient S Phase Induction in Growth Factor-Deprived Cells.
Previous work has pointed to the roles for both E2F and G1 cdk activity in the induction of S phase (17–23). To further explore the roles of E2F and cyclin E/cdk2, we have investigated the ability of cyclin E/cdk2 overexpression to induce S phase and the requirement for E2F activity in this process. To this end, we generated a series of recombinant adenoviruses encoding various cell-cycle regulatory proteins including E2F family members, cyclin E and cdk2, and the cdk inhibitor p21. Recombinant adenoviruses have the important property of being able to infect either quiescent or proliferating cells. Because the entire population of cells is infected, it allows for the biochemical analysis of endogenous mRNA, protein levels, or kinase activity, which are affected by the expression of protein(s) produced by the viruses.
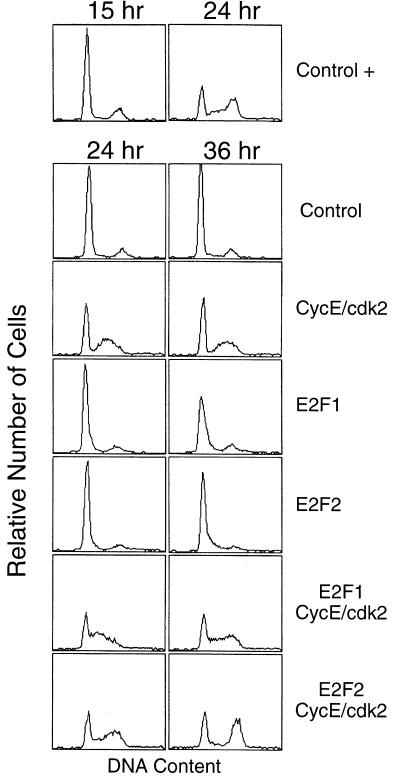
As shown by the fluorescence-activated cell sorting analyses in Fig. 1, the overexpression of cyclin E/cdk2 in quiescent REF52 fibroblasts led to an induction of S phase, as indicated by the increased number of cells with a greater than G1 DNA content. However, although the cyclin E/cdk2-expressing cells did enter S phase, there was not a significant accumulation of a G2 population. Under the conditions used in this experiment, the overexpression of either E2F1 or E2F2 resulted in a slight increase in S phase cells, as indicated by the shoulder in the fluorescence-activated cell sorting profile, but coexpression of E2F activity with cyclin E/cdk2, particularly E2F2, resulted in a more efficient S phase and a marked accumulation of cells with a G2 DNA content. In short, it appears that efficient induction of DNA replication can be achieved by a collaborative action of E2F and cyclin E/cdk2.
Figure 1.
E2F and cyclin E/cdk2 collaborate in S phase induction. Analysis of S phase induction with flow cytometry. Cells were starved and infected with Ad-Con (control, moi = 600 ffu per cell), Ad-E2F1 (moi = 100 ffu per cell), Ad-E2F2 (moi = 100 ffu per cell), and Ad-CycE and Ad-Cdk2 (moi = 200 ffu per cell and 300 ffu per cell, respectively) as indicated; the final moi for each sample was adjusted to 600 ffu per cell with Ad-Con. Cells were then incubated with fresh starvation medium or with medium containing 10% serum (+) and then harvested at the indicated times and processed for flow cytometry. The horizontal axis reflects relative DNA content, and the vertical axis represents cell number.
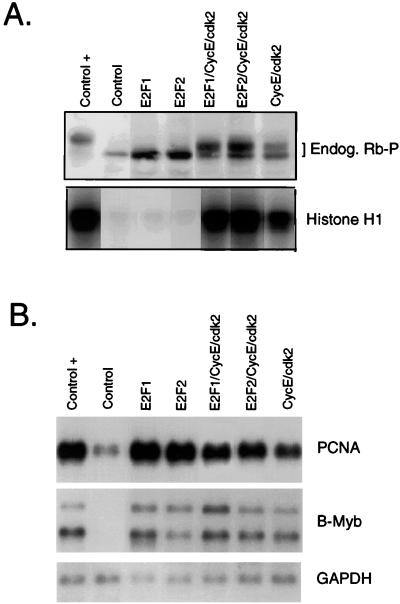
Given previous indications of a role for E2F activity in the induction of S phase from a quiescent state (8, 9), we measured the activation of E2F target genes in cells overexpressing cyclin E/cdk2. As seen in Northern analysis, the production of cyclin E/cdk2 activity resulted in the activation of a number of known E2F target genes (Fig. 2B). In addition, the induction of these genes by cyclin E/cdk2 was comparable to that seen after expression of E2F1 or E2F2 or by the addition of serum. It thus appears clear that cyclin E/cdk2 does lead to an increase in E2F activity, coincident with the induction of S phase. One possible mechanism for the induction of E2F activity would be phosphorylation of Rb or Rb family members, either relieving negative control of E2F activity or eliminating E2F-dependent repression. Indeed, as shown in Fig. 2A, cyclin E/cdk2 overexpression led to phosphorylation of endogenous Rb protein, as indicated by the reduced mobility of the Rb protein in an SDS gel. Although our previous work has suggested an inabil bity of endogenous cyclin E/cdk2 activity to mediate Rb phosphorylation, consistent with recent work suggesting a dependence on prior phosphorylation by a cyclin D-dependent kinase (33), we suspect that the overexpression achieved from the virus infections may at least partially overcome the normal control of phosphorylation.
Figure 2.
Overexpression of cyclin E/cdk2 induces Rb phosphorylation and E2F-dependent transcription. (A) Assay for Rb phosphorylation. Cells treated as in Fig. 1 were harvested in SDS/PAGE loading buffer at 24 hr postinfection, and Rb (Endog. Rb-P) was detected by using Western blotting (Upper). Alternatively, cells treated as in Fig. 1 were lysed in immunoprecipitation buffer, and the lysates were used for immunoprecipitation with antibodies specific for cyclin E, and the cyclin E-associated kinase assays were performed as described (42) by using histone H1 as a substrate (Lower). (B) E2F activity measured by using Northern analysis. Cells treated as in A were harvested at 24 hr postinfection and processed for Northern analysis by using specific probes for proliferating cell nuclear antigen (PCNA), B-Myb, and glyceraldehyde-3-phosphate dehydrogenase.
The fact that expression of either E2F1 or E2F2 activated the expression of E2F target genes to levels similar to those seen in either cells expressing cyclin E/cdk2 alone or in late G1 of serum stimulated cells (Fig. 2B) indicates that the ability of cyclin E/cdk2 to collaborate with E2F is not simply the result of enhancing E2F activity. Rather, this result strongly suggests a role for cyclin E/cdk2 activity that is independent of E2F.
Deregulated E2F Activity Can Suppress a cdk Deficiency.
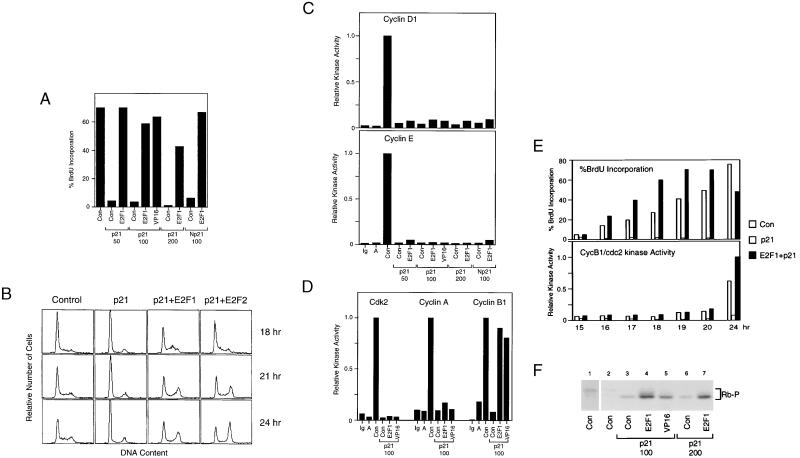
Our previous work has shown that E2F can induce S phase in the absence of G1 cdk activity (17). To further explore the relationship between E2F activity and G1 cdk activity in the induction of S phase, we have directly analyzed the events associated with the ability of E2F to bypass a G1 arrest mediated by the overexpression of the p21 cdk inhibitor. REF52 cells, released from a density arrest, were infected with an Ad-p21 virus (or an Ad-Np21 virus, which expresses a C-terminal deletion mutant of p21 that fails to interact with proliferating cell nuclear antigen) and then assayed for entry into S phase by BrdUrd incorporation. As shown in Fig. 3A, p21 effectively blocked the induction of S phase, and coinfection with an E2F1-expressing adenovirus bypassed the p21-induced block. Moreover, fluorescence-activated cell sorting analysis of the DNA content of cells expressing E2F1 or E2F2 in the presence of p21 revealed that the E2F-mediated bypass led to an accumulation of cells with a G2 content of DNA (Fig. 3B), indicating that replication had occurred. Overexpression of an E2F1–VP16 chimera, which lacks the capacity to bind Rb but retains the transcription activation potential of E2F1 (34), also efficiently bypassed the p21 block (Fig. 3A), indicating that the E2F1-mediated bypass is not simply due to Rb sequestration.
Figure 3.
E2F1 induces S phase in the absence of cdk activity. (A) E2F-mediated S phase induction in p21-arrested cells. REF52 cells were held at density arrest for 24 hr and infected with either Ad-Con (control virus with empty expression cassette) at a moi of 125 ffu per cell, Ad-E2F1 or Ad-E2F1/VP16 (AVP16′′) at an moi of 25 ffu per cell together with Ad-p21 (moi of 50, 100, or 200 ffu per cell as indicated) or Ad-Np21 (moi of 100 ffu per cell), and then replated at subconfluent densities. Cells were labeled with BrdUrd from 12 to 24 hr postinfection, and BrdUrd was detected by using indirect immunofluorescence. At least 300 nuclei were scored for BrdUrd immunofluorescence. (B) E2F induces DNA replication. REF52 cells were held at density arrest for 24 hr before infection with either Ad-Con (moi of 125 ffu per cell), Ad-p21 (moi of 100 ffu per cell), or Ad-p21 and Ad-E2F1 or Ad-E2F2 (moi of 100 and 25 ffu per cell, respectively). Cells were then replated at subconfluent densities, harvested at the indicated times postinfection, and processed for flow cytometry. The horizontal axis reflects relative DNA content and the vertical axis represents cell number. (C) E2F-mediated S phase induction in the absence of cyclin D- or cyclin E-associated kinase activity. Cells treated as in A were lysed in IP buffer, and the lysates were immunoprecipitated with control IgG antibodies (Ig) or antibodies against either cyclin D1 or cyclin E. The sample designated A represents lysates made from density-arrested cells immunoprecipitated with the corresponding specific antibodies. The kinase assays were performed as described (42) except glutathione S-transferase–Rb was used as the substrate. (D) E2F-mediated S phase induction in the absence of cdk2- or cyclin A-associated kinase activity. Cells treated as in A were lysed in IP buffer and the lysates immunoprecipitated with control IgG antibodies (Ig) or antibodies against either cdk2, cyclin A, or cyclin B1. The samples designated A represent lysates made from density-arrested cells immunoprecipitated with the corresponding specific antibodies. The kinase assays were performed as in C. (E) E2F-mediated S phase and cyclin B1-associated kinase induction. Density-arrested REF52 cells were infected with either Ad-Con (moi of 125 ffu per cell; open bar), Ad-p21 (moi of 100 ffu per cell, gray bar), or Ad-p21 and Ad-E2F1 (moi of 100 ffu per cell and 125 ffu per cell, respectively, black bar) and replated at subconfluent densities. Cells were either labeled with BrdUrd for 1 hr before fixation at the indicated times and processed as in A (Upper) or lysed in IP buffer at the indicated times and assayed for cyclin B1-associated kinase activity as in D (Lower). (F) Assay for Rb phosphorylation. Representative cell samples treated as in A were harvested in SDS/PAGE loading buffer, and Rb was detected by using Western blotting. To better visualize the Rb product in lane 2, we show a 3-fold longer exposure of this sample in lane 1.
The E2F1-mediated S phase induction was not accompanied by a restoration of cyclin D1-, cyclin E-, cyclin A-, or cdk2-associated kinase activities (Fig. 3 C and D). In contrast to G1 cdk activity, cyclin B1-dependent cdc2 kinase activity was restored in cells overexpressing E2F1 (Fig. 3D). This observation raised the possibility that the ability of cells to enter S phase, despite the lack of cdk2 activity, was caused by a compensatory action of cdc2. However, careful examination of the timing of DNA replication and cyclin B1-dependent kinase activation revealed that cells entered S phase several hours before the appearance of cyclin B1 kinase activity (Fig. 3E). Although the activation of cyclin B-dependent kinase in the absence of cdk2 activity is surprising in light of the requirement of cdk2 activity for cyclin B/cdc2 activation, (35), this may be the consequence of the E2F-dependent induction of cdc2 (26, 36). In any event, Rb phosphorylation, which normally occurs as cells are stimulated to grow and pass through G1, was completely blocked by p21, and Rb remained hypophosphorylated during the E2F1- or E2F1–VP16-mediated bypass of the p21 block (Fig. 3F). We conclude from this data that E2F overproduction in G1-arrested cells can induce S phase in the absence of detectable G1 cdk activity.
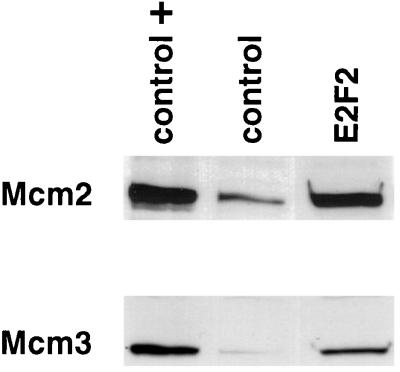
Various experiments suggest that relevant targets for G1 cdk-mediated phosphorylation are components of the replication initiation complex. Moreover, at least one recent report suggests a role for phosphorylation in facilitating the assembly of the pre-replicative complex (37). Because many of the genes encoding the proteins that form the replication initiation complex are known to be E2F targets (10), one possible explanation for the E2F-mediated bypass of a p21 block would be the ability of E2F to produce these proteins, possibly allowing an inefficient assembly of replication complexes. As shown in Fig. 4, the overexpression of E2F1 or E2F2 does indeed lead to an increase in the level of the Mcm2 and Mcm3 proteins, two components of the Mcm complex. Although these are but two examples, the fact that the majority of the genes encoding replication proteins are activated by E2F suggests that it is likely that the various components of the replication apparatus are indeed elevated in E2F-expressing cells.
Figure 4.
E2F overexpression induces Mcm protein accumulation. REF52 cells were starved and infected with Ad-Con (moi of 100 ffu per cell) or Ad-E2F2 (moi of 100 ffu per cell) as described. Cells were then incubated with fresh starvation medium or with medium containing 10% serum (control +) and then harvested 24 hr postinfection. Endogenous Mcm2 and Mcm3 proteins were detected by using Western blot analysis with antibodies specific for each protein.
Inhibition of Cyclin E/cdk2 Does Not Block S Phase Induction in Rb−/− Cells.
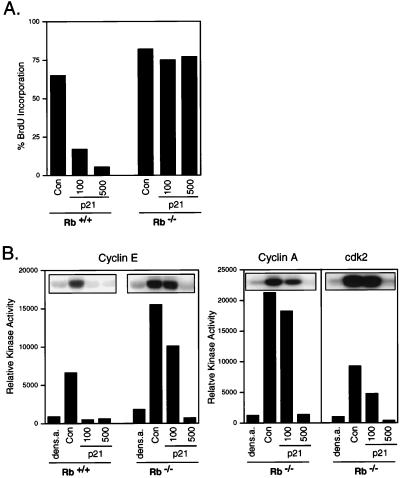
Finally, we have also assessed the ability of cells to proceed into S phase, in the absence of G1 cdk activity, under circumstances where endogenous E2F activity is deregulated as opposed to relying on the overproduction of exogenous protein. In particular, we have compared the effect of the p21 cyclin kinase inhibitor on mouse fibroblasts that are either wild-type for Rb or deficient in Rb. As expected, increasing doses of p21 blocked the ability of Rb+/+ cells to enter S phase, as measured by BrdUrd incorporation (Fig. 5A). In contrast, p21 did not inhibit S phase in the Rb−/− cells, even at the highest dose. In each case, there was a near-complete inhibition of cyclin E or A/cdk2 kinase activity at the higher dose of p21 (Fig. 5B). Thus, in the absence of Rb function, which leads to a loss of E2F control, cells can enter S phase despite the substantial inhibition of cyclin E/cdk2 activity, a result consistent with recent studies indicating an inefficient G1 arrest by p21 in Rb-negative cells (43).
Figure 5.
Inhibition of cyclin E/cdk2 does not block S phase in the absence of Rb. (A) Inhibition of S phase induction by p21. Mouse embryo fibroblasts (MEF) from wild-type (Rb+/+) or knockout (Rb−/−) embryos were held at density arrest for 24 hr and infected with either Ad-Con (control virus with empty expression cassette) at a moi of 125 ffu per cell or Ad-p21 (moi of 100 or 500 ffu per cell as indicated) and then replated at subconfluent densities. Cells were labeled with BrdUrd from 12 to 24 hr postinfection, and BrdUrd was detected by using indirect immunofluorescence. At least 300 nuclei were scored for BrdUrd immunofluorescence. (B) Inhibition of cdk activity by p21. Cells treated as in A or cells density-arrested for 24 hr were lysed in IP buffer and immunoprecipitated with antibodies against either cyclin E, cyclin A, or cdk2. The kinase assays were performed by using glutathione S-transferase–Rb as the substrate.
DISCUSSION
Various studies demonstrate the role for G1 cdk activity in the activation of DNA replication. Likewise, other work has documented a critical role for E2F activity in the activation of S phase, likely the consequence of activation of various genes encoding important DNA replication proteins. Although the relationship of E2F activity to cyclin E/cdk2 is complex, with each having the ability to affect the accumulation of the other, it also is true that the two activities play independent roles in activating S phase. For instance, recent experiments demonstrate that induction of E2F activity in Drosophila embryos is not sufficient to promote S phase in the absence of cyclin E/cdk2 activity (38). Other experiments using mammalian fibroblasts have demonstrated an ability of cyclin E/cdk2 to induce S phase in quiescent cells (39), including in the apparent absence of detectable E2F activity (40). Moreover, Xenopus egg extracts exhibit cell cycles lacking a G1 phase but still require cdk activity to start DNA replication (18–23), strongly implicating a functional interaction between G1 cdks and the replication machinery independent of transcription. These observations have led to the commonly held view that passage through the G1/S checkpoint requires the phosphorylation of a component(s) of the replication complex, thereby triggering the activity of the complex and thus the initiation of DNA replication. Nevertheless, it is also true that E2F can induce S phase in the absence of detectable G1 cdk activity, as shown by the experiments in this paper. Thus, if the initiation of DNA replication requires the phosphorylation of components of the replication complex under some conditions, such as in early Xenopus or Drosophila embryos, how then can initiation occur in the absence of this critical phosphorylation event in other situations? Stated otherwise, how can E2F overexpression suppress the phosphorylation deficiency?
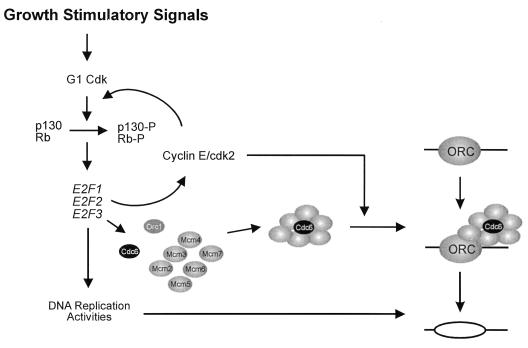
Given the substantial evidence linking cdk-mediated phosphorylation with the initiation of DNA replication, as well as the realization of a role for E2F in the activation of a large number of DNA replication components, including the majority of those that directly participate in replication initiation (10, 31), we propose that a critical E2F target interacts with the replication initiation complex in a G1 cdk-dependent manner but that overproduction of this target can compensate for the phosphorylation requirement. As one example, recent work has shown that the loading of Cdc45 onto chromatin requires cdk-mediated phosphorylation (37). As suggested by the schematic in Fig. 6, we propose that phosphorylation of a component of the ORC complex, or one of the ORC-associated factors such as Cdc6, the Mcm proteins, or Cdc45, might be necessary to allow the interaction of ORC with a protein that is activated by E2F, which could be the phosphorylation target itself, and that is essential for origin function. Under circumstances of significantly reduced levels of cyclin E/cdk2 activity, this interaction would become inefficient and thus would limit DNA replication. In this view, both E2F activity and cyclin E/cdk2 activity are necessary for allowing the initiation of DNA replication. In those instances where transcription is not required for S phase entry as in Xenopus egg extracts, a preexisting pool of the E2F targets would appear to suffice for DNA replication to ensue. It is also true that the role of cdk-mediated phosphorylation may be more complex because recent results from Xenopus in vitro assays suggests a role for cdk activity following the assembly of ORC–Mcm–Cdc6 complexes on chromatin (41).
Figure 6.
Collaboration of cyclin E/cdk2 with E2F in the induction of S phase. The figure depicts the interaction of the multisubunit complex known as ORC with a DNA replication origin. In addition to the Orc1 protein, the accumulation of additional components of the initiation complex, including each of the Mcms and Cdc6, depends on E2F activity. It is proposed that either the assembly of these components into a functional complex or the activation of an assembled complex is facilitated by cyclin E/cdk2-mediated phosphorylation, thereby triggering origin firing and the initiation of DNA replication.
Our experiments also show that the ability of E2F overexpression to induce S phase is reduced in the absence of growth factors. We can envision several possible explanations for this result. First, we cannot exclude the possibility that some G1 cdk activity is present during the E2F-dependent bypass in the presence of serum, which is not detectable in our assays and does not result in detectable Rb phosphorylation. Alternatively, other p21-resistant, growth factor-dependent kinases may compensate for the G1 cdk deficiency. For instance, the Dbf4/cdc7 kinase, which can phosphorylate various components of the origin complex and is required for the initiation of replication, could represent such a kinase. Indeed, it is possible that the formation of the origin complex is promoted by cdk-dependent phosphorylation, such that a deficiency in cdk activity is overcome by producing each of the interacting proteins, and Cdc7/Dbf4 would then play the role of activating an otherwise complete complex. Finally, it is possible that E2F-mediated activation of a critical target(s) becomes more difficult on prolonged growth factor deprivation such that sufficient levels of the target(s) cannot be achieved.
The model proposed in Fig. 6, whereby E2F is responsible for the accumulation of an activity essential for origin function in conjunction with the action of G1 cdk activity, would provide a tight connection between the decision to begin DNA replication and the necessary preparations for DNA replication. Given the fact that E2F appears to be responsible for the accumulation of many of the activities that provide the substrates for DNA replication, as well as various replication accessory factors, a direct role for an E2F target in the initiation event would ensure a coordinated process, with initiation not beginning before the accumulation of these necessary components.
Acknowledgments
We thank Mike Cook and Alan Fisher of the Duke Comprehensive Cancer Center Flow Cytometry Facility for the flow cytometric analysis. We also thank Peter Whyte, S. Van den Heuvel, and Robert Weinberg for the generous gift of plasmids and H. Kimura for a generous gift of the Mcm3 antibody. Finally, thanks to Kaye Culler for assistance in the preparation of the manuscript and Rachel Rempel for providing critical comments. J.D. was supported by a National Institutes of Health postdoctoral fellowship and G.L. by a fellowship from the Medical Research Council of Canada (MRC), J.R.N. is an Investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
- cdk
cyclin-dependent kinase
- ffu
focus-forming units
- moi
multiplicity of infection
References
- 1.Baldin V, Lukas J, Marcote M J, Pagano M, Bartek J, Draetta G. Genes Dev. 1993;7:812–821. doi: 10.1101/gad.7.5.812. [DOI] [PubMed] [Google Scholar]
- 2.Quelle D E, Ashmun R A, Shurtleff S A, Kato J-Y, Bar-Sagi D, Roussel M F, Sherr C J. Genes Dev. 1993;7:1559–1571. doi: 10.1101/gad.7.8.1559. [DOI] [PubMed] [Google Scholar]
- 3.Ohtsubo M, Theodoras A M, Schumacher J, Roberts J M, Pagano M. Mol Cell Biol. 1995;15:2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lukas J, Bartkova J, Rohde M, Strauss M, Bartek J. Mol Cell Biol. 1995;15:2600–2611. doi: 10.1128/mcb.15.5.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lukas J, Parry D, Aagaard L, Mann D J, Bartkov J, Strauss M, Peters G, Bartek J. Nature (London) 1995;375:503–506. doi: 10.1038/375503a0. [DOI] [PubMed] [Google Scholar]
- 6.Koh J, Enders G H, Dynlacht B D, Harlow E. Nature (London) 1995;375:506–510. doi: 10.1038/375506a0. [DOI] [PubMed] [Google Scholar]
- 7.Medema R H, Herrera R E, Lam F, Weinberg R A. Proc Natl Acad Sci USA. 1995;92:6289–6293. doi: 10.1073/pnas.92.14.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu C-L, Classon M, Dyson N, Harlow E. Mol Cell Biol. 1996;16:3698–3706. doi: 10.1128/mcb.16.7.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishizaki J, Nevins J R, Sullenger B. Nat Med. 1996;2:1386–1389. doi: 10.1038/nm1296-1386. [DOI] [PubMed] [Google Scholar]
- 10.Leone G, DeGregori J, Yan Z, Jakoi L, Ishida S, Williams R S, Nevins J R. Genes Dev. 1998;12:2120–2130. doi: 10.1101/gad.12.14.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson D G, Schwarz J K, Cress W D, Nevins J R. Nature (London) 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- 12.Qin X-Q, Livingston D M, Kaelin W G, Adams P D. Proc Natl Acad Sci USA. 1994;91:10918–10922. doi: 10.1073/pnas.91.23.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shan B, Lee W-H. Mol Cell Biol. 1994;14:8166–8173. doi: 10.1128/mcb.14.12.8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kowalik T F, DeGregori J, Schwarz J K, Nevins J R. J Virol. 1995;69:2491–2500. doi: 10.1128/jvi.69.4.2491-2500.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asano M, Nevins J R, Wharton R P. Genes Dev. 1996;10:1422–1432. doi: 10.1101/gad.10.11.1422. [DOI] [PubMed] [Google Scholar]
- 16.Brook A, Xie J-E, Du W, Dyson N. EMBO J. 1996;15:3676–3683. [PMC free article] [PubMed] [Google Scholar]
- 17.DeGregori J, Leone G, Ohtani K, Miron A, Nevins J R. Genes Dev. 1995;9:2873–2887. doi: 10.1101/gad.9.23.2873. [DOI] [PubMed] [Google Scholar]
- 18.Blow J J, Nurse P. Cell. 1990;62:855–862. doi: 10.1016/0092-8674(90)90261-c. [DOI] [PubMed] [Google Scholar]
- 19.Fang F, Newport J W. Cell. 1991;66:731–742. doi: 10.1016/0092-8674(91)90117-h. [DOI] [PubMed] [Google Scholar]
- 20.Strausfeld U P, Howell M, Rempel R, Maller J L, Hunt T, Blow J J. Curr Biol. 1994;4:876–883. doi: 10.1016/s0960-9822(00)00196-2. [DOI] [PubMed] [Google Scholar]
- 21.Chen J, Jackson P K, Kirschner M W, Dutta A. Nature (London) 1995;374:386–388. doi: 10.1038/374386a0. [DOI] [PubMed] [Google Scholar]
- 22.Yan H, Newport J. J Cell Biol. 1995;129:1–15. doi: 10.1083/jcb.129.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adachi Y, Laemmli U K. EMBO J. 1994;13:4153–4164. doi: 10.1002/j.1460-2075.1994.tb06733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwarz J K, Bassing C H, Kovesdi I, Datto M B, Blazing M, George S, Wang X-F, Nevins J R. Proc Natl Acad Sci USA. 1995;92:483–487. doi: 10.1073/pnas.92.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nevins J R. Methods Enzymol. 1980;65:768–785. doi: 10.1016/s0076-6879(80)65072-1. [DOI] [PubMed] [Google Scholar]
- 26.DeGregori J, Kowalik T, Nevins J R. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeGregori J, Leone G, Miron A, Jakoi L, Nevins J R. Proc Natl Acad Sci USA. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van den Heuvel S, Harlow E. Science. 1993;262:2050–2054. doi: 10.1126/science.8266103. [DOI] [PubMed] [Google Scholar]
- 29.Hinds P W, Mittnacht S, Dulic V, Arnold A, Reed S I, Weinberg R A. Cell. 1992;70:993–1006. doi: 10.1016/0092-8674(92)90249-c. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Graham C, Lacy S, Duncan A M V, Whyte P. Genes Dev. 1993;7:2366–2377. doi: 10.1101/gad.7.12a.2366. [DOI] [PubMed] [Google Scholar]
- 31.Ohtani K, DeGregori J, Leone G, herendeen D R, Kelly T J, Nevins J R. Mol Cell Biol. 1996;16:6977–6984. doi: 10.1128/mcb.16.12.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ikeda M-A, Jakoi L, Nevins J R. Proc Natl Acad Sci USA. 1996;93:3215–3220. doi: 10.1073/pnas.93.8.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lundberg A S, Weinberg R A. Mol Cell Biol. 1998;18:753–761. doi: 10.1128/mcb.18.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson D G, Cress W D, Jakoi L, Nevins J R. Proc Natl Acad Sci USA. 1994;91:12823–12827. doi: 10.1073/pnas.91.26.12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guadagno T M, Newport J W. Cell. 1996;84:73–82. doi: 10.1016/s0092-8674(00)80994-0. [DOI] [PubMed] [Google Scholar]
- 36.D’Urso G, Marraccino R L, Marshak D R, Roberts J M. Science. 1990;250:786–791. doi: 10.1126/science.2173140. [DOI] [PubMed] [Google Scholar]
- 37.Zou L, Stillman B. Science. 1998;280:593–596. doi: 10.1126/science.280.5363.593. [DOI] [PubMed] [Google Scholar]
- 38.Duronio R J, Brook A, Dyson N, O’Farrell P H. Genes Dev. 1996;10:2505–2513. doi: 10.1101/gad.10.19.2505. [DOI] [PubMed] [Google Scholar]
- 39.Connell-Crowley L, Elledge S J, Harper J W. Curr Biol. 1998;8:65–68. doi: 10.1016/s0960-9822(98)70021-1. [DOI] [PubMed] [Google Scholar]
- 40.Lukas J, Herzinger T, Hansen K, Moroni M C, Resnitzky D, Helin K, Reed S I, Bartek J. Genes Dev. 1997;11:1479–1492. doi: 10.1101/gad.11.11.1479. [DOI] [PubMed] [Google Scholar]
- 41.Hua X H, Newport J. J Cell Biol. 1998;140:271–281. doi: 10.1083/jcb.140.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeGregori J, Leone G, Ohtani K, Miron A, Nevins J R. Genes Dev. 1995;9:2873–2887. doi: 10.1101/gad.9.23.2873. [DOI] [PubMed] [Google Scholar]
- 43.Nicolescu A B, Chen X, Smeets M, Hengst L, Prives C, Reed S I. Mol Cell Biol. 1998;18:629–643. doi: 10.1128/mcb.18.1.629. [DOI] [PMC free article] [PubMed] [Google Scholar]