Abstract
The pyrimidine(6–4)pyrimidone photoproduct [(6–4) adduct] is one of the major photoproducts induced by UV irradiation of DNA and occurs at TpT sites. The (6–4) adduct is highly mutagenic and leads most often to a 3′ T → C transition with 85% replicating error frequency [LeClerc, J. E., Borden, A. & Lawrence, C. W. (1991) Proc. Natl. Acad. Sci. USA 88, 9685–9689]. To determine the origin of the specific 3′ T → C transition of the (6–4) adduct, we have used experimental NMR restraints and molecular dynamics to determine the solution structure of a (6–4)-lesion DNA decamer duplex that contains a mismatched base pair between the 3′ T residue and an opposed G residue. Normal Watson–Crick-type hydrogen bonding is retained at the 5′ T of the lesion site. The O2 carbonyl of the 3′ T residue forms hydrogen bonds with the imino and amino protons of the opposed G residue. This potential hydrogen bonding stabilizes the overall helix and restores the highly distorted conformation of the (6–4) adduct to the typical B-form-like DNA structure. This structural feature can explain the marked preference for the insertion of an A residue opposite the 5′ T and a G residue opposite the 3′ T of the (6–4) lesion during trans-lesion synthesis. Thus these insertions yield the predominant 3′ T → C transition.
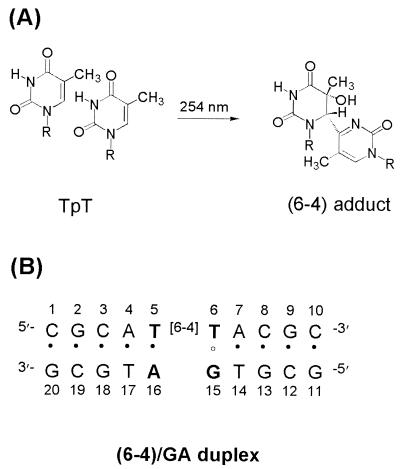
UV light irradiation of DNA produces a variety of photoproducts that cause mutations to be introduced in the DNA and ultimately the development of various cancers (1–4). The cis-syn cyclobutane pyrimidine dimer (cis-syn dimer) and pyrimidine(6–4)pyrimidone photoproduct [(6–4) adduct] (Fig. 1A) constitute the two major classes of UV-induced DNA photoproducts and occur at TpT sites in the DNA (5, 6). The cis-syn dimer, which is the most abundant photoproduct, has a much lower repair rate for the DNA repair enzymes than does the (6–4) adduct. The cis-syn dimer undergoes excision repair in vivo with a half-life of ≈24 hr, whereas the (6–4) adduct is repaired rapidly with a half-life of ≈4 hr (7). In vitro, the cis-syn dimer is repaired about nine times slower than the (6–4) adduct by the Escherichia coli uvr(A)BC endonuclease (8). The binding affinities of the DNA damage binding proteins (for example, uvrA subunit, human damaged DNA binding protein, and human replicating protein A) for the cis-syn dimer are also much lower than those for the (6–4) adduct (9, 10). However, cis-syn dimer displays a very low mutagenic rate (11–14). Structural studies of a DNA decamer duplex containing a cis-syn dimer suggest that the formation of Watson–Crick base pairs between the two T residues and the corresponding A residues could explain the low mutagenecity of the dimer (15).
Figure 1.
Chemical structures analyzed in this study. (A) Chemical structure of the (6–4) adduct. (B) DNA sequence context of the (6–4)/GA duplex.
Although the T-T (6–4) adduct is collectively 5- to 10-fold less abundant in UV-irradiated DNA and is more rapidly repaired than the cis-syn dimer, the (6–4) adduct is highly mutagenic and yields a specific mutation (13, 14, 16). In SOS-induced E. coli cells, adenine was found to be incorporated into the site opposite the 5′ T of the (6–4) adduct with a frequency of 95% (16). However, for the 3′ T, A was incorporated only 11% of the time, whereas G was incorporated 85% of the time (16). The marked preference for the insertion of G opposite the 3′ T of (6–4) adducts during trans-lesion DNA synthesis leads to a predominant 3′ T → C transition with 85% replicating error frequency (16). Structural studies of a DNA decamer duplex containing a (6–4) adduct opposite two A residues revealed that the O2 carbonyl of the 3′ T could not form a hydrogen bond with the amino proton of the opposite A residue (17). This result explains the high frequency of mutation occurring at the 3′ T site, but does not account for the origin of the specific 3′ T → C transition at the (6–4) lesion. A structural model that could explain the high mutagenic specificity of the (6–4) adduct was proposed (14, 16, 18). This model structure is based on base-pairing studies and thermodynamic data and involves a hydrogen bonding interaction between the 3′ T and opposing G residue. However, there have been no reports of a single-crystal or solution structure of a DNA duplex containing a (6–4) adduct in mutated sequence context. These types of structural studies thus are required to gain support for the proposed model of hydrogen bonding.
We have determined the three-dimensional solution structure of a DNA decamer duplex that contains a mismatched base pair between the 3′ T of the (6–4) adduct and an opposed G residue [designated by the (6–4)/GA duplex, Fig. 1B]. In this paper, the conformational influence of the 3′ T⋅G base pair in the (6–4)/GA duplex has been compared in detail with that of the 3′ T⋅A base pair in the (6–4)/AA duplex, which was established in our previous work (17). This structural comparison provides insight into the mechanism that determines base selection during trans-lesion synthesis and can account for the origin of the specific 3′ T → C transition of the (6–4) adduct.
MATERIALS AND METHODS
Sample Preparation.
The (6–4) adduct-containing DNA decamer was prepared by direct 254-nm UV irradiation of DNA oligomers in aqueous solution and purified as described (17, 19). The (6–4) adduct in the DNA decamer could be characterized by observing UV absorption band at 325 nm, and its homogeneity could be identified by using 1H-NMR spectroscopy. The (6–4)/GA duplex was prepared by dissolving the lesion-containing and complementary oligonucleotide strands (adjusted to a stoichiometric 1:1 ratio) in an aqueous solution containing 10 mM sodium phosphate (pH 6.6) and 200 mM NaCl.
NMR Experiments.
All NMR data sets generated with the (6–4)/GA duplex were collected with a Bruker DMX-600 spectrometer (Korea Basic Science Institute, Taejon). Details of the NMR experiments and data processing can be found in our earlier published studies of a photoproduct-containing DNA duplex (17, 19). Nuclear Overhauser effect (NOE) distance restraints from nonexchangeable protons were obtained from two-dimensional NOE spectroscopy (NOESY) experiments with mixing times of 50, 80, 160, and 300 ms in a D2O buffer solution. Exchangeable proton NOEs were determined by using NOESY spectra in H2O buffer with 120- and 250-ms mixing times. Watson–Crick-type hydrogen bonding restraints were imposed in each base pair, except the T6⋅G15 base pair.
Structure Calculation.
The structure of the (6–4)/GA duplex was calculated by using the program x-plor version 3.1 (20) with restrained molecular dynamics. We initially generated the normal A- and B-form starting structures with modification of the (6–4) adduct at the T5-T6 position. These structures were subjected to the restrained molecular dynamics and simulated annealing protocol. The first stage of computation began with energy minimization, followed by 10 ps of molecular dynamics at 800 K. The force constants for the distance restraints gradually were increased over 10 cycles of 1-ps dynamics. The final values of the force constants were 100 kcal mol−1⋅Å−2. The system subsequently was cooled to 300 K over 10 cycles of 0.5-ps dynamics followed by energy minimization. The final stage involved 15 ps of the restrained molecular dynamics at 300 K, and the structures subsequently were energy minimized. Twenty structures (14 of the B-form and six of the A-form initial structures) were chosen on the basis of the lowest NOE violations and total energies.
The averaged structure of the distance-refined structures next was optimized by using full relaxation matrix refinement (based on NOE intensity) with x-plor. NOE volumes from 277 cross peaks for each of the three mixing times of 50, 80, and 160 ms were used as restraints. Dynamics was undertaken at 300 K over 10 ps by using different seeds for initial velocity assignments. Ten runs of the calculations yielded well-converged structures. The helical parameters of the refined structures were calculated by using the program curves (21).
RESULTS
NMR Resonance Assignment.
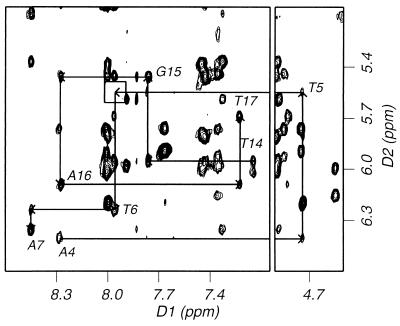
The nonexchangeable base and sugar protons of the nucleotides were assigned according to their intraresidue and sequential NOE connectivities. An example of the sequential NOE connectivities between base protons and their own and 5′ flanking sugar H1′ protons is outlined in an expanded NOESY contour plot in a D2O buffer solution (Fig. 2). Saturation of the 5–6 double bond of the T5 base by forming the (6–4) adduct led to the up-field shift of its H6 resonances (4.75 ppm). No sequential NOE cross peak was observed between T5-H1′ and T6-H6 protons (boxed in Fig. 2), indicating a substantial structural change in the vicinity of the (6–4) lesion.
Figure 2.
Expanded NOESY (300-ms mixing time) contour plot of the (6–4)/GA duplex in a D2O solution containing 10 mM sodium phosphate (pH 6.6) and 200 mM NaCl at 1°C. Shown is the sequential NOE connectivities between base protons and their own and 5′ flanking sugar H1′ protons for A4 to A7 and T14 to T17 (the central four base pairs).
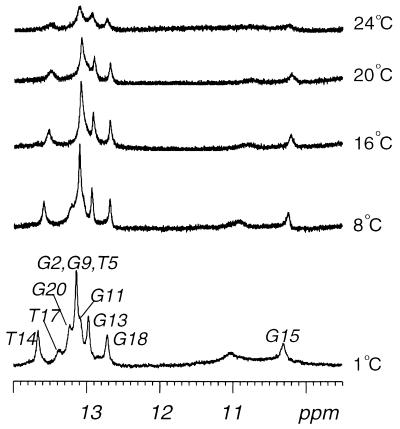
The exchangeable protons were assigned by analyzing NOESY data in an H2O buffer solution. The temperature-dependent imino proton spectra (9.5–14 ppm) in H2O buffer is plotted in Fig. 3. The most striking feature of these spectra is the persistence of the imino proton resonance at 10.30 ppm (assigned to G15) for experiments performed at 1–20°C, whereas all imino resonances of the (6–4)/AA duplex disappeared at 10°C (17). This result indicates that the greater stability achieved by substituting a G for an A opposite the 3′ T site of the (6–4) adduct is likely to result from the stabilizing hydrogen bonding between the 3′ T and the imino proton of the opposite G residue.
Figure 3.
Temperature dependence of the imino proton resonances of the 1H-NMR spectra for the (6–4)/GA duplex in an H2O buffer solution containing 10 mM sodium phosphate (pH 6.6) and 200 mM NaCl. The positions of nucleotides in the decamer duplex that give rise to the resonances are indicated. The experimental temperatures are shown on the right.
Structure Determination.
The solution structure of the (6–4)/GA duplex was calculated according to the protocols outlined in Materials and Methods. A total of 334 distance restraints were derived from NOE cross peaks. In addition, a number of 81 distance restraints for hydrogen bonds were imposed on Watson–Crick base pairs; however, neither base-pair planarity nor backbone dihedral angles were restrained. A converged subset of 20 structures refined by the restrained molecular dynamics were identified on the basis of low NOE violations and total energies. These structures exhibited pair-wise rms deviation (rmsd) values of 1.06 ± 0.35 Å for all heavy atoms (Table 1). Full relaxation matrix refinements of the averaged structure yielded a well-converged subset of ten refined structures. Ten superimposed refined structures of the (6–4)/GA duplex are plotted in Fig. 4A and exhibit pair-wise rmsd values of 0.41 ± 0.18 Å for all heavy atoms (Table 1).
Table 1.
Structure refinement statistics and analysis of (6-4)/GA–duplex
| Restrained molecular dynamics | |
| Number of distance restraints | 415 |
| Number of accepted structures | 20 |
| Pairwise rmsd, Å | 1.06 ± 0.35 |
| NOE violation energy | 212.20 ± 11.82 |
| Number NOE violations | |
| Greater than 0.3 Å | 0.40 ± 0.58 |
| Greater than 0.2 Å | 14.20 ± 1.53 |
| rmsd from restraints | 0.0715 ± 0.0020 |
| Relaxation matrix refinement | |
| Number of intensity restraints | 277 × 3 |
| Number of accepted structures | 10 |
| Pairwise rmsd, Å | 0.41 ± 0.18 |
| R1/6-factor | 0.0502 ± 0.0002 |
| x-plor energies, kcal/mol | |
| Total | −298.43 ± 2.56 |
| Bond length | −111.64 ± 1.08 |
| Bond angle | 452.13 ± 2.36 |
| Dihedral angle | 180.15 ± 1.69 |
| Improper angle | 22.55 ± 0.08 |
| Hydrogen bond | −177.81 ± 0.89 |
| Van der Waals | 1.86 ± 0.90 |
| Electrostatic | −1,209.33 ± 3.80 |
| Relaxation | 302.63 ± 1.37 |
| rmsd from ideal geometry | |
| Bond length | 0.0235 ± 0.0001 |
| Bond angle | 5.2405 ± 0.0218 |
| Improper angle | 6.1746 ± 0.0151 |
Figure 4.
Solution structure of the (6–4)/GA duplex. (A) Superimposed stereo view of 10 intensity refined structures of the (6–4)/GA duplex. (B) Stick view of the central four base pairs of the (6–4)/GA duplex. The sugar and backbone atoms are magenta. Two thymine bases (T5, T6) of the (6–4) adduct are red, and their opposite G15 and A16 bases are yellow and green, respectively. The flanking base pairs are cyan. The hydrogen atoms are excluded.
Structural calculation showed that the (6–4) covalent linkage did cause a significant bend in DNA helix of the (6–4)/GA duplex (overall bending of 27.4 ± 1.5°). DNA unwinding was not observed in the (6–4)/GA duplex, so that the helical periodicity is 10.09 ± 0.09 bp per turn. However, the DNA helix was unwound by ≈30° at the (6–4) lesion site in the (6–4)/AA duplex. This finding indicates that the 3′ T⋅G base pairing of the (6–4) lesion reverses the helical unwinding rather than the bending caused by forming the (6–4) lesion.
Conformational Feature at the (6–4) Lesion Site.
The T5⋅A16 base pair of the (6–4) lesion in the (6–4)/GA duplex showed Watson–Crick base pairing like the (6–4)/AA duplex; however, judging from the weak NOE cross peak between the A16-H2 and T5-imino proton, the structure of the 5′ T⋅A base pair is distorted (Fig. 5). The calculated structure of the (6–4)/GA duplex shows that the T6⋅G15 base pair is stabilized by hydrogen bonds that form between the imino and amino protons of G15 and the O2 carbonyl of T6 (heteroatom separations of 3.09 ± 0.10 and 2.79 ± 0.03 Å, respectively), and base stacking between the G15 and A16 bases opposite the (6–4) adduct (Fig. 5A). The hydrogen bonding feature in the T6⋅G15 base pair is supported by the weak NOE cross peak between the imino and hydrogen-bonded amino proton (7.84 ppm) of G15. Solution structure also showed that the lone pair on N3 of T6 is not accessible for hydrogen bonding. This result is in contrast to the proposed base-pairing scheme (16), in which the lone pair of N3 plays a role. The solution structure of the (6–4)/AA duplex showed that the hydrogen bonding between the O2 carbonyl of T6 and the A16 amino proton and base stacking between the A15 and A16 bases are unfavorable, so that an interior loop structure is formed at the 3′ T site of the (6–4) adduct (Fig. 5B). This observation would account for the low thermal stability of the (6–4)/AA duplex (22). The disappearance of the interior loop structure as results of the T6⋅G15 base pairing can account for the slow exchange of the G15 imino proton and the greater thermal stability of the (6–4)/GA duplex. This thermal stability is in agreement with thermodynamics studies (18, 22).
Figure 5.
Comparative ball and stick view of two base pairs of the (6–4) adduct (A) in the (6–4)/GA duplex and (B) in the (6–4)/AA duplex. Balls are colored by using the accepted atomic color representation: white, hydrogen; green, carbon; blue, nitrogen; red, oxygen. The dotted lines indicate the hydrogen bonds determined by the program insight ii.
The glycosidic bond torsion angles of both T residues (T5: χ = −91.0 ± 0.4°; T6: χ = −48.5 ± 2.9°) of the (6–4) lesion were similar to those in the (6–4)/AA duplex. However, sugar puckers of these residues were significantly different in the two duplexes. The C1′-exo sugar pucker (pseudo-rotation P = 139.8 ± 0.8°) at T5 and the C4′-endo (P = 223.6 ± 3.3°) at the T6 were observed in the (6–4)/GA duplex, whereas both T residues in the (6–4)/AA duplex adopt N-type sugar puckers. Although backbone conformation at T6-A7 is severely distorted in the (6–4)/AA duplex, the corresponding region of the (6–4)/GA duplex did not deviate from the B-DNA conformation. This structural feature is consistent with the observation of the sequential NOEs between A7-H8 and the T6 sugar protons in the (6–4)/GA duplex. Such NOEs are absent from the structural analysis of the (6–4)/AA duplex (23). This result implies that the 3′ T⋅G base pair diminishes dramatically the distortion of the backbone conformation between the (6–4) lesion and its 3′ flanking region.
The G15 and A16 bases opposite the (6–4) adduct showed distinct stacking features with respect to their adjacent bases (Fig. 4B). The T14-G15 step was distorted in the stacking interaction, whereas the A16-T17 step was well stacked. Nonplanarity and the high propeller twist of the T5 base caused a significant disruption of the A4⋅T17 base pair (base pair buckle of 18.5 ± 1.4°, propeller twist of 12.0 ± 1.2°, and opening of −12.0 ± 0.6°). This destabilization was confirmed by the observed temperature dependence of the imino resonance (Fig. 3) and the absence of a diagonal peak of the T17 imino proton in 250-ms NOESY spectrum in H2O buffer. Taken together, these results indicate that the 3′ T⋅G base pairing of the (6–4) adduct increases the thermal stability of the DNA helix, especially the 3′ flanking region of the (6–4) lesion.
DISCUSSION
The (6–4) adduct is both highly mutagenic (error frequency of >91%) and highly specific with respect to the mutations induced (3′ T → C transition in >93% of case) under SOS conditions in E. coli (16). It has been suggested that the nucleotide substitution induced by the (6–4) adduct results from the misinstructive rather than noninstructive property of the distorted template (12). The term misinstructive is used to indicate the existence of physical interactions between a template and an incoming dNTP (16). Our structural studies reveal that these physical interactions play a crucial role in determining the nature of the dNTP incorporated opposite the 3′ T of the (6–4) lesion. Specifically, we propose that the potential hydrogen bonding at the 3′ T⋅G base pair explains in part the high specificity and frequency of mutations induced by the (6–4) adduct. We also found that the 3′ T⋅G base pair increases the thermal stability of the duplex region (formed between the primer and the damaged template) and diminishes the conformational distortion of the template [produced between the (6–4) lesion and its 3′ flanking side]. The greater thermal stability and lower distortion of the template facilitate incorporation of a G residue opposite the 3′ T during trans-lesion synthesis. Therefore, we conclude that the 3′ T → C transition is a misinstructive mutation caused by the important physical features described above.
The 5′ T of the (6–4) lesion has been shown to encode primarily a T residue (95%) (16), whereas the corresponding abasic site (the prototypical noninstructional lesion) encodes a T residue with much lower specificity (77%) (24). This observation confirms the idea that the 5′ T of the (6–4) lesion is instructive rather than noninstructive and is supported by structural studies of the (6–4) adduct-containing DNA duplexes that have either a 3′ T⋅A base pair (17) or a 3′ T⋅G base pair (herein). These structural studies reveal that the 5′ T maintains standard Watson–Crick hydrogen bonding interactions with an A residue in the complementary strand.
The (6–4) adducts are repaired about nine times faster than cis-syn dimers by the E. coli uvr(A)BC endonuclease (8). These excision rates correlate with the photoproduct binding affinities of the uvrA subunit [DNA binding subunit of the E. coli uvr(A)BC endonuclease], which binds the (6–4) adduct- and cis-syn dimer-containing DNA duplexes with relative binding affinities of 9:1 (9). It has been suggested that binding of the uvrA subunit to damaged DNA depends on the greater degree of DNA unwinding caused by the lesions (25). This hypothesis is supported by the observed high binding affinity of uvrA for the psoralen-thymine monoadduct (9), which causes unwinding of the DNA duplex, but does not cause bending (26). However, human damaged DNA-binding protein (DDB), which binds to DNA photoproducts with an affinity similar to that of the uvrA subunit, has no measurable affinity for the psoralen-thymine monoadduct (9). This finding implies that the binding of human DDB to damaged DNA does not depend on the amount of DNA unwinding caused by the lesion. It is reported that the activity of the human excinuclease for the (6–4) adducts is not affected by substitution of a G residue opposite the 3′ T site (27). The structural basis of absent helical unwinding in our model of the (6–4)/GA duplex suggests that the binding affinity of human DDB and repair rate of human excinuclease for the DNA damage is not correlated with the helical unwinding caused by the lesion but some other structural features.
In summary, by using experimental NMR restraints and molecular dynamics, we have demonstrated the structural features unique to the 3′ T⋅G base pair of the (6–4)/GA duplex in solution. The O2 carbonyl of the 3′ T of the (6–4) lesion forms the stable hydrogen bonds with the imino and amino protons of an opposed G. This stable hydrogen bonding increases the thermal stability of the overall helix and restores the highly distorted conformation of the (6–4) lesion site in the (6–4)/AA duplex to the typical B-form-like structure. This result demonstrates that the greater stability of G over A opposite the 3′ T of the (6–4) lesion may facilitate the misincorporation of a G during trans-lesion synthesis and lead to the highly specific 3′ T → C mutation at this site.
Acknowledgments
We thank Dr. Chaejoon Cheong for helpful discussions. This work was supported by the Korea Science and Engineering Foundation (Grant 981-0304-024-2) and the Korea Science and Engineering Foundation through the Center for Molecular Catalysis at Seoul National University.
ABBREVIATIONS
- (6–4) adduct
pyrimidine(6–4)pyrimidone photoproduct
- cis-syn dimer
cis-syn cyclobutane pyrimidine dimer
- NOE
nuclear Overhauser effect
- NOESY
NOE spectroscopy
- rmsd
rms deviation
Footnotes
Data deposition: The atomic coordinate of the (6–4)/GA duplex has been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1cfl).
References
- 1.Brash D E. Photochem Photobiol. 1988;48:59–66. doi: 10.1111/j.1751-1097.1988.tb02786.x. [DOI] [PubMed] [Google Scholar]
- 2.Mitchell D L. Photochem Photobiol. 1988;48:51–57. doi: 10.1111/j.1751-1097.1988.tb02785.x. [DOI] [PubMed] [Google Scholar]
- 3.Ananthaswamy H N, Pierceall W E. Photochem Photobiol. 1990;52:1119–1136. doi: 10.1111/j.1751-1097.1990.tb08452.x. [DOI] [PubMed] [Google Scholar]
- 4.Brash D E, Rudolph J A, Simon J A, Lin A, McKenna G J, Baden H P, Halperin A J, Potén J. Proc Natl Acad Sci USA. 1991;88:10124–10128. doi: 10.1073/pnas.88.22.10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitchell D L, Nairn R S. Photochem Photobiol. 1989;49:805–819. doi: 10.1111/j.1751-1097.1989.tb05578.x. [DOI] [PubMed] [Google Scholar]
- 6.Pfeifer G P. Photochem Photobiol. 1997;65:270–283. doi: 10.1111/j.1751-1097.1997.tb08560.x. [DOI] [PubMed] [Google Scholar]
- 7.Mitchell D L, Haipek C A, Clarkson J M. Mutat Res. 1985;143:109–112. doi: 10.1016/s0165-7992(85)80018-x. [DOI] [PubMed] [Google Scholar]
- 8.Reardon J T, Nichols A F, Keeney S, Smith C A, Taylor J-S, Linn S, Sancar A. J Biol Chem. 1993;268:21301–21308. [PubMed] [Google Scholar]
- 9.Svoboda D L, Smith C A, Taylor J-S, Sancar A. J Biol Chem. 1993;268:10694–10700. [PubMed] [Google Scholar]
- 10.Burns J L, Guzder S N, Sung P, Prakash S, Prakash L. J Biol Chem. 1996;271:11607–11610. doi: 10.1074/jbc.271.20.11607. [DOI] [PubMed] [Google Scholar]
- 11.Banerjee S K, Christensen R B, Lawrence C W, LeClerc J E. Proc Natl Acad Sci USA. 1988;85:8141–8145. doi: 10.1073/pnas.85.21.8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibbs P E M, Kilbey B J, Banerjee S K, Lawrence C W. J Bacteriol. 1993;175:2607–2612. doi: 10.1128/jb.175.9.2607-2612.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibbs P E M, Borden A, Lawrence C W. Nucleic Acids Res. 1995;23:1919–1922. doi: 10.1093/nar/23.11.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith C A, Wang M, Jiang N, Che L, Zhao X, Taylor J-S. Biochemistry. 1996;35:4146–4154. doi: 10.1021/bi951975c. [DOI] [PubMed] [Google Scholar]
- 15.Kim J-K, Patel D J, Choi B-S. Photochem Photobiol. 1995;62:44–50. doi: 10.1111/j.1751-1097.1995.tb05236.x. [DOI] [PubMed] [Google Scholar]
- 16.LeClerc J E, Borden A, Lawrence C W. Proc Natl Acad Sci USA. 1991;88:9685–9689. doi: 10.1073/pnas.88.21.9685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J-K, Choi B-S. Eur J Biochem. 1995;228:849–854. doi: 10.1111/j.1432-1033.1995.tb20331.x. [DOI] [PubMed] [Google Scholar]
- 18.Fujiwara Y, Iwai S. Biochemistry. 1997;36:1544–1550. doi: 10.1021/bi9619942. [DOI] [PubMed] [Google Scholar]
- 19.Lee J-H, Hwang G-S, Kim J-K, Choi B-S. FEBS Lett. 1998;428:269–274. doi: 10.1016/s0014-5793(98)00508-0. [DOI] [PubMed] [Google Scholar]
- 20.Brünger A T. x-plor. New Haven, CT: Yale Univ. Press; 1992. , Version 3.1. [Google Scholar]
- 21.Lavery R, Sklenar H. J Biomol Struct Dyn. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 22.Jing Y, Kao J F-L, Taylor J-S. Nucleic Acids Res. 1998;26:3845–3853. doi: 10.1093/nar/26.16.3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hwang G-S, Kim J-K, Choi B-S. Eur J Biochem. 1996;235:359–365. doi: 10.1111/j.1432-1033.1996.00359.x. [DOI] [PubMed] [Google Scholar]
- 24.Jiang N, Taylor J-S. Biochemistry. 1993;32:472–481. doi: 10.1021/bi00053a011. [DOI] [PubMed] [Google Scholar]
- 25.Oh E Y, Grossman L. Nucleic Acids Res. 1986;14:8557–8571. doi: 10.1093/nar/14.21.8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spielmann H P, Dwyer T J, Sastry S S, Hearst J E, Wemmer D E. Proc Natl Acad Sci USA. 1995;92:2345–2349. doi: 10.1073/pnas.92.6.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mu D, Tursun M, Duckett D R, Drummond J T, Mondrich P, Sancar A. Mol Cell Biol. 1997;17:760–769. doi: 10.1128/mcb.17.2.760. [DOI] [PMC free article] [PubMed] [Google Scholar]