Abstract
The nuclear pore complex (NPC) and its relationship to the nuclear envelope (NE) was characterized in living cells using POM121–green fluorescent protein (GFP) and GFP-Nup153, and GFP–lamin B1. No independent movement of single pore complexes was found within the plane of the NE in interphase. Only large arrays of NPCs moved slowly and synchronously during global changes in nuclear shape, strongly suggesting mechanical connections which form an NPC network. The nuclear lamina exhibited identical movements. NPC turnover measured by fluorescence recovery after photobleaching of POM121 was less than once per cell cycle. Nup153 association with NPCs was dynamic and turnover of this nucleoporin was three orders of magnitude faster. Overexpression of both nucleoporins induced the formation of annulate lamellae (AL) in the endoplasmic reticulum (ER). Turnover of AL pore complexes was much higher than in the NE (once every 2.5 min). During mitosis, POM121 and Nup153 were completely dispersed and mobile in the ER (POM121) or cytosol (Nup153) in metaphase, and rapidly redistributed to an immobilized pool around chromatin in late anaphase. Assembly and immobilization of both nucleoporins occurred before detectable recruitment of lamin B1, which is thus unlikely to mediate initiation of NPC assembly at the end of mitosis.
Keywords: nuclear pore complex; nuclear envelope; confocal microscopy; FRAP; live cell imaging
Introduction
The nuclear envelope (NE)* creates distinct nuclear and cytoplasmic compartments in eukaryotic cells. It consists of two concentric membranes in direct continuity with the ER (Gerace and Burke, 1988). In higher eukaryotes, the NE is stabilized by the nuclear lamina, a tight meshwork of intermediate filament proteins underlying the inner nuclear membrane (for review see Gruenbaum et al., 2000). The double membrane is perforated by nuclear pores, large protein complexes that form aqueous channels and create the only connection between inner and outer nuclear membrane. Biochemical and genetic studies have contributed evidence for molecular interactions between all major structural components of the NE (membranes, pores, lamins, and heterochromatin), suggesting the nuclear periphery is a highly cross-linked system (for review see Goldberg and Allen, 1995; Wilson, 2000).
The nuclear pore complex (NPC) is the best studied component of the NE. In embryonic and cancer cells, pore complexes are also found in cytoplasmic annulate lamellae (AL), dense stacks of ER-derived membranes containing pores in high densities (for review see Kessel, 1992). The function of AL is unclear to date. A depot for excess amounts of nucleoporins before their degradation has been suggested for Drosophila embryos (Stafstrom and Staehelin, 1984). Alternatively, in embryogenesis AL could serve as a vital reservoir of maternal nucleoporins required for subsequent cell generations (Cordes et al., 1995) and an analogous function can be envisaged in rapidly growing cells. Immunologically, AL pore complexes (ALPCs) are very similar to NPCs and even nucleocytoplasmic transport factors are targeted to AL (Cordes et al., 1997). Pore complex structure has been determined by electron microscopy and reviewed extensively (Pante and Aebi, 1995; Allen et al., 2000). From a central spoke ring embedded in the nuclear membranes emanate eight cytoplasmic and nuclear filaments, the latter joined by a distal ring to form the nuclear basket. A wealth of biochemical and genetic data on the protein subunits (nucleoporins) of NPCs has become available recently (Belgareh and Doye, 1999; Ryan and Wente, 2000), producing a basically complete inventory of yeast (Rout et al., 2000) and a steadily growing list of vertebrate nucleoporins.
Despite extensive information about the molecular properties of the NPC, little is known about its dynamic characteristics in living cells. In vivo studies have only begun to characterize the dynamics of some NE components, such as the nuclear membrane (Ellenberg et al., 1997; Haraguchi et al., 2000) and nuclear lamins (Broers et al., 1999; Moir et al., 2000), with most emphasis on postmitotic assembly of the nucleus (for review see Collas and Courvalin, 2000). The only in vivo data on the dynamics of the NPC is from budding yeast and suggests a high mobility of NPCs in the NE (Belgareh and Doye, 1997; Bucci and Wente, 1997).
Here, we analyze the NPC in intact mammalian cells. Two well-characterized nucleoporins, POM121 and Nup153, were tagged with multiple copies of GFP to visualize them at low, nontoxic expression levels. POM121 is an integral membrane protein localized to the central spoke ring complex and believed to participate in anchoring it in the nuclear membrane (Hallberg et al., 1993). Nup153 is a soluble nucleoporin localized to the nuclear basket of the NPC (Cordes et al., 1993; Sukegawa and Blobel, 1993) and implicated in several transport processes (Bastos et al., 1996; Shah and Forbes, 1998; Ullman et al., 1999), potentially in a dynamic manner (Nakielny et al., 1999). Recently, Nup153 has also been suggested to interact directly with B type lamins in Xenopus egg extracts (Smythe et al., 2000). With this system, we used confocal microscopy and fluorescence photobleaching techniques to characterize dynamic properties of the NPC, such as turnover of the complex in NE and ER, its mobility in the NE, and its postmitotic assembly.
Results
Noninvasively visualizing single NPCs with multiple GFP-tagged nucleoporins
In this study we have tagged two well-characterized nucleoporins, POM121 (Hallberg et al., 1993) and Nup153 (Sukegawa and Blobel, 1993) with multiple copies (Zaal et al., 1999) of green fluorescent protein (GFP) to visualize them at low, nontoxic expression levels. Fusions of POM121 and Nup153 to single GFPs have been shown to incorporate into the NPC (Söderqvist et al., 1997; Pante et al., 2000). POM121-EG/YFP3 and EGFP2/3-Nup153 specifically labeled NPCs and were indistinguishable from the single-tagged proteins in all aspects except brightness. NPC number or distribution was not changed by expression of these chimeras as shown by high resolution confocal microscopy. We found an NPC density of ∼3.5 ± 0.5/μm2 and total number of NPCs per nucleus of 1660 ± 190 (n = 5; Fig. 1, A and E) . This was identical to NPCs counted in untransfected NRK cells by immunofluorescence using mAb 414 or anti-p62 antibodies (data not shown) and is in the range reported previously by ultrastructural and immunofluorescence analysis (Maul et al., 1980; Kubitscheck et al., 1996).
Figure 1.
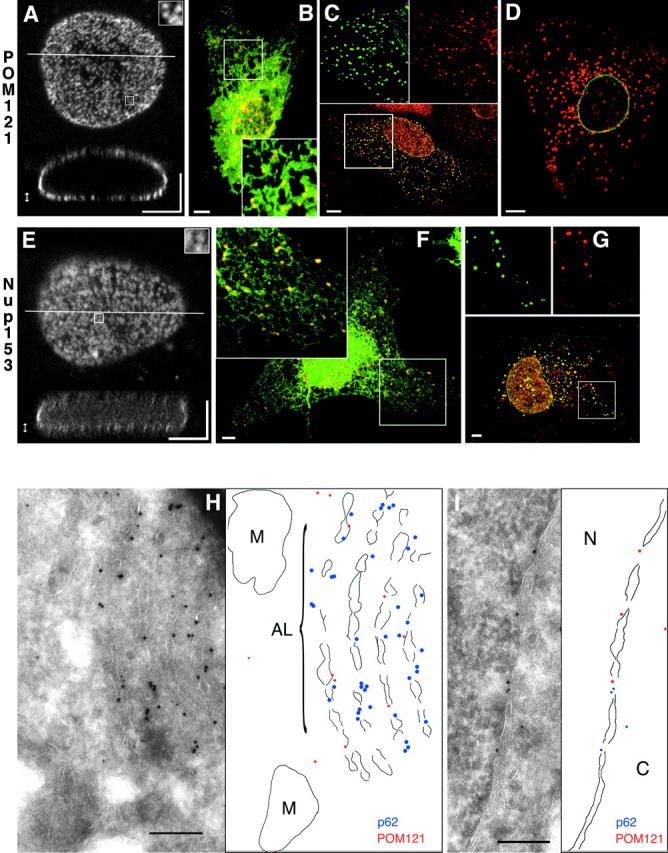
POM121-GFP and GFP-Nup153 label single NPCs and induce AL in live cells. (A–G) Images are confocal z-stacks (A and E) or single sections (B–D, F, and G). (A) Three-dimensional reconstruction of a live NRK cell nucleus expressing POM121-EGFP3. Top, maximum intensity projected of optical sections of the lower nuclear surface (double-pointed arrow). Inset enlarges 1 μm2 to show labeling of single NPCs. Bottom, xz slice at the line indicated in the top panel. (B) Live PtK2 cell coexpressing POM121-YFP3 (red) and SRβ-ECFP (green). AL labeled by POM121 are in direct contact to or colocalize with ER tubules and sheets as marked by SRβ. (C) HeLa cell expressing POM121-EGFP3 (green), fixed and stained for the endogenous nucleoporin p62 (red). Since the p62 antibody did not cross-react in PtK2 cells, HeLa cells were used for this experiment. AL containing POM121 colocalize with p62 antibodies (yellow in the merged lower panel, compare patterns in the split inset images). (D) Live PtK2 cell coexpressing ECFP–lamin B1 (green) and POM121-YFP3 (red). AL labeled by POM121 do not contain lamin B1. (E) Three-dimensional reconstruction of a live NRK cell nucleus expressing EGFP3-Nup153. Projections as in A. (F) Live COS7 cell coexpressing EGFP3-Nup153 (red) and SRβ-ECFP (green). COS7 cells were used for better ER morphology than NRK cells (compare with B). Yellow shows colocalization of AL with ER tubules. (G) COS7 cell expressing EGFP3-Nup153 (red) fixed and stained for the en-
dogenous nucleoporin POM121 (red). AL containing Nup153 also label with anti-POM121 antibodies (yellow in the merged lower panel, compare patterns in the split inset images). Some POM121-positive structures do not contain Nup153. (H and I) Cryoimmuno double labeling electron micrographs from HeLa cells expressing POM121-EGFP3. Membrane boundaries are outlined next to the micrographs. Labeled are rat POM121 (10-nm gold) and human p62 (5-nm gold). POM121 and p62 colocalize on fenestrated membrane stacks (AL) in H and nuclear pores in I. M, mitochondria; N, nucleus; C, cytoplasm. Bars: (A–G) 5 μm; (H and I) 200 nm.
Overexpression of nucleoporins induces the formation of AL
Expression of EGFP2/3-Nup153 and POM121-EGFP3 induced the formation of cytoplasmic clusters (Fig. 1, B–D, F, and G) reminiscent of AL. This occurred at expression levels that did not change nuclear morphology and NPC distribution. Cytoplasmic clusters appeared in PtK2 cells that endogenously do not contain AL as well as in NRK, HeLa, and COS7 cells that normally display AL at a low frequency (Cordes et al., 1996). To ensure that these structures were not unspecific aggregates we performed in vivo double labeling of POM121-YFP3 and EGFP3-Nup153 with SRβ-ECFP, an ER marker, and ECFP-lamin B1, a component of the nuclear lamina, as well as immunofluorescence with antibodies against endogenous nucleoporins. Cytoplasmic structures induced by overexpression of either POM121-EGFP3/YFP3 or EGFP2/3-Nup153 were always in contact with ER membranes (Fig. 1, B and F), colocalized with endogenous nucleoporins p62 or POM121 by immunofluorescence (Fig. 1, C and G), and never contained lamin B1 (Fig. 1 D). The same results were obtained with both nucleoporins (not all data shown). At high cytotoxic expression levels, both nucleoporins induced the intranuclear membrane stacks (Bastos et al., 1996) or bodies (Soderqvist et al., 1996) described previously. Only cells with the NPC/AL localization characteristic of low expression were used for further experiments.
To characterize AL ultrastructurally, cryoimmunogold double labeling electron microscopy was performed on HeLa cells transiently expressing POM121-EGFP3. Overexpressed POM121 was detected by a rat-specific antibody, and endogenous p62 with a monoclonal antibody. AL appeared as dense stacks of fenestrated membranes in which POM121-EGFP3 and p62 were colocalized (Fig. 1 H), as they did on gaps in the NE representing nuclear pores (Fig. 1 I). Thus, overexpression of single nucleoporins can induce the formation of AL, which recruit several endogenous nucleoporins and are always associated with ER membranes. AL induction by POM121 has in the meantime been confirmed biochemically in an independent study (Imreh and Hallberg, 2000).
POM121 turns over very slowly in NPCs, but quite rapidly in AL
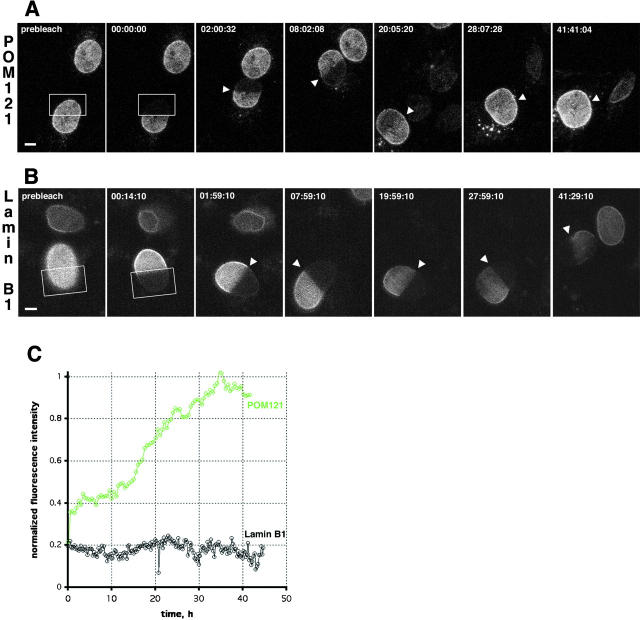
We next investigated how stably POM121 was associated with the NPC by FRAP (Ellenberg and Lippincott-Schwartz, 1997). FRAP can be used to measure the diffusional mobility and immobile fraction (IF) of GFP-tagged proteins in live cells (Ellenberg et al., 1997), or the binding of such proteins to a complex (Houtsmuller et al., 1999; McNally et al., 2000). Initial FRAP experiments of transiently expressing cells showed little recovery of POM121-EGFP3 into a photobleached box after 30 min (an IF of >95%; data not shown). Therefore fluorescence recovery was followed for a total of 42 h after the photobleach with confluent, stably transfected NRKPOM121-YFP3 cells (Fig. 2 A and Video 1). Complete recovery was reached only after 35 h with a t 1/2 of ∼20 h (Fig. 2 C), and complete recovery in log phase–growing cells could not be detected before the next cell division (data not shown). POM121 can thus serve as a marker to put a lower limit on the turnover of the NPC itself, whose behavior has to reflect that of its slowest exchanging components. To investigate if other structural components of the NE share the slow turnover of NPCs as a whole, we examined the nuclear lamina. FRAP experiments were performed on cells expressing a tagged B type lamin, EGFP–lamin B1. No significant recovery was detected up to 45 h after the photobleach in nondividing confluent interphase cultures (Fig. 2 B and Video 2).
Figure 2.
Turnover of NPCs and the lamina measured by FRAP. (A) FRAP of NRKPOM121-YFP3 cells. The boxed area was photobleached to background levels. Recovery was monitored immediately after the bleach and every 30 min by taking a stack of five confocal images after autofocussing. Representative maximum intensity projections are shown. Arrowheads mark the boundary between bleached and nonbleached regions. Time, hh:mm:ss. See Video 1 for the entire sequence. (B) FRAP experiment similar to A of NRK cells transiently expressing EGFP–lamin B1. Single confocal sections 0.5 μm above the coverslip surface were acquired every 15 min after autofocussing. Times, scale, and arrowheads as in A. See Video 2 for the entire sequence. (C) Plot shows fluorescence recovery of POM121 (green) and lamin B1 (black) in the bleached regions. Bleached and nonbleached half of the nuclei were tracked manually to measure mean intensities. Values were background subtracted and then normalized to total loss of fluorescence. Bars, 5 μm. Online supplemental material (Videos 1 and 2) is available at http://www.jcb.org/cgi/content/full/200101089/DC1.
The low turnover of the NPC in the nuclear membrane prompted us to investigate pore complex turnover in AL by FRAP analysis, since ALPCs appeared similar to NPCs by cryoimmunoelectron microscopy (Fig. 1, H and I). A significant portion of AL associated POM121-EGFP3 exchanged completely after only ∼8 min, with a t 1/2 of ∼76 ± 15 s (n = 6), i.e., three orders of magnitude faster than in NPCs, which showed no exchange during this time (Fig. 3 A and supplemental data). However, there was a significant IF of AL-associated POM121 varying between 50–70% (see online supplemental Fig. S1, available at http://www.jcb.org/cgi/content/full/200101089/DC1).
Figure 3.
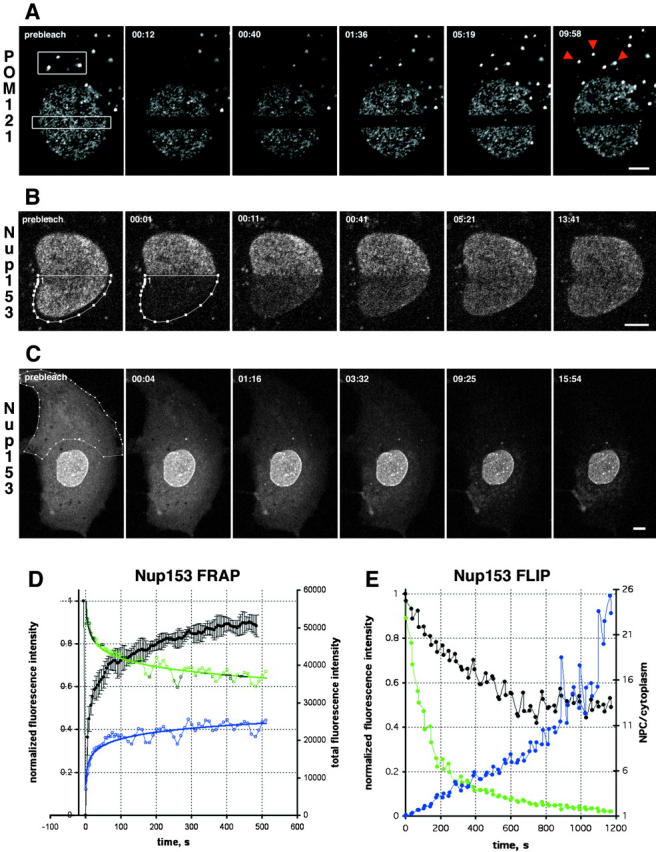
Turnover of nucleoporins within NPCs and ALPCs measured by FRAP. (A) FRAP of NRK cells expressing POM121-EGFP3 transiently. Boxed regions were photobleached and fluorescence recovery of ALPCs and NPCs was followed immediately after the bleach and then every 28 s in confocal sections. Arrowheads indicate the bleached AL. Note difference in recovery of AL and NPCs. Time, mm:ss. For quantitation see online supplemental Fig. S1 A. (B) FRAP of NRK cells expressing EGFP2-Nup153 transiently. Recovery was monitored immediately after the bleach and then every 5 s. Time and scale as in A. (C) FLIP of PtK2 cells expressing EGFP2-Nup153 transiently. Outlined region was photobleached repetitively 30 times every 30 s. Before and after each bleach the depletion of fluorescence was monitored in a confocal section. Time and scale as in A. (D) Plots of recovery in the bleached half and equilibration between bleached and nonbleached half of a Nup153 FRAP similar to B. Average mean intensities of the bleached region (black, left Y-axis) and standard deviation (n = 4). Data was normalized for total loss of fluorescence. Change of total fluorescence from the nonbleached (green) and bleached half (blue, both right Y-axis) measured for the experiment shown in B. (E) Plots of EGFP2-Nup153 depletion from the nucleus and cytoplasm for the FLIP in C. Average nuclear (black), cytoplasmic (green, both left Y-axis), and ratio of NPC/cytoplasmic fluorescence (blue, right Y-axis) is shown. Data was normalized to total loss of fluorescence. Note that nuclear fluorescence remains constant after initial loss of overlapping cytoplasmic signal, whereas cytoplasmic fluorescence is reduced to background levels. Bars, 5 μm. Online supplemental material (Fig. S1) is available at http://www.jcb.org/cgi/content/full/200101089/DC1.
Nup153 rapidly exchanges between NPCs
Having observed the stable association of POM121 with NPCs, we determined the binding time of Nup153 to NPCs and ALPCs. FRAP revealed complete recovery of EGFP2-Nup153 fluorescence in NPCs within 10 min with a t 1/2 of 15 ± 2 s (n = 4) (Fig. 3, B and D) and an IF of ∼10%. To test if the NH2-terminal GFP tag did not alter the ability of Nup153 to interact with the NPC, we also examined a fusion of GFP to the COOH terminus of Nup153. Nup153-EGFP's behavior was indistinguishable from the NH2-terminal constructs in FRAP experiments (see online supplemental Fig. S1, available at http://www.jcb.org/cgi/content/full/200101089/DC1), recovering with a t 1/2 of 15 ± 9 s and an IF of ∼15% (n = 6). In ALPCs, the IF of GFP2-Nup153 rose up to 40%, but recovery kinetics were unchanged (data not shown). In the NE, the unbleached half of the nucleus lost fluorescence with the same kinetics and amplitude as the bleached half recovered (Fig. 3 D, compare blue and green curves), suggesting that recovery was from the unbleached nuclear pool (Fig. 1 E). However, biochemical studies have suggested that Nup153 can reach the cytoplasmic face of the NPC and might actually be a shuttling protein (Nakielny et al., 1999). Fluorescence loss in photobleaching (FLIP) experiments were performed to test whether the nuclear pool could communicate with the cytoplasmic (soluble and AL) pool (Fig. 3 C). Cycles of 30 photobleaches that completely depleted all Nup153 from the cytoplasm were not able to deplete the NPC pool of the protein over a 15-min period (Fig. 3, C and E). Thus, the nuclear pool is diffusionally isolated from the cytoplasmic pool over periods of 20 min. In summary, photobleaching reveals that Nup153 continuously binds to and dissociates from the NPC from a freely diffusing nuclear pool, whereas POM121 remains tightly associated to NPCs throughout the lifetime of these structures, which as a whole are not turned over once assembled after mitosis.
NPCs and B type lamins form tightly connected elastic networks in mammalian cells
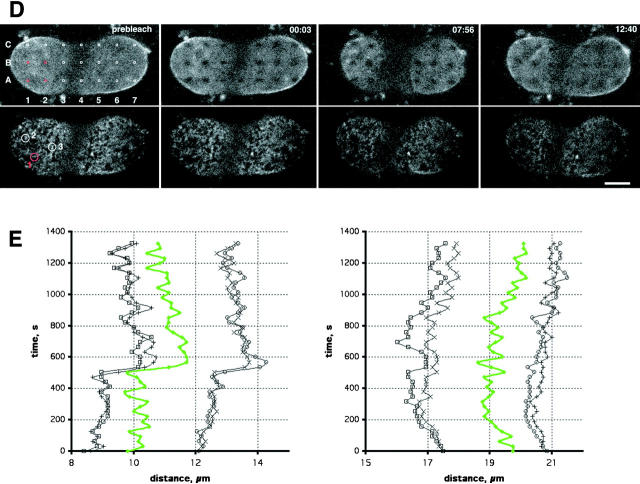
FRAP experiments of POM121 also revealed a stable boundary between bleached and nonbleached nuclear regions suggesting individual NPCs undergo little independent movement. (Figs. 2 A and 3 A). To investigate this further, we performed time-lapse imaging of the lower nuclear surface at single pore resolution (Fig. 4 A and Video 3). Tracking sets of individual NPCs showed no independent movement (Fig. 4 B). Rather, large arrays of NPCs moved in synchronous waves and the relative position of individual NPCs remained constant (Fig. 4 A and Video 3). This behavior was consistent with a spatially constrained two-dimensional network of pores. The slowly turned over lamina (Fig. 2, B and C) was a good candidate to connect mechanically such a network. To investigate this, double label time-lapse tracking experiments with NRK cells coexpressing ECFP–lamin B1 and POM121-YFP3 were performed. To track movement on the smooth surface of the lamina, we selectively photobleached a pattern of 21 0.9 × 0.6 μm landmarks into the ECFP-lamin B1–labeled lamina and followed movements of NPCs and the lamin pattern for 30 min (Fig. 4, D and E). NPC and lamin movement were strictly correlated, consistent with lamins and NPCs being part of the same network. That this network was elastic became evident in landmark-tracking experiments on migrating cells expressing only EGFP-lamin B1 (Fig. 4 C and Video 4). Folds passing through the lower nuclear surface distorted the grid only temporarily. The grid always relaxed back to its original position at the end of cellular movement (Fig. 4 C and Video 4). Supplemental videos are available at http://www.jcb.org/cgi/content/full/200101089/DC1.
Figure 4.
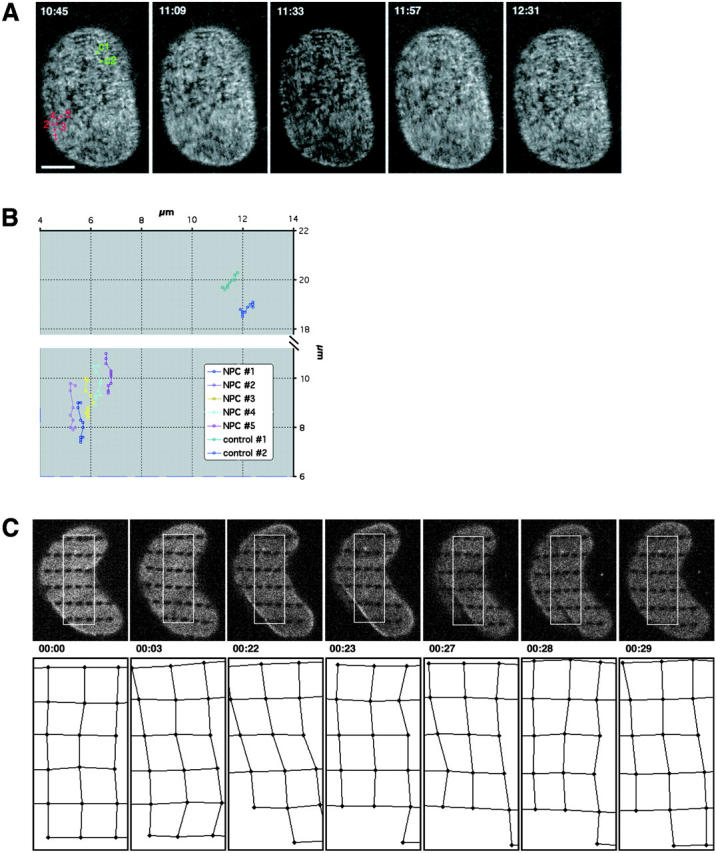
Tracking of NPC and lamina movement in interphase. (A) Time-lapse of a NRK cell expressing POM121-EGFP3 transiently. The lower nuclear surface was followed in a single confocal section every 2 s for a total of 30 min on a real-time confocal microscope. Sequence was averaged with a running window of five frames. Representative frames show numbered NPCs in a time window of ∼2 min used for tracking in B. Time, mm:ss. NPC movement is difficult to appreciate in still images; see Video 3 for the complete sequence. (B) Tracking of NPC movement. NPCs labeled #1–5 in A in a region of local NE movement are tracked together with two NPCs labeled C1 and C2 in A, which reside in an area of little movement and serve to illustrate global nuclear drift. Note the parallel and synchronous tracks of the NPCs. (C) Pattern FRAP of an NRK cell nucleus transiently expressing EGFP–Lamin B1. 33 1 × 0.5-μm regions were photobleached in the lower nuclear surface. Movement of landmarks was followed in single confocal sections every minute for 30 min (top row). Marks in the boxed area were used to track elastic deformations of the lamin lattice. Global cellular movement was corrected with two reference points. Relative position changes are shown in the bottom row as a network connecting the center of the bleach marks. Time, h:mm:ss; horizontal box length, 6 μm. (D) Pattern FRAP of a NRK nucleus transiently coexpressing POM121-YFP3 and ECFP–lamin B1. The outlined 21 0.9 × 0.6-μm regions were photobleached selectively in the lamina using a 413-nm Kr laser line. Movement of the lamina landmarks and the unbleached NPCs was then followed in single double-labeled confocal sections every 31 s for 30 min. Representative frames show distortion of nuclear shape by cell migration (note lamina folding at 07:56 and 12:40). Marks and NPCs used for tracking in E are red. Time, mm:ss. See Video 4 to better appreciate lamina elasticity. (E) Tracking of NPCs and lamina bleachmarks. Exemplary time-space tracks are shown for NPC #1 (green) and lamin grid marks A1–B2 (black) over 22 min in x, t and y, t plots. Global nuclear drift was normalized using to B5 and B7 marks. Note correlation between the x and y lamina and NPC movement. NPCs #2 and 3 and the surrounding marks behaved identically (not shown). Bars, 5 μm. Online supplemental material (Videos 3 and 4) is available at http://www.jcb.org/cgi/content/full/200101089/DC1.
In vivo reassembly of NPCs after mitosis
Having characterized the dynamics of NPCs in interphase we next investigated their biogenesis after mitosis. Four-dimensional (4-D) confocal time-lapse microscopy was used to follow cells from metaphase to G1 expressing either POM121-YFP3, EGFP2-Nup153, or ECFP–lamin B1. No structures that could have reflected intact NPCs, ALs, or lamin filaments were detected in metaphase, consistent with the complete disassembly of nuclear structure in mitosis. POM121, a nuclear membrane protein, was found equilibrated with the ER, whereas both Nup153 and lamin B1 were dispersed homogeneously in the cytosol (Fig. 5 A). The nucleoporins POM121 and Nup153 started to concentrate around chromatin in anaphase B, ∼4 min after metaphase/anaphase transition, which served as a temporal reference to compare different cells (Fig. 5, B and C) at the same time as nuclear membrane assembly marked by LBR-GFP (data not shown; Ellenberg et al., 1997). POM121 and Nup153 rapidly concentrated around chromatin, depleting their ER/cytoplasmic pool almost entirely by the end of telophase (Fig. 5, B and C). ALs, which were present in the POM121-YFP3–expressing cells before mitosis, reformed only much later with the onset of cytokinesis (Fig. 5 C, top). Since NPCs appeared to be linked to the lamina in interphase nuclei, we reasoned that lamins might play an important role in pore complex assembly. To our surprise, confocal time-lapse analysis of coexpressed ECFP–lamin B1 and POM121-YFP3 revealed that lamin B1 recruitment to chromosomes occurred ∼5 min (Fig. 5, C and D) after POM121 recruitment. Concentration of lamin B1 around chromatin could only be detected in late telophase/early cytokinesis (Fig. 5, B and C), a stage when chromatin is already sealed by a pore containing membrane (Fig. 5, B and C). To confirm that the GFP-tagged proteins reflected the behavior of the endogenous proteins, double immunofluorescence of POM121 and lamins was performed in untransfected NRK cells. As seen in vivo, POM121 assembly took place in early anaphase, whereas lamins were recruited to chromatin only in telophase (see online supplemental Fig. S2, available at http://www.jcb.org/cgi/content/full/200101089/DC1).
Figure 5.
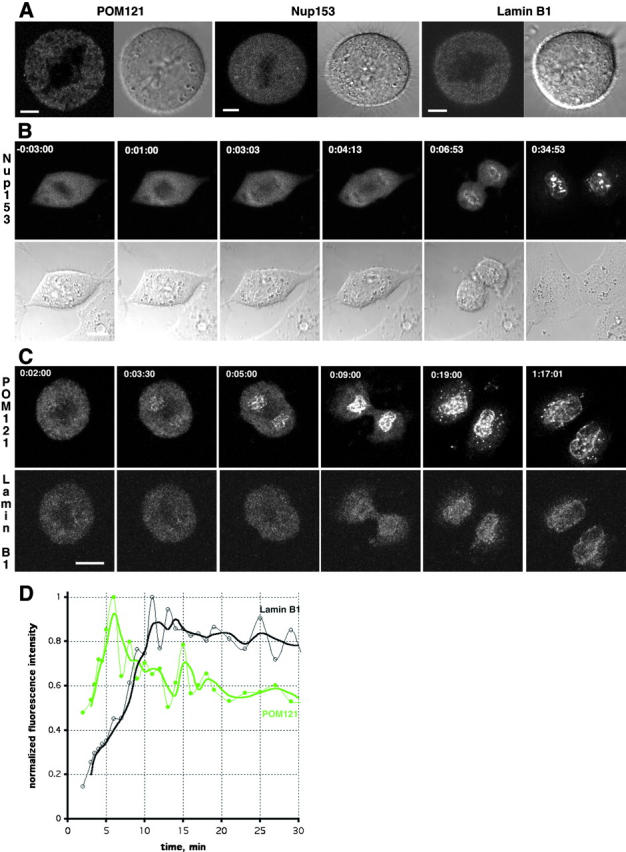
Recruitment of nucleoporins and lamin B1 during nuclear assembly. (A) In vivo localization of transiently expressed POM121-YFP3, EGFP3-Nup153, and ECFP–lamin B1 in metaphase NRK cells. Shown are confocal sections and DIC images centered on the metaphase plate. Note reticular pattern for POM121 and diffuse distribution of Nup153 and lamin B1. (B) 4-D sequence (10 slices every 2.5 μm acquired every 10–60 s depending on the dynamics of the cell cycle stage) of an NRK cell transiently expressing EGFP2-Nup153. Transparent projection of the four slices containing chromosomes (top) and DIC images highlighting the position of the chromosomes (bottom) are shown. Recruitment of Nup153 on the surface of chromosomes starts at ∼4 min after metaphase to anaphase transition. Time, h:mm:ss normalized to meta/anaphase transition equals 0. (C) 4-D double label sequence (six slices every 3 μm acquired every 30–120 s depending on the dynamics of the cell cycle stage) of an NRK cell transiently coexpressing POM121-YFP3 (top) and ECFP–lamin B1 (bottom). Shown are maximum intensity projections of the z-slices containing chromosomes. POM121 recruitment starts ∼3.5 min after meta/anaphase transition, whereas lamin B1 is only seen at 9 min (see D). Time, h:mm:ss normalized to meta/anaphase transition equals 0. (D) Plot of mean fluorescence intensity of the reforming NE for POM121 (green) and lamin B1 (black) shown in B. Before visible accumulation of lamin, nuclear rim areas were identified by POM121 localization. Data was background subtracted and normalized for bleaching during the time series. Lines are moving averages of two frames. Note the delay of ∼5 min between peak concentration/area for POM121 versus lamin B1. Bars: (A) 5 μm; (B and C) 10 μm. Online supplemental material (Fig. S2) is available at http://www.jcb.org/cgi/content/full/200101089/DC1.
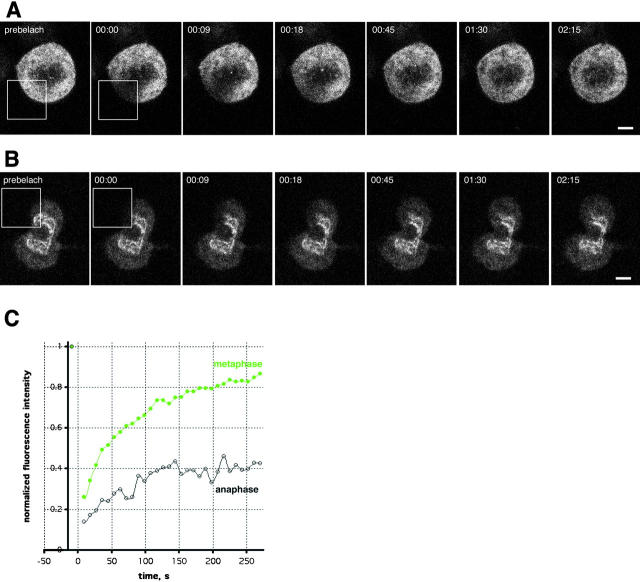
Next, we investigated whether assembling NPCs were tightly bound to the chromatin surface in the absence of B type lamins by FRAP analysis of POM121. In metaphase, POM121-EGFP3 diffused freely in the interconnected membranes of the ER with an apparent diffusion constant of DPOM121 = 0.25 μm2/s and an IF <10% (Fig. 6 C), a value comparable to ER membrane proteins (Nehls et al., 2000). Strikingly, performing FRAP experiments in the same cell ∼6 min later during nuclear assembly we found the chromatin-associated pool of POM121-EGFP3 strongly immobilized. In spite of the flux of new material into the growing pronuclei, 60% of POM121 in these structures was immobile over several minutes as the cell progressed into telophase (Fig. 6 B). This occurred well before lamin B1 became detectable around chromosomes (Fig. 5, C and D).
Figure 6.
POM121 is immobilized during nuclear assembly in anaphase. FRAP of a NRK cell transiently expressing POM121-EGFP3. (A) Boxed area was photobleached to background levels in metaphase. Recovery was monitored every 9 s in a single image acquired with open pinhole. Note rapid recovery of fluorescence into the ER as the cell reaches metaphase/anaphase transition (02:15). (B) FRAP of the same cell as in A ∼6 min after anaphase onset. Note the absence of complete recovery during the transition to telophase. Times, hh:mm:ss. (C) Plot of recovery in the bleached areas shown in A and B for metaphase (green)and anaphase (black). Data was normalized to total loss of fluorescence, time 0 corresponds to the midpoint of the bleach. Note the difference in IFs 4 min after the bleach. Bars, 5 μm.
Discussion
Visualizing the dynamics of single protein complexes in living cells
In this study, the dynamics of single NPCs and small clusters of NPCs were visualized with POM121-EGFP3 and EGFP2/3-Nup153 in living cells. This has so far been achieved only in brightly labeled fixed specimens by immunofluorescence (Kubitscheck et al., 1996). It is likely that POM121 is present in 16–24 copies in the NPC, based on the estimated abundance of yeast transmembrane nucleoporins (Rout et al., 2000). At steady state, probably no more than 24 copies of POM121-EGFP3 are present in one NPC, resulting in 48–72 GFPs easily detectable above background with confocal microscopes (Patterson et al., 1997). This approach should be very useful for future studies of NPCs and other large protein complexes in live cells.
Induction of AL by expressing a single nucleoporin
We have found that expression of either Nup153 or POM121 could induce the formation of AL in cell lines normally devoid of pore complexes in the ER. For Nup153, this is the first report of its presence in AL. Both Nup153 and POM121 have been reported to be absent from endogenous AL by immunofluorescence analysis (Cordes et al., 1996; Ewald et al., 1996) but see Imreh and Hallberg (2000). This difference is most likely explained by inaccessibility of epitopes in tightly packed AL structures, consistent with our finding of a high IF of both nucleoporins in AL. The formation of ALPCs by the expression of a single nucleoporin could suggest a function in pore complex self-assembly. For POM121 and gp210 (Wozniak and Blobel, 1992), such a function has been hypothesized (Goldberg et al., 1997). However, our results with Nup153 suggest that the formation of AL is not unique to transmembrane nucleoporins. Interestingly, two other soluble nucleoporins we have investigated, Nup107 and Nup133, did not induce AL over many expression levels but remained diffusely cytoplasmic (unpublished data). AL induction may thus represent an assay for a class of nucleoporins involved in NPC self-assembly. In this context, it is interesting to note that a recent study in yeast observed profound effects on membrane organization by overexpression of Nup53 (Marelli et al., 2001).
Turnover of pore complexes in the NE
We measured the turnover of subunits in single NPCs by FRAP, an issue that has not been studied previously. FRAP essentially measures the off-rate of the bleached nucleoporin if free fluorescent nucleoporin is available and the system is in steady state. The assumption of steady state is reasonable several days posttransfection or in stable cell lines (used for POM121), where expression levels are constant and do not change the number or distribution of NPCs. New NPC insertion, which would also lead to recovery in interphase, is corrected in our measurements because it affects both bleached and unbleached regions equally. To our surprise we found POM121 to be stably associated with the NPC throughout interphase. This is consistent with POM121 functioning to tether the mostly soluble NPC to the nuclear membrane. It also puts a lower limit on the lifetime of the entire NPC in mammalian cells, which is thus essentially stable for the entire duration of interphase. The behavior of POM121 is in contrast to many quite dynamic membrane proteins that we have investigated with FRAP in previous studies (Ostlund et al., 1999; Zaal et al., 1999; Nehls et al., 2000). FRAP of Nup153 measured a 3–4 orders of magnitude higher turnover (t 1/2 = 15 s), with no significant IF remaining bound at the NPC, strikingly demonstrating that stable association is not a general property of nucleoporins. The combined results from our FRAP and FLIP experiments suggest that Nup153 constantly associates and dissociates from the nucleoplasmic side of the NPC and can exchange between NPCs by rapid diffusion in the nucleoplasm. The diffuse nucleoplasmic signal we observed in GFP-Nup153–expressing cells supports this notion. Bleached regions of Nup153 always recovered uniformly, demonstrating that recovery was not limited by diffusion. The inability of cytoplasmic FLIPs to deplete nuclear Nup153 argues against shuttling of the protein, as was suggested from biochemical studies (Nakielny et al., 1999). However, due to the limited resolution of CLSM we cannot exclude that Nup153 becomes exposed to the cytoplasmic face of the NPC without ever completely dissociating from it. Turnover in the NPC is one important criterion by which to assign functions to different classes of nucleoporins. We propose that structural proteins that make up the backbone of the NPC should have a low turnover that reflects the lifetime of the complex. On the other hand, nucleoporins that are loosely associated to the NPC and might mediate dynamic transport events can be expected to have many associations/dissociations during the lifetime of the complex.
We also investigated the turnover of the nuclear lamina by FRAP analysis of GFP–lamin B1. The absence of significant turnover over 45 h in nondividing cells is consistent with measurements of lamins A, C, and B over short times in interphase by other groups (Broers et al., 1999; Moir et al., 2000). In the cells we have studied, pore complexes and the nuclear lamina, once they are formed during postmitotic biogenesis they are essentially stable until cells enter the next mitosis. This is consistent with an anchoring function of lamins for NPCs in the NE, proposed previously from ultrastructural studies (Akey, 1989; Goldberg and Allen, 1993). The direct interaction between B type lamins and Nup153 that has been suggested by studies in Xenopus extracts (Smythe et al., 2000) seems unlikely to play a role in this anchoring, since the turnover of these two proteins differs by more than three orders of magnitude in interphase and Nup153 has no significant IF at the NPC.
Turnover of pore complexes in the ER
The turnover of POM121 was two orders of magnitude higher in AL in the ER than in NPCs. If we consider the low turnover of POM121 in NPCs as a hallmark of a structural/membrane anchoring function, ALPCs are apparently much less permanently assembled in the ER than in the NE. It is possible that the absence of an underlying lamina in the ER contributes to this difference in pore complex stability. Another clue to AL structure came from the observation that both POM121 and Nup153 have significant IFs in ALs. There are two possible explanations for these findings. (a) The tightly stacked pores in the convoluted ER membranes make the inner ALPCs relatively inaccessible to exchange via the membrane (POM121, IF ∼60%) and to a lesser degree via the cytoplasm (Nup153, IF ∼30%). (b) The mobile fraction represents incompletely assembled pore complexes on the periphery of ALs, whereas the IF represents the behavior of fully assembled ALPCs that are identical to NPCs. However, the latter possibility seems unlikely, because it is inconsistent with the increase in IF observed for Nup153 in AL compared with NPCs and we thus favor explanation a. This would support the view of AL function as a dynamic storage compartment for nucleoporins. In many oocytes ALs provide new nucleoporins for NPC assembly in several generations of embryonic cells. Since ∼50% of NPC assembly takes place in interphase (Maul et al., 1972; Winey et al., 1997) ALPCs would have to make their subunits available to de novo NPC assembly much more frequently than once per cell cycle.
Immobile NPCs are part of a NE network
We also investigated whether the NPC could undergo movement within the plane of the NE. This was especially interesting in the light of reports from budding yeast suggesting high mobility of NPCs following karyogamy (Bucci and Wente, 1997) or in a nucleoporin mutant background (Belgareh and Doye, 1997). Using single NPC tracking combined with landmark-bleaching experiments, we found that lamins and NPCs are part of one stable network in the nuclear periphery. It is tempting to speculate that lamins are responsible for anchoring and evenly distributing NPCs in the NE of interphase cells. This view is supported by pore clustering induced in lamin mutants in Drosophila and C. elegans (Lenz-Bohme et al., 1997; Liu et al., 2000). We observed an extraordinary stability of the peripheral network, both in terms of the turnover of its structural proteins as well as restrictions to movements within the two-dimensional plane of the NE. However, the NE did undergo elastic deformations during cellular movements. It will be interesting to reinvestigate the dynamics of yeast NPCs in wild-type cells not undergoing karyogamy, especially since recent studies on the NPC-associated proteins Mlp1 and Mlp2 suggested the stable tethering of telomeres to the nuclear periphery (Galy et al., 2000).
NPCs in mitosis
Our finding of a very stable protein network in the interphase NE prompted us to analyze how it reassembled after cell division. NPCs and ALs were completely disassembled in metaphase, with POM121 freely diffusing in ER membranes. This behavior is very reminiscent of our studies on lamin B receptor (Ellenberg et al., 1997) and reinforces the concept that nuclear membranes do not fragment in mitosis but are absorbed by the intact membrane network of the ER. Nup153 and lamin B1 did not show any morphological membrane association in metaphase but were dispersed in the cytoplasm. This is in contrast to early reports suggesting that B type lamins are associated with mitotic membranes (Gerace and Blobel, 1980; Stick et al., 1988). This discrepancy could be due to differences in cell lines or methods: ultrastructure and cell fractionation in the early studies versus live cell imaging in our studies. However, we can not exclude that a small fraction of membrane-associated EGFP–lamin B1 would be masked by an excess of soluble protein in our cells. POM121 and Nup153 were recruited to chromosomes early in anaphase, consistent with studies in fixed cells (Bodoor et al., 1999; Haraguchi et al., 2000). Direct in vivo comparison of POM121 and lamin B1, as well as immunofluorescence of the endogenous proteins, revealed that B type lamins are absent during the initial NPC assembly that already lead to immobilization of nucleoporins. This is again consistent with studies in fixed cells (Chaudhary and Courvalin, 1993; Bodoor et al., 1999). Therefore, B type lamins are unlikely to mediate the initial attachment of nuclear (pre-)pores to chromatin in stoichiometric amounts. However, due to the detection limits of light microscopy, we cannot exclude a function of a small fraction of nuclear lamins in NPC assembly. Our data is in apparent contradiction to a recent study in fixed cells, where lamin B staining appeared to precede staining by mAb 414 (recognizing several nucleoporins, including p62 and Nup153; Moir et al., 2000). However, when those authors checked with specific anti-Nup153 antibodies, they also found the nucleoporin to precede lamin B during nuclear assembly.
This study provides the first in vivo characterization of the dynamic properties of NPCs in mammalian cells. As a consequence of our data, one can view the NPC as a major structural component of a stable, crosslinked NE protein network comprised of lamins, inner nuclear membrane proteins, and peripheral heterochromatin. Reflecting the properties of this system, NPCs are organized in an elastic network and are immobile in the plane of the nuclear membrane. In addition, individual pore complexes turn over several subunits very slowly, making the whole complex stable for the entire duration of interphase.
Materials and methods
Antibodies and cell culture
Mouse anti-p62 antibody was from Transduction Laboratories, mAb 414 was a gift from Mary Dasso (The National Institutes of Health), mouse antilamin antibody R19 was a gift from Georg Krohne (University of Würzburg, Germany). Rabbit polyclonal anti-POM121 (rat) was made against the COOH-terminal 141 amino acids of rat POM121; it specifically recognizes the rat and monkey but not the human protein (Imreh and Hallberg, 2000). Secondary antibodies were from Southern Biotechnology Associates, Inc. (TRITC) or Molecular Probes (Alexa 488). HeLa, NRK, COS7 and PtK2, and cells were grown as described (Ellenberg et al., 1997) in LabTekII-chambered No. 1 coverglasses (LabTek). NRK cells stably expressing POM121-YFP3 (NRKPOM121-YFP3) were selected according to standard protocols and maintained at 0.5 mg/ml G418. For imaging, medium was changed to DME without phenol red supplemented with 25 mM HEPES-KOH, pH 7.3, 20% FCS. Transfection was with FuGene 6 (Roche).
DNA constructs
The following fusions to GFP and its spectral variants were used in this study: pEGFP2-Nup153, pEGFP3-Nup153, pNup153-EGFP, POM121-EGFP3, POM121-YFP3 (Imreh and Hallberg, 2000), ECFP–lamin B1, EGFP–lamin B1, lamin B receptor–GFP (Ellenberg et al., 1997), and SRβ-ECFP. All constructs are based on pEGFP-N/C-X vectors (CLONTECH Laboratories, Inc.) and were made by placing the indicated number of EGFP, EYFP, or ECFP ORFs in frame at the COOH or NH2 terminus of the full length cDNAs of the proteins of interest. PCR-generated constructs were verified by sequencing. For detailed description of the cloning strategies, see online supplemental material available at http://www.jcb.org/cgi/content/full/200101089/DC1.
Confocal microscopy, immunofluorescence, and image analysis
Live cells were imaged at 37°C maintained by an air stream incubator (ASI 400; Nevtek) in conjunction with an objective heater (Bioptechs) on several confocal microscope systems. Time-lapse at single NPC resolution using EGFP (Fig. 4 A) was on an UltraView real time spinning disk confocal microscope (PerkinElmer). For immunofluorescence, cells were prepared as described (Sciaky et al., 1997), except that fixation was for 10 min in −20°C MeOH followed by 5 min at −20°C in acetone. Images were acquired on a ZEISS LSM 510 using 488 nm in conjunction with a BP 505–550 for EGFP/Alexa 488, and 543 nm in conjunction with a LP 560 for rhodamine. The same system was used for three-dimensional reconstructions at single NPC resolution using EGFP.
Time-lapse of confocal z-stacks (4-D imaging) was on a custom built ZEISS LSM510 described in the online supplemental information available at http://www.jcb.org/cgi/content/full/200101089/DC1. Images were assembled in Adobe Photoshop® 6.0. Quantitative image analysis was performed either in NIH Image or the LSM 2.5 SP2 software (ZEISS). Average intensities of regions of interests were exported into Microsoft Excel, background subtracted, and plotted. Other normalizations are described in the figure legends.
Photobleaching experiments
FRAP was performed essentially as described on a ZEISS LSM 410 (Ellenberg et al., 1997) or 510 (Zaal et al., 1999), as well as on a custom built confocal microscope at the European Molecular Biology Laboratory, the compact confocal camera (CCC), and the custom made ZEISS LSM 510 using 413 nm for ECFP and 488 nm for EGFP. For long term NPC/lamin turnover measurements, z-stacks were acquired after the photobleach in 15 or 30 min intervals after computer-controlled autofocussing on the coverglass surface by macros written for the LSM 410 or CCC software (Salmon et al., 1999). Grid photobleaching was performed on a ZEISS LSM 510. Average intensities of FRAP experiments were corrected for the total fluorescence lost during the photobleach and the bleaching that occurred during scanning of recovery. Calculations of apparent diffusion constant for POM121 in mitotic ER was based on quantitative stripbleach experiments as described (Ellenberg et al., 1997).
Electron microscopy
HeLa Cells transfected with POM 121-EGFP3 were fixed in 4% formaldehyde in 0.1 M phosphate buffer at pH 7.4 overnight, then scraped gently from the tissue culture dish and pelleted. The cell pellet was rinsed in phosphate buffer, resuspended in gelatin, and cryoprotected in 2.3 M sucrose at 4°C overnight. Ultrathin cryosections for immunolabeling were obtained as described (Liou et al., 1996). Sections were incubated with antibodies to POM121 and p62 as described (Griffiths, 1993), then incubated with protein A–gold (Utrecht University, Utrecht, Netherlands) using 5 and 10 nm sizes, respectively. Sections were viewed and photographed with a transmission electron microscope (CM 10; Philips). Unfortunately, we were not able to obtain specific labeling with these antibodies in a resin-embedded specimen that would have provided better ultrastructural resolution of the fenestrated cytoplasmic membranes.
Online supplemental material
Detailed description of the DNA constructs used in this study and the microscope setup used for multicolor GFP imaging are available at http://www.jcb.org/cgi/content/full/200101089/DC1. The movies alluded to in the results and figure legends are available at the same address. Video 1 shows a long-term FRAP of POM121 (Fig. 2 A), Video 2 a long-term FRAP of lamin B1 (Fig. 2 B), Video 3 NPC movement in the interphase nucleus (Fig. 4 A), and Video 4 lamina elasticity in a pattern FRAP (Fig. 4 C).
Acknowledgments
The authors would like to thank Howard Worman (Columbia University, New York, NY) for the gift of the human lamin B1 cDNA; Brian Burke (University of Calgary, Calgary, Canada) for the gift of human Nup153 cDNA; Georg Krohne (University of Würzburg, Würzburg, Germany) for the gift of lamin R19 antibody; Theresa Ward (National Institutes of Health, Cell Biology and Metabolism Branch, Bethesda, MD) for help with the SRβ-ECFP plasmid; and Ernst Stelzer, Nick Salmon, and James Jonkman (European Molecular Biology Laboratory, Heidelberg, Germany) for help with macroprogramming for the CCC operating software and for improvements of the CCC microscope. In addition we would like to thank Valérie Doye, Iain Mattaj, Elisa Izaurralde, and Gwen Rabut for critical reading of the manuscript.
J. Beaudouin was supported by a fellowship of the European Molecular Biology Laboratory's international Ph.D. program.
The online version of this paper contains supplemental material.
Footnotes
Abbreviations used in this paper: 4-D, four-dimensional; AL, annulate lamellae; ALPC, annulate lamellae pore complex; CCC, compact confocal camera; CLSM, confocal laser scanning microscopy; FLIP, fluorescence loss in photobleaching; GFP, green fluorescent protein; IF, immobile fraction; NE, nuclear envelope; NPC, nuclear pore complex.
References
- Akey, C.W. 1989. Interactions and structure of the nuclear pore complex revealed by cryo-electron microscopy. J. Cell Biol. 109:955–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, T.D., J.M. Cronshaw, S. Bagley, E. Kiseleva, and M.W. Goldberg. 2000. The nuclear pore complex: mediator of translocation between nucleus and cytoplasm. J. Cell Sci. 113:1651–1659. [DOI] [PubMed] [Google Scholar]
- Bastos, R., A. Lin, M. Enarson, and B. Burke. 1996. Targeting and function in mRNA export of nuclear pore complex protein Nup153. J. Cell Biol. 134:1141–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh, N., and V. Doye. 1997. Dynamics of nuclear pore distribution in nucleoporin mutant yeast cells. J. Cell Biol. 136:747–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh, N., and V. Doye. 1999. Yeast and vertebrate nuclear pore complexes: evolutionary conserved, yet divergent macromolecular assemblies. Protoplasma. 209:133–143. [Google Scholar]
- Bodoor, K., S. Shaikh, D. Salina, W.H. Raharjo, R. Bastos, M. Lohka, and B. Burke. 1999. Sequential recruitment of NPC proteins to the nuclear periphery at the end of mitosis. J. Cell Sci. 112:2253–2264. [DOI] [PubMed] [Google Scholar]
- Broers, J.L., B.M. Machiels, G.J. van Eys, H.J. Kuijpers, E.M. Manders, R. van Driel, and F.C. Ramaekers. 1999. Dynamics of the nuclear lamina as monitored by GFP-tagged A-type lamins. J. Cell Sci. 112:3463–3475. [DOI] [PubMed] [Google Scholar]
- Bucci, M., and S. Wente. 1997. In vivo dynamics of the nuclear pore complexes in yeast. J. Cell Biol. 136:1185–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary, N., and J.-C. Courvalin. 1993. Stepwise reassembly of the nuclear envelope at the end of mitosis. J. Cell Biol. 122:295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collas, I., and J.C. Courvalin. 2000. Sorting nuclear membrane proteins at mitosis. Trends Cell Biol. 10:5–8. [DOI] [PubMed] [Google Scholar]
- Cordes, V.C., S. Reidenbach, A. Kohler, N. Stuurman, R. van Driel, and W.W. Franke. 1993. Intranuclear filaments containing a nuclear pore complex protein. J. Cell Biol. 123:1333–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes, V.C., S. Reidenbach, and W.W. Franke. 1995. High content of a nuclear pore complex protein in cytoplasmic annulate lamellae of Xenopus oocytes. Eur. J. Cell Biol. 68:240–255. [PubMed] [Google Scholar]
- Cordes, V.C., S. Reidenbach, and W.W. Franke. 1996. Cytoplasmic annulate lamellae in cultured cells: composition, distribution, and mitotic behavior. Cell Tissue Res. 284:177–191. [DOI] [PubMed] [Google Scholar]
- Cordes, V.C., H.R. Rackwitz, and S. Reidenbach. 1997. Mediators of nuclear protein import target karyophilic proteins to pore complexes of cytoplasmic annulate lamellae. Exp. Cell Res. 237:419–433. [DOI] [PubMed] [Google Scholar]
- Ellenberg, J., and J. Lippincott-Schwartz. 1997. Fluorescence photobleaching techniques. Cells: A Laboratory Manual. Vol. 2. D. Spector, R. Goldman, and L. Leinwand, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 79.1–79.23.
- Ellenberg, J., E.D. Siggia, J.E. Moreira, C.L. Smith, J.F. Presley, H.J. Worman, and J. Lippincott-Schwartz. 1997. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J. Cell Biol. 138:1193–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald, A., U. Kossner, U. Scheer, and M.C. Dabauvalle. 1996. A biochemical and immunological comparison of nuclear and cytoplasmic pore complexes. J. Cell Sci. 109:1813–1824. [DOI] [PubMed] [Google Scholar]
- Galy, V., J.C. Olivo-Marin, H. Scherthan, V. Doye, N. Rascalou, and U. Nehrbass. 2000. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature. 403:108–112. [DOI] [PubMed] [Google Scholar]
- Gerace, L., and G. Blobel. 1980. The nuclear envelope lamina is reversibly depolymerized during mitosis. Cell. 19:277–287. [DOI] [PubMed] [Google Scholar]
- Gerace, L., and B. Burke. 1988. Functional organization of the nuclear envelope. Annu. Rev. Cell Biol. 4:335–374. [DOI] [PubMed] [Google Scholar]
- Goldberg, M.W., and T.D. Allen. 1993. The nuclear pore complex: three-dimensional surface structure revealed by field emission, in-lens scanning electron microscopy, with underlying structure uncovered by proteolysis. J. Cell Sci. 106:261–274. [DOI] [PubMed] [Google Scholar]
- Goldberg, M.W., and T.D. Allen. 1995. Structural and functional organization of the nuclear envelope. Curr. Opin. Cell Biol. 7:301–309. [DOI] [PubMed] [Google Scholar]
- Goldberg, M.W., C. Wiese, T.D. Allen, and K.L. Wilson. 1997. Dimples, pores, star-rings, and thin rings on growing nuclear envelopes: evidence for structural intermediates in nuclear pore complex assembly. J. Cell Sci. 110:409–420. [DOI] [PubMed] [Google Scholar]
- Griffiths, G. 1993. Fine Structure Immunocytochemistry. Springer-Verlag, Berlin. 459 pp.
- Gruenbaum, Y., K.L. Wilson, A. Harel, M. Goldberg, and M. Cohen. 2000. Review: nuclear lamins--structural proteins with fundamental functions. J. Struct. Biol. 129:313–323. [DOI] [PubMed] [Google Scholar]
- Hallberg, E., R.W. Wozniak, and G. Blobel. 1993. An integral membrane protein of the pore membrane domain of the nuclear envelope contains a nucleoporin-like region. J. Cell Biol. 122:513–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi, T., T. Koujin, T. Hayakawa, T. Kaneda, C. Tsutsumi, N. Imamoto, C. Akazawa, J. Sukegawa, Y. Yoneda, and Y. Hiraoka. 2000. Live fluorescence imaging reveals early recruitment of emerin, LBR, RanBP2, and Nup153 to reforming functional nuclear envelopes. J. Cell Sci. 113:779–794. [DOI] [PubMed] [Google Scholar]
- Houtsmuller, A.B., S. Rademakers, A.L. Nigg, D. Hoogstraten, J.H. Hoeijmakers, and W. Vermeulen. 1999. Action of DNA repair endonuclease ERCC1/XPF in living cells. Science. 284:958–961. [DOI] [PubMed] [Google Scholar]
- Imreh, G., and E. Hallberg. 2000. An integral membrane protein from the nuclear pore complex is also present in the annulate lamellae: implications for annulate lamella formation. Exp. Cell Res. 259:180–190. [DOI] [PubMed] [Google Scholar]
- Kessel, R.G. 1992. Annulate lamellae: a last frontier in cellular organelles. Int. Rev. Cytol. 133:43–120. [DOI] [PubMed] [Google Scholar]
- Kubitscheck, U., P. Wedekind, O. Zeidler, M. Grote, and R. Peters. 1996. Single nuclear pores visualized by confocal microscopy and image processing. Biophys. J. 70:2067–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz-Bohme, B., J. Wismar, S. Fuchs, R. Reifegerste, E. Buchner, H. Betz, and B. Schmitt. 1997. Insertional mutation of the Drosophila nuclear lamin Dm0 gene results in defective nuclear envelopes, clustering of nuclear pore complexes, and accumulation of annulate lamellae. J. Cell Biol. 137:1001–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou, W., H.J. Geuze, and J.W. Slot. 1996. Improving structural integrity of cryosections for immunogold labeling. Histochem. Cell Biol. 106:41–58. [DOI] [PubMed] [Google Scholar]
- Liu, J., T.R. Ben-Shahar, D. Riemer, M. Treinin, P. Spann, K. Weber, A. Fire, and Y. Gruenbaum. 2000. Essential roles for caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression, and spatial organization of nuclear pore complexes. Mol. Biol. Cell. 11:3937–3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marelli, M., C.P. Lusk, H. Chan, J.D. Aitchison, and R.W. Wozniak. 2001. A link between the synthesis of nucleoporins and the biogenesis of the nuclear envelope. J. Cell Biol. 153:709–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul, G.G., H.M. Maul, J.E. Scogna, M.W. Lieberman, G.S. Stein, B.Y. Hsu, and T.W. Borun. 1972. Time sequence of nuclear pore formation in phytohemagglutinin-stimulated lymphocytes and in HeLa cells during the cell cycle. J. Cell Biol. 55:433–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul, G.G., L.L. Deaven, J.J. Freed, G.L. Campbell, and W. Becak. 1980. Investigation of the determinants of nuclear pore number. Cytogenet. Cell Genet. 26:175–190. [DOI] [PubMed] [Google Scholar]
- McNally, J.G., W.G. Muller, D. Walker, R. Wolford, and G.L. Hager. 2000. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science. 287:1262–1265. [DOI] [PubMed] [Google Scholar]
- Moir, R.D., M. Yoon, S. Khuon, and R.D. Goldman. 2000. Nuclear lamins A and B1. Different pathways of assembly during nuclear envelope formation in living cells. J. Cell Biol. 151:1155–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny, S., S. Shaikh, B. Burke, and G. Dreyfuss. 1999. Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J. 18:1982–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehls, S., E.L. Snapp, N.B. Cole, K.J. Zaal, A.K. Kenworthy, T.H. Roberts, J. Ellenberg, J.F. Presley, E. Siggia, and J. Lippincott-Schwartz. 2000. Dynamics and retention of misfolded proteins in native ER membranes. Nat. Cell Biol. 2:288–295. [DOI] [PubMed] [Google Scholar]
- Ostlund, C., J. Ellenberg, E. Hallberg, J. Lippincott-Schwartz, and H.J. Worman. 1999. Intracellular trafficking of emerin, the emery-dreifuss muscular dystrophy protein. J. Cell Sci. 112:1709–1719. [DOI] [PubMed] [Google Scholar]
- Pante, N., and U. Aebi. 1995. Toward a molecular understanding of the structure and function of the nuclear pore complex. Int. Rev. Cytol. 162B:225–255. [DOI] [PubMed] [Google Scholar]
- Pante, N., F. Thomas, U. Aebi, B. Burke, and R. Bastos. 2000. Recombinant Nup153 incorporates in vivo into Xenopus oocyte nuclear pore complexes. J. Struct. Biol. 129:306–312. [DOI] [PubMed] [Google Scholar]
- Patterson, G.H., S.M. Knobel, W.D. Sharif, S.R. Kain, and D.W. Piston. 1997. Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophys. J. 73:2782–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, K.M., E.K. Chan, B.J. Grant, K.F. Sullivan, E.M. Tan, and C.A. Glass. 1990. In vitro posttranslational modification of lamin B cloned from a human T-cell line. Mol. Cell. Biol. 10:2164–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout, M.P., J.D. Aitchison, A. Suprapto, K. Hjertaas, Y. Zhao, and B.T. Chait. 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148:635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, K.J., and S.R. Wente. 2000. The nuclear pore complex: a protein machine bridging the nucleus and cytoplasm. Curr. Opin. Cell Biol. 12:361–371. [DOI] [PubMed] [Google Scholar]
- Salmon, N.J., S. Lindek, and E.H.K. Stelzer. 1999. Databases for microscopes and microscopical images. In Handbook on Computer Vision and Applications. Vol. 2. B. Jähne, P. Haußecker, and P. Geissler, editors. Academic Press, San Diego, CA. 907–926.
- Sciaky, N., J. Presley, C. Smith, K.J. Zaal, N. Cole, J.E. Moreira, M. Terasaki, E. Siggia, and J. Lippincott-Schwartz. 1997. Golgi tubule traffic and the effects of brefeldin A visualized in living cells. J. Cell Biol. 139:1137–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, S., and D.J. Forbes. 1998. Separate nuclear import pathways converge on the nucleoporin Nup153 and can be dissected with dominant-negative inhibitors. Curr. Biol. 8:1376–1386. [DOI] [PubMed] [Google Scholar]
- Smythe, C., H.E. Jenkins, and C.J. Hutchison. 2000. Incorporation of the nuclear pore basket protein nup153 into nuclear pore structures is dependent upon lamina assembly: evidence from cell-free extracts of Xenopus eggs. EMBO J. 19:3918–3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderqvist, H., W.Q. Jiang, N. Ringertz, and E. Hallberg. 1996. Formation of nuclear bodies in cells overexpressing the nuclear pore protein POM121. Exp. Cell Res. 225:75–84. [DOI] [PubMed] [Google Scholar]
- Söderqvist, H., G. Imreh, M. Kihlmark, C. Linnman, N. Ringertz, and E. Hallberg. 1997. Intracellular distribution of an integral nuclear pore membrane protein fused to green fluorescent protein--localization of a targeting domain. Eur. J. Biochem. 250:808–813. [DOI] [PubMed] [Google Scholar]
- Stafstrom, J.P., and L.A. Staehelin. 1984. Are annulate lamellae in the Drosophila embryo the result of overproduction of nuclear pore components? J. Cell Biol. 98:699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stick, R., B. Angres, C.F. Lehner, and E.A. Nigg. 1988. The fates of chicken nuclear lamin proteins during mitosis: evidence for a reversible redistribution of lamin B2 between inner nuclear membrane and elements of the endoplasmic reticulum. J. Cell Biol. 107:397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukegawa, J., and G. Blobel. 1993. A nuclear pore complex protein that contains zinc finger motifs, binds DNA, and faces the nucleoplasm. Cell. 72:29–38. [DOI] [PubMed] [Google Scholar]
- Ullman, K.S., S. Shah, M.A. Powers, and D.J. Forbes. 1999. The nucleoporin nup153 plays a critical role in multiple types of nuclear export. Mol. Biol. Cell. 10:649–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, K.L. 2000. The nuclear envelope, muscular dystrophy and gene expression. Trends Cell Biol. 10:125–129. [DOI] [PubMed] [Google Scholar]
- Winey, M., D. Yarar, T.H. Giddings, Jr., and D.N. Mastronarde. 1997. Nuclear pore complex number and distribution throughout the Saccharomyces cerevisiae cell cycle by three-dimensional reconstruction from electron micrographs of nuclear envelopes. Mol. Biol. Cell. 8:2119–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak, R.W., and G. Blobel. 1992. The single transmembrane segment of gp210 is sufficient for sorting to the pore membrane domain of the nuclear envelope. J. Cell Biol. 119:1441–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaal, K.J.M., C.L. Smith, R.S. Polishchuk, N. Altan, N.B. Cole, J. Ellenberg, K. Hirschberg, J.F. Presley, T.H. Roberts, E. Siggia, R.D. Phair, and J. Lippincott-Schwartz. 1999. Golgi membranes are absorbed into and reemerge from the ER during mitosis. Cell. 99:589–601. [DOI] [PubMed] [Google Scholar]