Abstract
The formation of the active spliceosome, its recruitment to active areas of transcription, and its role in pre-mRNA splicing depends on the association of a number of multifunctional serine/arginine-rich (SR) proteins. ZNF265 is an arginine/serine-rich (RS) domain containing zinc finger protein with conserved pre-mRNA splicing protein motifs. Here we show that ZNF265 immunoprecipitates from splicing extracts in association with mRNA, and that it is able to alter splicing patterns of Tra2-β1 transcripts in a dose-dependent manner in HEK 293 cells. Yeast two-hybrid analysis and immunoprecipitation indicated interaction of ZNF265 with the essential splicing factor proteins U1-70K and U2AF35. Confocal microscopy demonstrated colocalization of ZNF265 with the motor neuron gene product SMN, the snRNP protein U1-70K, the SR protein SC35, and with the transcriptosomal components p300 and YY1. Transfection of HT-1080 cells with ZNF265–EGFP fusion constructs showed that nuclear localization of ZNF265 required the RS domain. Alignment with other RS domain–containing proteins revealed a high degree of SR dipeptide conservation. These data show that ZNF265 functions as a novel component of the mRNA processing machinery.
Keywords: zinc finger protein; RS domain; SR proteins; RNA processing; nuclear localization; renin
Introduction
Gene transcription and pre-mRNA splicing are dynamic and highly coordinated processes that occur in a spatially organized manner in the nucleus (Singer and Green, 1997). Splicing takes place in the spliceosome, a large RNA–protein complex composed of various small nuclear ribonucleoprotein particles (snRNPs),* and many other protein factors that include members of the highly conserved serine/arginine-rich (SR) protein family. SR proteins, by RNA–protein and protein–protein interactions, coordinate the passage of the spliceosomal complex though the splicing reaction (reviewed in Fu, 1995; Manley and Tacke, 1996; Cáceres et al., 1997). SR protein recruitment to active areas of transcription and RNA processing involves their signature arginine/serine-rich (RS) domains and an interaction with RNA polymerase II through its COOH-terminal domain (Yuryev et al., 1996; Du and Warren, 1997; Kim et al., 1997; Misteli and Spector, 1999).
RS domains mediate protein–protein interactions with other general splicing factors during the formation of the spliceosome. By yeast two-hybrid for example, interactions of the SR proteins SC35 and SF2/ASF with both U1-70K and U2AF35 have been documented, the latter two proteins functionally binding to the 5′ and 3′ splice sites, respectively, in early splicing complexes (Wu and Maniatis, 1993; Cao and Garcia-Blanco, 1998). Binding of SR proteins to exonic splicing enhancers generally stimulates splicing (Sun et al., 1993; Dirksen et al., 1994; Liu et al., 1998, 2000; reviewed in Blencowe, 2000), but antagonism of splice site recognition has also been observed (Labourier et al., 1999; Barnard and Patton, 2000). Many of the functions of SR proteins are facilitated by a meshwork of interacting factors that promote the passage of the splicing reaction and participate in postsplicing processes such as mRNA transport, which appears to be coupled to splicing (Cáceres et al., 1998; Belshan et al., 2000).
ZNF265 (formally termed “Zis”) is a zinc finger– and RS domain–containing protein (Karginova et al., 1997; Adams et al., 2000) that was first identified, along with renin, because of its modulated expression in differentiating renal juxtaglomerular cells (Karginova et al., 1997); it is now known to be expressed by most tissues, especially early in development (Adams et al., 2000). We have also found that the nuclear magnetic resonance solution structure of the zinc fingers accords with RNA binding (Plambeck, C.A., D.J. Adams., L. van der Weyden., J.P. Mackay, and B.J. Morris. 22nd Ann. Conf. Org. Express. Genome. 2001. Abstr. 2–28). Therefore, we explored the function of ZNF265 by demonstrating its localization within cells, identifying the other proteins that it binds to in splicing complexes, and showing its potential to modulate alternative splicing in cells.
Results and discussion
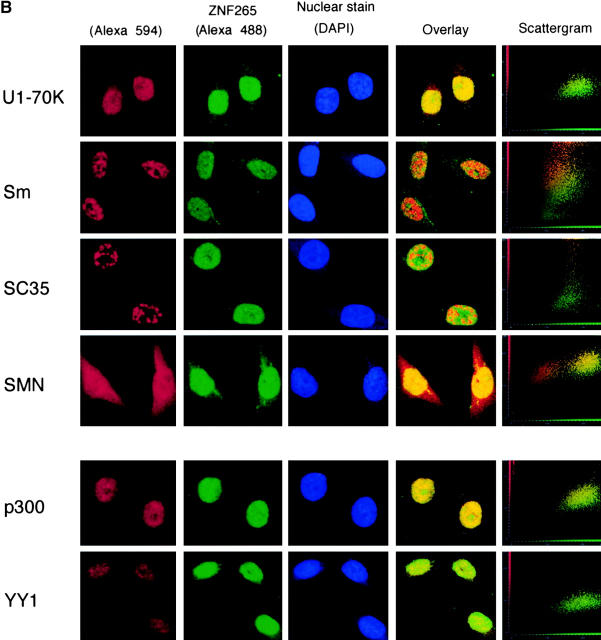
Using a polyclonal ZNF265 antibody (Fig. 1 A) and antibodies directed against specific components of the spliceosome, we observed nuclear colocalization of ZNF265 with the survival of motor neuron (SMN) protein, the authentic SR protein SC35 (at the periphery of the SC35-staining aggregates), and the snRNP protein U1-70K, but none with the common snRNP protein antigen Sm (Fig. 1 B). As expected, SMN showed some cytoplasmic localization (Pagliardini et al., 2000), but this did not overlap with the trace amount of cytoplasmic ZNF265 localization (Fig. 1 B). ZNF265 also colocalized with the transcription factors YY1 and p300 (Fig. 1 B), both of which have been shown to colocalize within active transcriptional compartments and, in the case of p300, with RNA polymerase II (Bannister and Kouzarides, 1996; Ogryzko et al., 1996; Yang et al., 1996; von Mikecz et al., 2000). These colocalizations are consistent with a role for ZNF265 in transcription and/or splicing. In this regard, ZNF265 may be cotranscriptionally recruited with RNA polymerase II to pre-mRNA transcripts, as has been reported for other RS domain–containing proteins (Corden and Patturajan, 1997).
Figure 1.
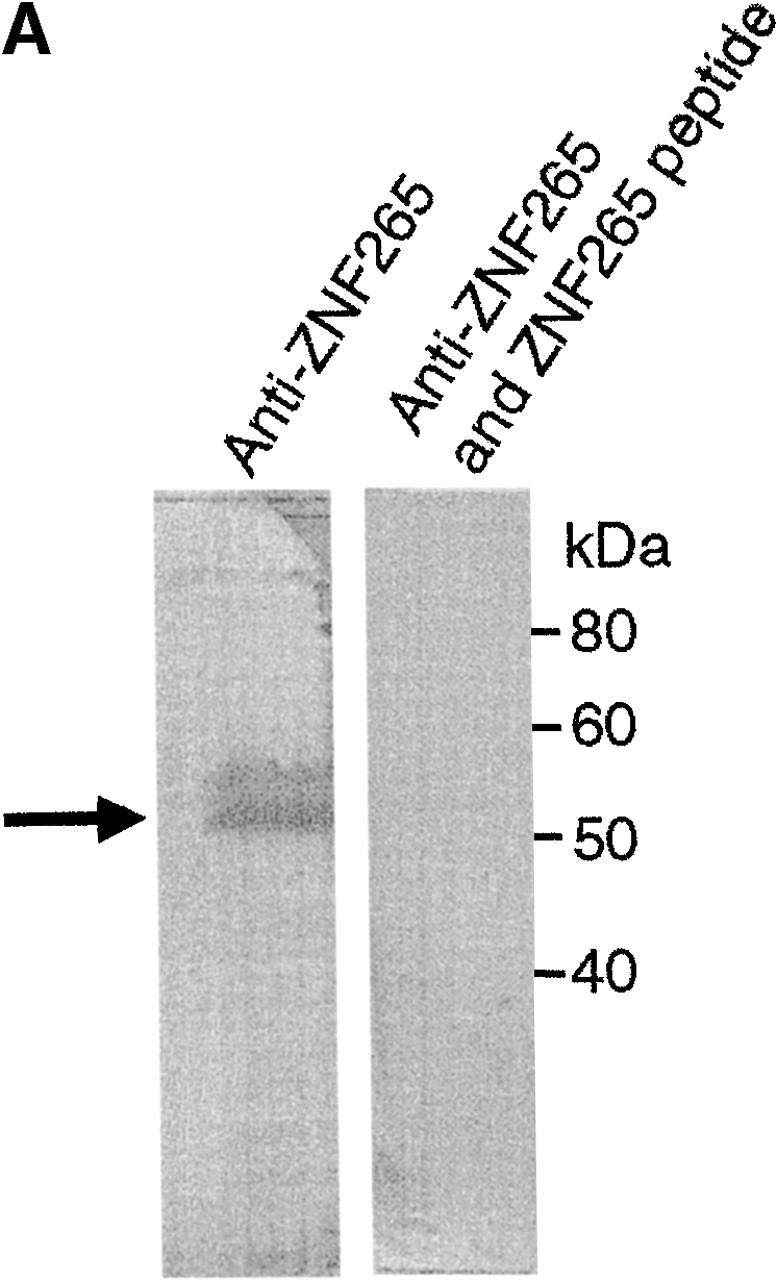
Subcellular colocalization of endogenous ZNF265 with endogenous nuclear factors. (A) Immunoblotting assay demonstrates specific recognition of ZNF265 by the polyclonal ZNF265 antibody (the arrow shows a 55-kD band), which was competed by ZNF265 oligopeptide antigen (2.5 μg/ml) in three replicate experiments. (B) Subcellular localization of various protein factors. Fixed Calu-6 cells were exposed to: (1st column) monoclonal antibodies against splicing factors U1-70K, Sm antigen, SC35, SMN, or transcriptosomal factors p300 and YY1, in respective rows, before incubation with Alexa 594 anti–mouse IgG (red); (2nd column) staining with anti-ZNF265 and detection with Alexa 488–conjugated anti–rabbit IgG (green); (3rd column) DAPI staining of nucleus (blue); (4th column) digital overlay of Z-series projections shown in columns 1 and 2 to demonstrate colocalization (yellow); (5th column) scattergrams of the overlayed projection shown in column 4. Each row represents the same field (width × height = 60 × 60 μm), acquired using three-channel confocal microscopy.
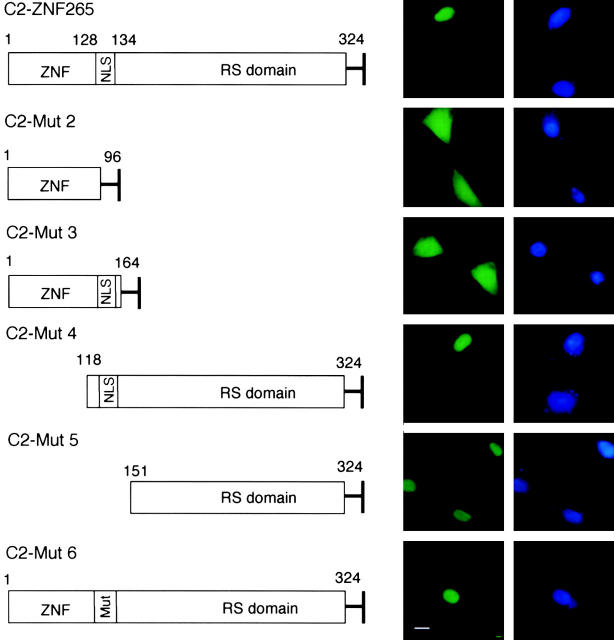
To determine the region of ZNF265 necessary for its nuclear localization, cDNA expression plasmids were generated from which specific domains were deleted. Compared with the nuclear localization of the wild-type ZNF265 fusion protein (C2-ZNF265), fusions containing the zinc finger with (C2-Mut3) or without (C2-Mut2) the putative nuclear localization signal (NLS) showed a predominantly cytoplasmic distribution (Fig. 2) . In contrast, nuclear localization was preserved when the RS domain was retained, either with (C2-Mut4) or without (C2-Mut5) the NLS. Consistent with this observation, nuclear localization was not affected by mutation of the NLS (C2-Mut6). Thus, nuclear localization is dictated by the RS domain of ZNF265, consistent with the behavior of other RS proteins such as SC35 (Hedley et al., 1995), SF2/ASF, SRp20, and 9G8 (Cáceres et al., 1997, 1998).
Figure 2.
Role of the RS domain of ZNF265 in nuclear localization. (Left) EGFP fusion protein constructs used for the expression of ZNF265. Wild-type ZNF265 sequence (1st row) and 5 mutant sequences (2nd–6th row) were used. (Right) EGFP fluorescence (green) and DAPI (blue) detection in HT-1080 cells at 48 h posttransfection. Bar, 10 μm.
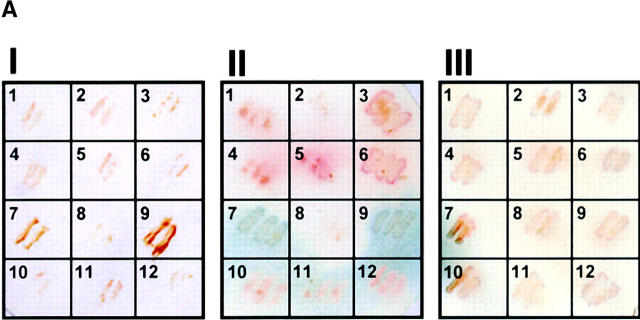
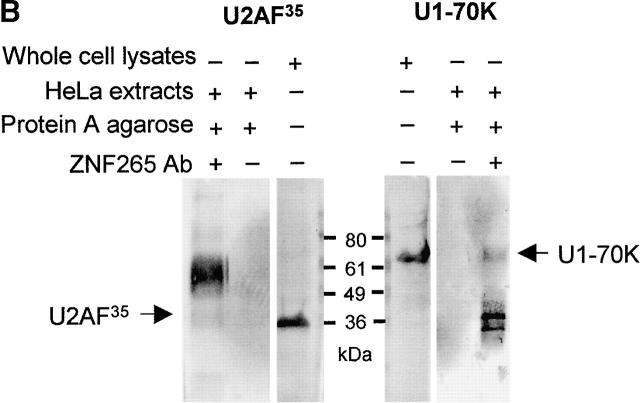
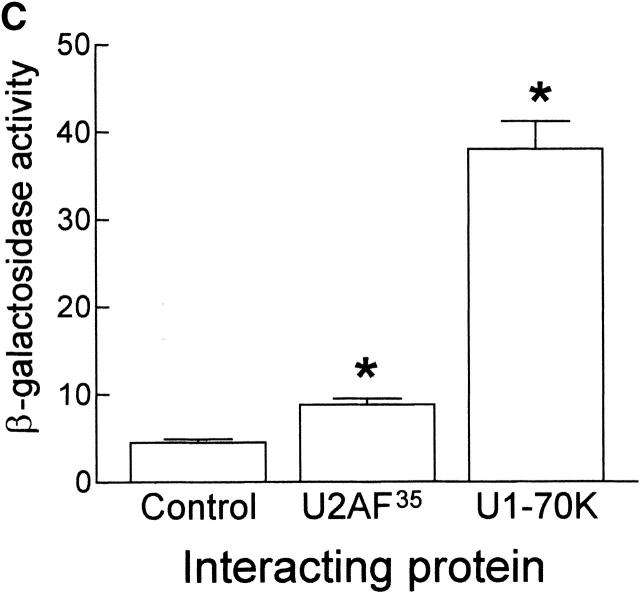
To test whether ZNF265 could interact with other RS domain–containing proteins we conducted a yeast two-hybrid screen against representative spliceosomal proteins that included many with RS domains, namely U1-70K, U2AF35, U2AF65, SC35, p80 Coilin, WT1, 9G8, SF2/ASF, SRp20, SRp30c, and SRp40. Interaction was seen with U1-70K and U2AF35, as determined by growth on SD-L-W-A-H plates, and the production of a blue precipitate on a β-gal filter assay (Fig. 3 A). Interaction of ZNF265 with U1-70K and U2AF35 was confirmed by coimmunoprecipitation (Fig. 3 B). Liquid β-gal assay, which provides a semiquantitative estimation of interaction strength, showed that ZNF265 interacted more strongly with U1-70K than with U2AF35 (Fig. 3 C). A U1-70K cDNA clone was also isolated in a yeast two-hybrid screen against a human fetal brain cDNA library using ZNF265 as “bait.” Analysis of this clone revealed that residues 180–437 of U1-70K were responsible for mediating the interaction of U1-70K with the RS domain of ZNF265 (unpublished data). It is notable that this region contains the residues necessary for the binding of SF2/ASF to U1-70K (Cao and Garcia-Blanco, 1998). Several cDNA clones for the SR protein kinase Clk1 were also isolated from this screen. Because ZNF265 contains the Clk1 consensus phosphorylation site R/KXR/KXR/KXSXXR (Colwill et al., 1996; Moeslein et al., 1999), there may be a role for phosphorylation in the regulation of ZNF265.
Figure 3.
Interaction of ZNF265 with the essential spliceosomal factors U1-70K and U2AF35. (A) Activation-domain plasmids (pACT) p80 coilin (1), 9G8 (2), SC35 (3), SRp20 (4), SRp30c (5), SRp40 (6), U1-70K (7), U2AF65 (8), U2AF35 (9), WT1 (10), ASF/SF2 (11), and negative control SNF4 (12), were transformed into AH109 yeast containing the pGBK-ZNF265 binding-domain plasmid, cultured on SD-L-W-A-H plates, and transferred to filters. The ability of the yeast containing pGBK-ZNF265 and either pACT-U1-70K or pACT-U2AF35 to grow on autotrophic media (I: brown) and produce β-gal (II: blue) was observed. The inability of yeast containing pACT plasmids alone to produce β-gal (III) was shown as a control. (B) Results of coimmunoprecipitation performed using anti-ZNF265 to pulldown U1-70K and U2AF35 in association with ZNF265 from HeLa cell nuclear extracts. Immunoprecipitates were analyzed by Western blotting using antibodies against U1-70K or U2AF35 (arrow points to band of predicted size). (C) Relative strength of interaction of ZNF265 with U2AF35 or U1-70K, shown as β-gal activity relative to that for interaction of T-antigen with p53 (mean ± SE, n = 4, *P < 0.0001). (Control) β-gal activity of AH109 yeast containing pGBK-ZNF265 alone.
The fact that ZNF265 interacts with U1-70K and U2AF35 points to its early commitment to the spliceosome, as the latter factors are necessary for the first detectable association between splice sites during formation of the E complex (Michaud and Reed, 1993; Wu and Maniatis, 1993; Xiao and Manley, 1998). Based on the composition of affinity-purified E complex, this association has been proposed to occur though direct or indirect interaction of U1snRNP and U2AF35 bound to the 5′ and 3′ splice sites, respectively (Michaud and Reed, 1993). One model has suggested that SF2/ASF or SC35 was subsequently able to form a bridge between U1-70K and U2AF35 (Wu and Maniatis, 1993). In contrast to the interaction of SF2/ASF and SC35 with U1-70K and U2AF35 (Wu and Maniatis, 1993), our finding that ZNF265 interacts more strongly with U1-70K than with U2AF35 (Fig. 3 C) suggests earlier binding of ZNF265 to U1-70K, as opposed to U2AF35, during recruitment to the spliceosome.
Alignment of the RS domain of ZNF265 with that of other RS domain–containing spliceosomal proteins (Fig. 4) showed strong SR dipeptide conservation; this was particularly evident between ZNF265, SC35, and SRp40. The aligned region of SC35 contains the putative RS domain NLS, RRRRRSRSRSRSRSRSRSRSRYSRSKSRSR-TRSRSRSTSKSRS (Hedley et al., 1995).
Figure 4.
Conservation of serine and arginine residues in ZNF265 and other RS domain proteins. Alignment of the RS domain of ZNF265 (NP_005446) with RS domains of the spliceosomal proteins SF2/ASF (NP_008855), SC35 (A42634), SRp20 (NP_003008), SRp40 (S59042), SRp55 (S59043), SRp75 (A48133), U1-70K (A25707), U2AF35 (Q01081), U2AF65 (NP_009210), 9G8 (A57198), and p54 (XP_001835). Sequence alignment was performed using “Pileup” and “Prettybox” (Australian Genomic Information Service). Residues conserved in the majority of the aligned proteins are shaded. Numbers indicate the amino acid position of each protein.
The specific recognition of splice sites within pre-mRNA precursors has been proposed as a control point for splicing. RS domain proteins could play a major role in such regulation, as they are required for the early recognition of splice sites during spliceosome assembly. Thus, the structural features of ZNF265, its nuclear localization, and its association with U1-70K and U2AF35 prompted us to investigate a functional role for this protein in pre-mRNA splicing.
In vitro splicing reactions showed that ZNF265 is immunoprecipitated in a complex that includes spliced mRNA (Fig. 5 A). This result indicates that ZNF265 binds directly or indirectly to mRNA, but much less to pre-mRNA. This property is shared with other splicing factors, such as SF2/ASF and RNPS1 (Hanamura et al., 1998; Mayeda et al., 1999), both of which synergistically stimulate general splicing. Here we show in splicing assays in cultured cells that ZNF265 can regulate alternative splicing in a concentration-dependent manner (Fig. 5 B). Namely, overexpression of ZNF265 resulted in exclusion of exons 2 and 3 from the Tra2-β1 pre-mRNA, which led to an increase in the production of the β3 alternatively spliced isoform. Our in vivo splicing result suggests that ZNF265 may have the ability to antagonize the alternative splicing activity of SR proteins on Tra2-β1 pre-mRNA. Splicing factor SR protein-mediated antagonism of alternative 5′ splice site selection has been reported for human hnRNP A1 protein in that hnRNP A1 causes activation of distal alternative 5′ splice site and exon exclusion in vitro and in vivo (Mayeda and Krainer, 1992; Mayeda et al., 1993; Cáceres et al., 1994; Yang et al., 1994). In contrast to hnRNP A1 that does not cause inhibition of general constitutive splicing, we have shown that addition of recombinant ZNF265 to SR protein-deficient HeLa cell S100 extracts supplemented with recombinant SF2/ASF may antagonize constitutive splicing of a β-globin pre-mRNA substrate and repress its splicing (our unpublished data). In Drosophila, RSF1 protein antagonizes and represses splicing by binding to SF2/ASF and preventing it from interacting with U1-70K (Labourier et al., 1999). It is possible that ZNF265 may also interfere with SF2/ASF-mediated constitutive splicing by binding directly to U1-70K.
Figure 5.
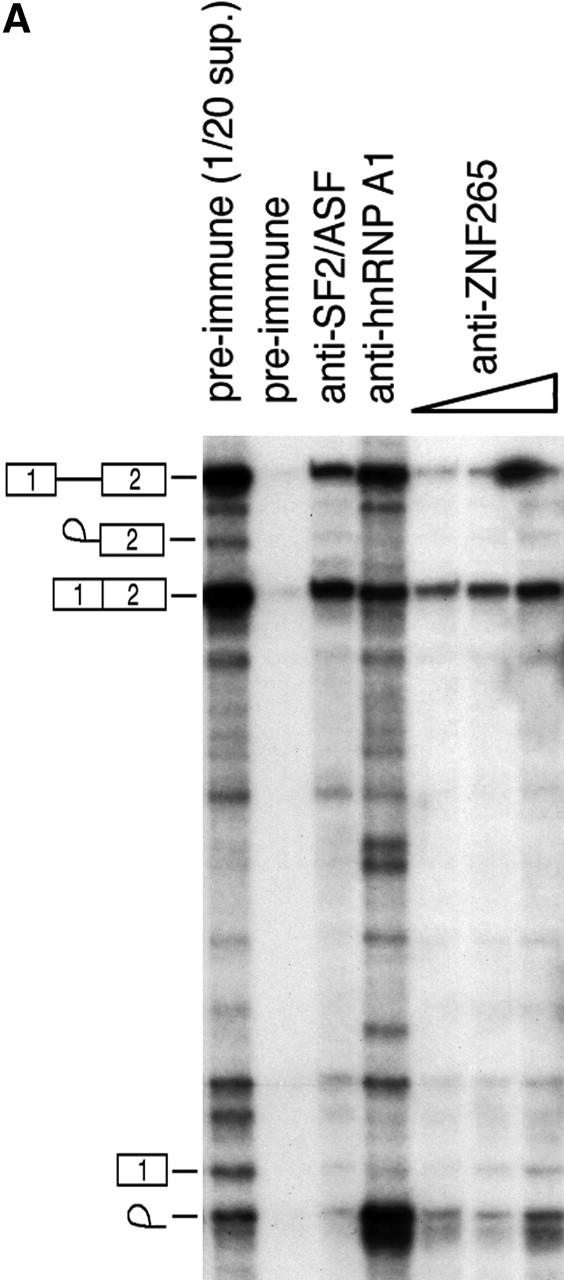
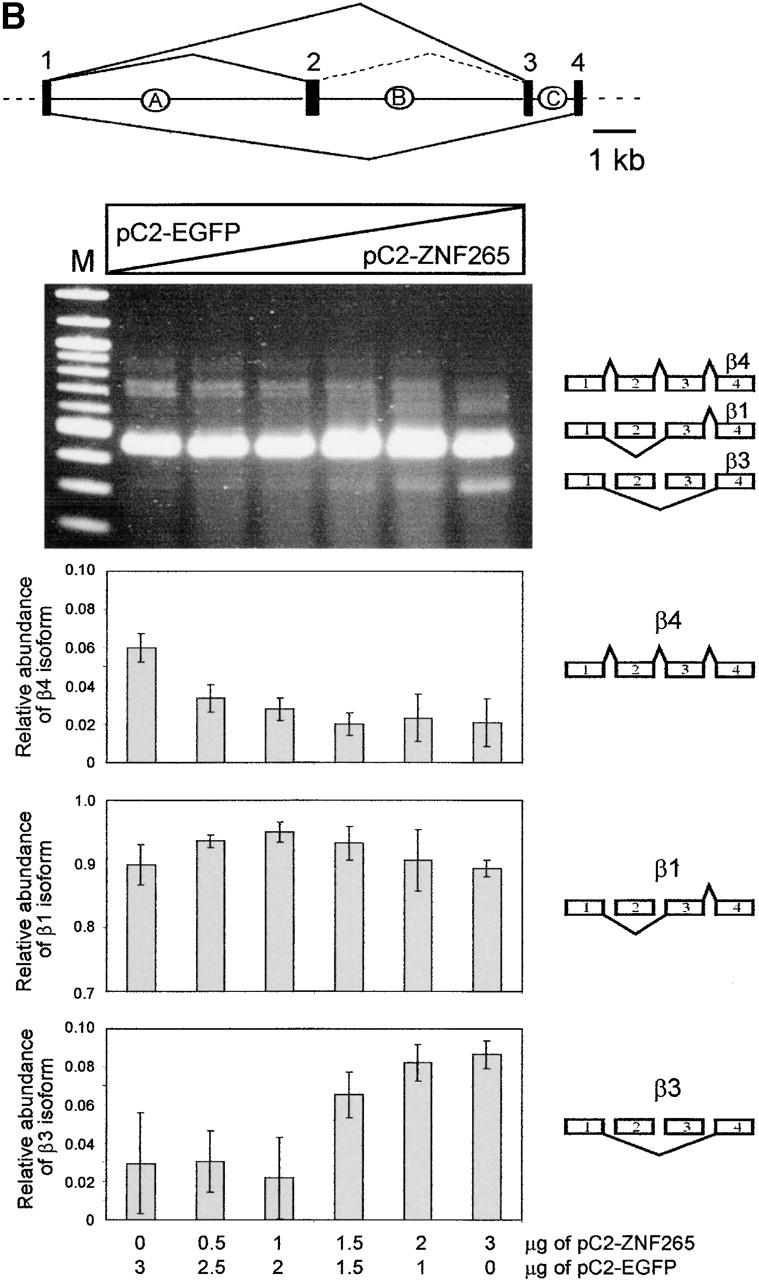
Function of ZNF265 in the spliceosome. (A) In vitro splicing reactions were performed using labeled β-globin pre-mRNA and immunoprecipitated with the indicated antibodies, pre-immune serum, anti-SF2/ASF/anti-hnRNP A1, and increasing amounts of anti-ZNF265 (shown by triangle), immobilized on protein G-Sepharose. The immunoprecipitated complexes were washed extensively and RNA was extracted and analyzed by denaturing PAGE followed by autoradiography. 1/20 of total RNA recovered from the supernatant of an immunoprecipitation with control preimmune serum reflects the initial relative abundance of predicted pre-mRNA, intermediates and products, which are schematically depicted on left hand side. (B) Ability of ZNF265 to stimulate exon exclusion of alternatively spliced Tra-β1 pre-mRNA. At top is schematic diagram of the Tra2-β1mini- gene construct and splice products (introns: A, B, C; exons: 1, 2, 3, 4). HEK 293 cells were transfected with 3 μg of total plasmid DNA and, as indicated below abscissa, an increasing proportion of the expression plasmid C2-ZNF265. Representative ethidium bromide stained gel is shown, with schematic diagram of β1, β3, and β4 isoforms detected by this assay depicted on the right. (M) 100-bp marker. Relative abundance of the β4, β1, and β3 isoforms from each lane (mean ± SD) from three experiments is shown in the panels.
The suggestion that ZNF265 binds directly to mRNA is supported by the association of the zinc finger region of the Xenopus homologue C4SR with cyclin 1B mRNA (Ladomery et al., 2000). Furthermore, our nuclear magnetic resonance studies of the first zinc finger of ZNF265 indicate a structure capable of binding zinc ions, which in turn induce conformational changes in the finger to expose RNA binding side chains (Plambeck, C.A., D.J. Adams, L. van der Weyden, J.P. Mackay, and B.J. Morris. 22nd Ann. Conf. Org. Express. Genome. 2001. Abstr. 2–28).
In conclusion, we have shown that ZNF265 colocalizes with the spliceosome, associates with mRNA and essential splicing factors U1-70K and U2AF35, and can regulate alternative splicing of the Tra2-β1 pre-mRNA. Therefore, ZNF265 is a functional component of the RNA processing machinery.
Materials and methods
Cell culture
Cell lines were obtained from The American Type Culture Collection (ATCC). Calu-6 cells (ATCC HTB-56) were cultured at 37°C, 5% CO2 in MEM (GIBCO BRL) as described previously (van der Weyden et al., 2000). HT-1080 human fibrosarcoma cells (ATCC CCL 121), HeLa cervical carcinoma cells (ATCC CCL 2), HepG2 hepatocellular carcinoma cells (ATCC HB 8065), and HEK293 (ATCC CRL 1573) were maintained at 37°C, 5% CO2 in DME (GIBCO BRL) supplemented with 10% FCS (GIBCO BRL), penicillin/streptomycin (5,000 U/ml; GIBCO BRL).
Antibodies
ZNF265 polyclonal antibodies (produced for us by Alpha Diagnostics) were generated by inoculating New Zealand white rabbits with a keyhole limpet hemocyanin–tagged peptide (CEDEDLSKYKLDEDED) corresponding to amino acids 160–174 of ZNF265 (GenBank/EMBL/DDBJ accession no. NP005446). Antiserum was affinity purified by column chromatography using antigen peptide immobilized on Sepharose-4B beads. Monoclonal SC35, p300, and YY1 antibodies were from PharMingen, Calbiochem, and Santa Cruz Biotechnology, Inc., respectively. Monoclonal SMN antibody (clone 11F3) was provided by Dr. G.E. Morris (MIRC Biotechnology Group, North East Wales Institute, Wrexham, UK), monoclonal Sm antibody (Lerner et al., 1981) was provided by Dr. A.I. Lammond (University of Dundee, Dundee, UK), monoclonal hnRNP-AI antibody (mAb9H10) was provided by Dr. G. Dreyfuss (Howard Hughes Medical Institute, University of Pennsylvania, Philadelphia, PA), and polyclonal U2AF35 (Zuo and Maniatis, 1996) and monoclonal U1-70K (Wu and Maniatis, 1993) were provided by Dr. T. Maniatis (Harvard University, Boston, MA). The SF2/ASF (mAb96) antibody used has been described previously (Hanamura et al., 1998). Secondary antibodies used were: Alexa Fluor 488–conjugated goat anti–mouse IgG (Molecular Probes), Alexa Fluor 594–conjugated goat anti–rabbit IgG (Molecular Probes), alkaline phosphatase–conjugated rabbit anti–mouse IgG (Sigma-Aldrich), and alkaline phosphatase–conjugated goat anti–rabbit IgG (Sigma-Aldrich).
Fluorescence, indirect immunofluorescence, and imaging
In preparation for visualization of fluorescence, cells were cultured on Lab-Tek chamber slides (Nunc) and fixed with 2% paraformaldehyde in situ. The cells were then permeablized with 0.5% (vol/vol) Triton X-100 in PBS for 1 h before being blocked overnight with 5% (vol/vol) goat serum in PBS. After sequential 45 min incubations at 37°C with the primary and secondary antibodies, the cells were stained with 300 nM DAPI (Molecular Probes) before being mounted with DABCO in PBS (Johnson et al., 1982). For localization studies using the ZNF265–enhanced green fluorescent protein (EGFP) fusion constructs, HT-1080 cells were transfected using SuperFect (QIAGEN) on Lab-Tek chamber slides and then fixed, DAPI stained, and mounted (as described above). Two-channel fluorescent images of the cells were acquired using a 12-bit cooled CCD camera (Sensicam) attached to an epifluorescent microscope (model E800; Nikon). For ZNF265 colocalization studies, three-channel fluorescent images were acquired on a Two Photon Imaging System (TCS MP; Leica) combined with a confocal microscope (TCS SP; Spectral). Both Alexa 488 and Alexa 594 were excited using the 488 line of an argon laser, whereas DAPI was excited in two-photon mode. Sequential scan Z-sections were obtained at 0.2 μm intervals, and projections through the Z-stack were created using confocal software (Leica). Colocalization scatter diagrams were created to confirm and quantify visual interpretation of single Z-sections and digitally projected images. All immunofluorescent images shown are representative of three independent experiments in which >95% of at least 500 cells assessed exhibited the same morphological pattern.
Plasmid constructs and subcloning
Full-length ZNF265 cDNA was amplified by RT-PCR and subcloned into pGEM T-Easy (Promega) to create the plasmid pZNFA (Adams et al., 2000). pZNFA was used as a template for PCR using primers ZNF-5′A (ctcgagtatgtcgaccaagaatttccgactc), which incorporates a 5′ XhoI site, and ZNF-3′A (cgcgttcgaagctctcccatatg). The resulting fragment was subcloned into pEGFP-C2 vector (CLONTECH Laboratories, Inc.) to generate C2-ZNF265, an in-frame fusion protein between ZNF265 and EGFP. PCR was used to obtain various domains of ZNF265 as EGFP fusion clones using C2-EGFP–pEGFP-C2. Plasmid constructs and PCR primers used were: C2-Mut2 (first 96 amino acids of ZNF265), primers ZNF-5′A and ZNF-3′B (ttatttagcatactttggagtatta); C2-Mut3 (zinc finger domains and NLS), primers ZNF-5′A and ZNF-3′C (gatcttcatccttcatccctc); C2-Mut4 (NLS and RS domains), primers ZNF-5′B (ctcgagagaatctgatggtgaatatgatg) and ZNF-3′A; C2-Mut5 (RS domain alone), primers ZNF-5′C, and ZNF-3′B (ctcgagagaatcagagggagaagaagagg). PCR (PCR Supermix High Fidelity; GIBCO BRL) consisted of 35 cycles of 95, 60, and 68°C for 1 min each, followed by 10 min at 68°C. C2-Mut6 (residues, RKKKK→AAAAA) was generated from C2-ZNF265 using primers NLS5′ (gatggtgaatatgatgagtttggagctgcagcggcagcatacagagggaaagcagttgg) and NLS3′ (ccaactgctttccctctgtatgctgccgctgcagctcca-aactcatcatattcaccatc), which destroyed the core sequence of this putative NLS. PCR was as above except that annealing was at 50°C, followed by digestion with 30 U of DpnI for 3 h. Clones were sequenced to confirm that the mutation had been introduced correctly. The ZNF265 yeast two-hybrid construct (pGBKT7-ZNF265) was generated by subcloning a BamHI-PstI fragment of the 1-kb PCR product amplified using the primers ZNF-3′A and ZNF-5′D (ggatccttatgtcgaccaagaatttccgagt), with clone ZNFA as template, into pGBKT7 vector (CLONTECH Laboratories, Inc.). DNA binding domain (pAct-BD) plasmids were provided by Drs. N. Hastie, R. Davies (MRC, Edinburgh, UK), and D. Elliot (University of Newcastle, Newcastle, UK) (Davies et al., 1998; Elliott et al., 2000).
Western blot analyses and immunoprecipitation
Calu-6, HT-1080, and HepG2 cells (4 × 105) were washed with PBS before being scraped off the flask and electrophoresed through a 12% SDS-PAGE gel (Laemmli, 1970). Proteins were then electroblotted onto polyvinylfluoride membrane (Gelman), blocked overnight in 5% skim milk, and incubated with ZNF265 antibody (1:250) for 2.5 h at 22°C, followed by goat anti–rabbit IgG (1:30,000) for another 2.5 h. Immunocomplexes were visualized by incubation of membrane in BCIP-NBT solution (FAST tablets; Sigma-Aldrich) for 5 min. For immunoprecipitations, HeLa cell nuclear extracts (100 μg/ml) (Dignam et al., 1983) were incubated with ZNF265 antibody in 1 ml TNE buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 0.15 M NaCl, 0.05% Nonidet P-40) for 2 h at 22°C. Protein G agarose (50% vol/vol in PBS; Roche) was added and the samples were rocked for 2 h at 22°C before being washed in TNE buffer. The samples were then analysed by Western blotting (as described above) using a rabbit anti-U2AF35 antibody (1:350) or mouse anti–U1-70K antibody (1:250).
Yeast culture and two-hybrid assay
Budding Saccharomyces cerevisiae strain AH109 (James et al., 1996) were cultured in YPAD (1% wt/vol yeast extract, 2% wt/vol peptone, 0.003% adenine, 2% wt/vol dextrose) and transformed sequentially (Gietz and Schiestl, 1991), first with pGBKT7–ZNF265, then with the appropriate activation domain plasmid. The yeast were then plated onto synthetic defined medium deficient in leucine, tryptophan, histidine, and adenine (SD-L-W-H-A), supplemented with kanamycin (30 μg/ml), and incubated at 30°C for 1 wk. The transformants were then analyzed for β-galactosidase (β-gal) expression by both filter assay (Turner and Crossley, 1998) and liquid assay (Galacton-Light Plus Kit; Tropix) (Asoh et al., 1998). Screening of the fetal brain cDNA library with ZNF265 as “bait” used S. cerevisiae strain AH109 (James et al., 1996) and Y187 (Harper et al., 1993). The bait ZNF265 plasmid was transformed into AH109, and the library plasmids were supplied pretransformed into Y187 (MATCHMAKER System 3; CLONTECH Laboratories, Inc.). At least 3.5 × 106 individual library plasmids were screened based on the ability of the transformed yeast to grow on SD-L-W-A-H plates and express β-gal. Library plasmids from 40 positive interactions were rescued from the yeast and sequenced.
In vitro and in vivo splicing and immunoprecipitation of splicing complexes
m7GpppG-capped 32P-labeled pre-mRNA substrates were made by runoff transcription of linearized template DNA with SP6 RNA polymerase (Mayeda et al., 1999). β-Globin plasmid pSP64-HβΔ6, used for in vitro transcription, has been described previously (Krainer et al., 1984). HeLa cell nuclear extracts were made as reported previously (Mayeda and Krainer, 1999). Immunoprecipitation of the products of in vitro splicing reactions was performed according to Hanamura et al.'s method (1998). The RNA products of the immunoprecipitates were analyzed by electrophoresis on a 5.5% polyacrylamide/7 M urea gel, followed by autoradiography.
Additional splicing assays were performed essentially as described in Stoss et al. (1999). Human transformer-2-β1 (Tra2-β1) minigene (Nayler et al., 1998) and C2-ZNF265 expression plasmids were transfected into HEK293 cells. After RNA isolation and reverse transcription (Hartmann et al., 1999), PCR to amplify minigene products was performed thus: 35 cycles of 94°C for 15 s, 65°C for 20 s, and 72°C for 40 s (Daoud et al., 1999). The resulting splicing pattern was quantified using the Herolab EASY system.
Acknowledgments
We gratefully acknowledge the assistance of the following: Michelle Pedler in transfection and antibody staining, Dr. Guy Cox (Electron Microscope Unit, University of Sydney, Sydney, Australia) for help with confocal microscopy, Drs. Tom Maniatis, Derek Kennedy, Gideon Dreyfuss, Angus Lamond, and Glen Morris for donation of antibodies, and Drs. Rachel Davies, Nick Hastie, and David Elliot for yeast two-hybrid constructs. We wish to express our gratitude to the reviewers of this paper for many helpful comments and suggestions that significantly improved the manuscript.
This work was funded by grants from the Australian Research Council (to B.J. Morris), the Lucille P. Markey Trust (to A. Mayeda), the Deutsche Forschungsgemeinschaft (to S. Stamm), and the National Health and Medical Research Council of Australia (to J.E.J. Rasko).
Footnotes
Abbreviations used in this paper: EGFP, enhanced green fluorescent protein; NLS, nuclear localization signal; RS, arginine/serine-rich; SMN, survival of motor neuron; snRNP, small nuclear ribonucleoprotein particle; SR, serine/arginine-rich.
References
- Adams, D.J., L. van der Weyden, A. Kovacic, F.J. Lovicu, N.G. Copeland, D.J. Gilbert, N.A. Jenkins, P.A. Ioannou, and B.J. Morris. 2000. Chromosomal localization and characterization of the mouse and human zinc finger protein 265 gene. Cytogenet. Cell Genet. 88:68–73. [DOI] [PubMed] [Google Scholar]
- Asoh, S., K. Nishimaki, R. Nanbu-Wakao, and S. Ohta. 1998. A trace amount of the human pro-apoptotic factor Bax induces bacterial death accompanied by damage of DNA. J. Biol. Chem. 273:11384–11391. [DOI] [PubMed] [Google Scholar]
- Bannister, A.J., and T. Kouzarides. 1996. The CBP co-activator is a histone acetyltransferase. Nature. 384:641–643. [DOI] [PubMed] [Google Scholar]
- Belshan, M., G.S. Park, P. Bilodeau, C.M. Stoltzfus, and S. Carpenter. 2000. Binding of equine infectious anemia virus rev to an exon splicing enhancer mediates alternative splicing and nuclear export of viral mRNAs. Mol. Cell. Biol. 20:3550–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard, D.C., and J.G. Patton. 2000. Identification and characterization of a novel serine–arginine-rich splicing regulatory protein. Mol. Cell. Biol. 20:3049–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe, B.J. 2000. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 25:106–110. [DOI] [PubMed] [Google Scholar]
- Cáceres, J.F., S. Stamm, D.M. Helfman, and A.R. Krainer. 1994. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 265:1706–1709. [DOI] [PubMed] [Google Scholar]
- Cáceres, J.F., T. Misteli, G.R. Screaton, D.L. Spector, and A.R. Krainer. 1997. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol. 138:225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres, J.F., G.R. Screaton, and A.R. Krainer. 1998. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 12:55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, W., and M.A. Garcia-Blanco. 1998. A serine/arginine-rich domain in the human U1 70k protein is necessary and sufficient for ASF/SF2 binding. J. Biol. Chem. 273:20629–20635. [DOI] [PubMed] [Google Scholar]
- Colwill, K., T. Pawson, B. Andrews, J. Prasad, J.L. Manley, J.C. Bell, and P.I. Duncan. 1996. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO (Eur. Mol. Biol. Organ.) J. 15:265–275. [PMC free article] [PubMed] [Google Scholar]
- Corden, J.L., and M. Patturajan. 1997. A CTD function linking transcription to splicing. Trends Biochem. Sci. 22:413–416. [DOI] [PubMed] [Google Scholar]
- Daoud, R., M. Berzaghi, F. Siedler, M. Hübener, and S. Stamm. 1999. Activity-dependent regulation of alternative splicing patterns in the rat brain. Eur. J. Neurosci. 11:788–802. [DOI] [PubMed] [Google Scholar]
- Davies, R.C., C. Calvio, E. Bratt, S.H. Larsson, A.I. Lamond, and N.D. Hastie. 1998. WT1 interacts with the splicing factor U2AF65 in an isoform-dependent manner and can be incorporated into spliceosomes. Genes Dev. 12:3217–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam, J.D., R.M. Lebovitz, and R.G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirksen, W.P., R.K. Hampson, Q. Sun, and F.M. Rottman. 1994. A purine-rich exon sequence enhances alternative splicing of bovine growth hormone pre-mRNA. J. Biol. Chem. 269:6431–6436. [PubMed] [Google Scholar]
- Du, L., and S.L. Warren. 1997. A functional interaction between the COOH-terminal domain of RNA polymerase II and pre-mRNA splicing. J. Cell Biol. 136:5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott, D.J., C.F. Bourgeois, A. Klink, J. Stevenin, and H.J. Cooke. 2000. A mammalian germ cell-specific RNA-binding protein interacts with ubiquitously expressed proteins involved in splice site selection. Proc. Natl. Acad. Sci. USA. 97:5717–5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, X.D. 1995. The superfamily of arginine/serine-rich splicing factors. RNA. 1:663–680. [PMC free article] [PubMed] [Google Scholar]
- Gietz, R.D., and R.H. Schiestl. 1991. Applications of high efficiency lithium acetate transformation of intact yeast cells using single-stranded nucleic acids as carrier. Yeast. 7:253–263. [DOI] [PubMed] [Google Scholar]
- Harper, J.W., G.R. Adami, N. Wei, K. Keyomarsi, and S.J. Elledge. 1993. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 75:805–816. [DOI] [PubMed] [Google Scholar]
- Hartmann, A.M., O. Nayler, F.W. Schwaiger, A. Obermeier, and S. Stamm. 1999. The interaction and colocalisation of SAM68 with the splicing associated factor YT521-B in nuclear dots is regulated by the Src family kinase p59fyn. Mol. Biol. Cell. 10:3909–3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanamura, A., J.F. Cáceres, A. Mayeda, B.R. Franza, and A.R. Krainer. 1998. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA. 4:430–444. [PMC free article] [PubMed] [Google Scholar]
- Hedley, M.L., H. Amrein, and T. Maniatis. 1995. An amino acid sequence motif sufficient for subnuclear localization of an arginine/serine-rich splicing factor. Proc. Natl. Acad. Sci. USA. 92:11524–11528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, P., J. Halladay, and E.A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 144:1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, G.D., R.S. Davidson, K.C. McNamee, G. Russell, D. Goodwin, and E.J. Holborow. 1982. Fading of immunofluorescence during microscopy: a study of the phenomenon and its remedy. J. Immunol. Methods. 55:231–242. [DOI] [PubMed] [Google Scholar]
- Karginova, E.A., E.S. Pentz, I.G. Kazakova, V.F. Norwood, R.M. Carey, and R.A. Gomez. 1997. Zis: a developmentally regulated gene expressed in juxtaglomerular cells. Am. J. Physiol. 273:F731–F738. [DOI] [PubMed] [Google Scholar]
- Kim, E., L. Du, D.B. Bregman, and S.L. Warren. 1997. Splicing factors associate with hyperphosphorylated RNA polymerase II in the absence of pre-mRNA. J. Cell Biol. 136:19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer, A.R., T. Maniatis, B. Ruskin, and M.R. Green. 1984. Normal and mutant human β-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 36:993–1005. [DOI] [PubMed] [Google Scholar]
- Labourier, E., H.M. Bourbon, I.E. Gallouzi, M. Fostier, E. Allemand, and J. Tazi. 1999. Antagonism between RSF1 and SR proteins for both splice-site recognition in vitro and Drosophila development. Genes Dev. 13:740–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladomery, M., R. Marshall, L. Arif, and J. Sommerville. 2000. C4SR, a novel zinc-finger protein with SR-repeats, is expressed during early development of Xenopus. Gene. 256:293–302. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227:680–685. [DOI] [PubMed] [Google Scholar]
- Lerner, E.A., M.R. Lerner, C.A. Janeway, Jr., and J.A. Steitz. 1981. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc. Natl. Acad. Sci. USA. 78:2737–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H.X., M. Zhang, and A.R. Krainer. 1998. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 12:1998–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H.X., S.L. Chew, L. Cartegni, M.Q. Zhang, and A.R. Krainer. 2000. Exonic splicing enhancer motif recognized by human SC35 under splicing conditions. Mol. Cell. Biol. 20:1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley, J.L., and R. Tacke. 1996. SR proteins and splicing control. Genes Dev. 10:1569–1579. [DOI] [PubMed] [Google Scholar]
- Mayeda, A., and A.R. Krainer. 1992. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 68:365–375. [DOI] [PubMed] [Google Scholar]
- Mayeda, A., and A.R. Krainer. 1999. Mammalian in vitro splicing assays. Meth. Mol. Biol. 118:315–321. [DOI] [PubMed] [Google Scholar]
- Mayeda, A., D.M. Helfman, and A.R. Krainer. 1993. Modulation of exon skipping and inclusion by heterogeneous nuclear ribonucleoprotein A1 and pre-mRNA splicing factor SF2/ASF. Mol. Cell. Biol. 13:2993–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda, A., J. Badolato, R. Kobayashi, M.Q. Zhang, E.M. Gardiner, and A.R. Krainer. 1999. Purification and characterization of human RNPS1: a general activator of pre-mRNA splicing. EMBO (Eur. Mol. Biol. Organ.) J. 18:4560–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud, S., and R. Reed. 1993. A functional association between the 5′ and 3′ splice site is established in the earliest prespliceosome complex (E) in mammals. Genes Dev. 7:1008–1020. [DOI] [PubMed] [Google Scholar]
- Misteli, T., and D.L. Spector. 1999. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol. Cell. 3:697–705. [DOI] [PubMed] [Google Scholar]
- Moeslein, F.M., M.P. Myers, and G.E. Landreth. 1999. The CLK family kinases, CLK1 and CLK2, phosphorylate and activate the tyrosine phosphatase, PTP-1B. J. Biol Chem. 274:26697–26704. [DOI] [PubMed] [Google Scholar]
- Nayler, O., C. Cap, and S. Stamm. 1998. Human transformer-2-β gene: complete nucleotide sequence, chromosomal localization and generation of a tissue specific isoform. Genomics. 53:191–202. [DOI] [PubMed] [Google Scholar]
- Ogryzko, V.V., R.L. Schiltz, V. Russanova, B.H. Howard, and Y. Nakatani. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 87:953–959. [DOI] [PubMed] [Google Scholar]
- Pagliardini, S., A. Giavazzi, V. Setola, C. Lizier, M. Di Luca, S. DeBiasi, and G. Battaglia. 2000. Subcellular localization and axonal transport of the survival motor neuron (SMN) protein in the developing rat spinal cord. Hum. Mol. Genet. 9:47–56. [DOI] [PubMed] [Google Scholar]
- Singer, R.H., and M.R. Green. 1997. Compartmentalization of eukaryotic gene expression: causes and effects. Cell. 91:291–294. [DOI] [PubMed] [Google Scholar]
- Stoss, O., P. Stoilov, A.M. Hartmann, O. Nayler, and S. Stamm. 1999. The in vivo minigene approach to analyze tissue-specific splicing. Brain Res. Brain Res. Protoc. 4:383–394. [DOI] [PubMed] [Google Scholar]
- Sun, Q., A. Mayeda, R.K. Hampson, A.R. Krainer, and F.M. Rottman. 1993. General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes Dev. 7:2598–2608. [DOI] [PubMed] [Google Scholar]
- Turner, J., and M. Crossley. 1998. Cloning and characterization of mCtBP2, a co-repressor that associates with basic Kruppel-like factor and other mammalian transcriptional regulators. EMBO (Eur. Mol. Biol. Organ.) J. 17:5129–5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Weyden, L., D.J. Adams, and B.J. Morris. 2000. Capacity for purinergic control of renin promoter via P2Y11 receptor and cAMP pathways. Hypertension. 36:1093–1098. [DOI] [PubMed] [Google Scholar]
- von Mikecz, A., S.S. Zhang, M. Montminy, E.M. Tan, and P. Hemmerich. 2000. CREB-binding protein (CBP)/p300 and RNA polymerase II colocalize in transcriptionally active domains in the nucleus. J. Cell Biol. 150:265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J.Y., and T. Maniatis. 1993. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 75:1061–1070. [DOI] [PubMed] [Google Scholar]
- Xiao, S.H., and J.L. Manley. 1998. Phosphorylation-dephosphorylation differentially affects activities of splicing factor ASF/SF2. EMBO (Eur. Mol. Biol. Organ.) J. 17:6359–6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X., M.R. Bani, S.J. Lu, S. Rowan, Y. Ben-David, and B. Chabot. 1994. The A1 and A1B proteins of heterogeneous nuclear ribonucleoparticles modulate 5′ splice site selection in vivo. Proc. Natl. Acad. Sci. USA. 91:6924–6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X.J., V.V. Ogryzko, J. Nishikawa, B.H. Howard, and Y. Nakatani. 1996. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1a. Nature. 382:319–324. [DOI] [PubMed] [Google Scholar]
- Yuryev, A., M. Patturajan, Y. Litingtung, R.V. Joshi, C. Gentile, M. Gebara, and J.L. Corden. 1996. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc. Natl. Acad. Sci. USA. 93:6975–6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, P., and T. Maniatis. 1996. The splicing factor U2AF35 mediates critical protein-protein interactions in constitutive and enhancer-dependent splicing. Genes Dev. 10:1356–1368. [DOI] [PubMed] [Google Scholar]