Abstract
The transcriptional regulator CtrA controls several key cell-cycle events in Caulobacter crescentus, including the initiation of DNA replication, DNA methylation, cell division, and flagellar biogenesis. CtrA is a member of the response regulator family of two component signal transduction systems. Caulobacter goes to great lengths to control the time and place of the activity of this critical regulatory factor during the cell cycle. These controls include temporally regulated transcription and phosphorylation and spatially restricted proteolysis. We report here that ctrA expression is under the control of two promoters: a promoter (P1) that is active only in the early predivisional cell and a stronger promoter (P2) that is active in the late predivisional cell. Both promoters exhibit CtrA-mediated feedback regulation: the early P1 promoter is negatively controlled by CtrA, and the late P2 promoter is under positive feedback control. The CtrA protein footprints conserved binding sites within the P1 and P2 promoters. We propose that the P1 promoter is activated after the initiation of DNA replication in the early predivisional cell. The ensuing accumulation of CtrA results in the activation of the P2 promoter and the repression of the P1 promoter late in the cell cycle. Thus, two transcriptional feedback loops coupled to cell cycle-regulated proteolysis and phosphorylation of the CtrA protein result in the pattern of CtrA activity required for the temporal and spatial control of multiple cell-cycle events.
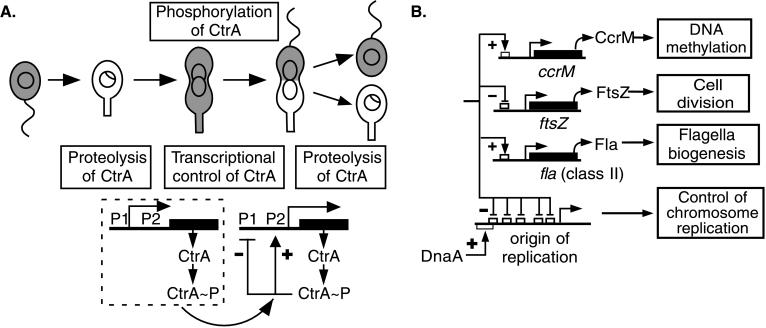
The cell cycle of the bacterium Caulobacter crescentus is accompanied by morphological transitions that produce an asymmetric predivisional cell. On cell division, one pole yields a swarmer cell, and the other pole yields a nonmotile stalked cell (Fig. 1B), each with different cell fates (1). The progeny- stalked cell immediately initiates DNA replication, whereas DNA replication is repressed in the progeny swarmer cell until later in the cell cycle when the swarmer cell differentiates into a new stalked cell (2, 3). Thus, the swarmer-to-stalked cell transition is coincident with the initiation of DNA replication. A critical function of the CtrA response regulator is to bind to and repress the origin of replication in the swarmer cell and thus control the time of initiation of DNA replication (4).
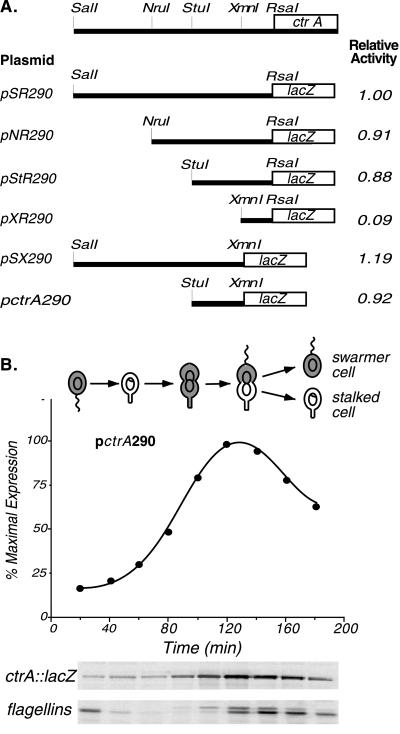
Figure 1.
(A) Deletion analysis to define the functional ctrA promoter region. Fragments of the region upstream of ctrA were fused to a promoterless lacZ reporter in pRKlac290. β-Galactosidase activity (16) was measured in mid-log phase wild-type cultures bearing the plasmids shown. The activity of pSR290, defined here as 1.0, was 7,560 Miller units. The ctrA promoter was localized to a 190-bp StuI to XmnI fragment (pctrA290). (B) Temporal expression of the ctrA promoter. The Caulobacter cell cycle is shown schematically. The gray shading marks the presence of CtrA. The theta and ring structures within the cells represent replicating DNA and nonreplicating DNA, respectively. As wild-type cells bearing pctrA290 progressed through the cell cycle, samples were pulse labeled with [35S]methionine at the indicated times, and β-galactosidase and flagellin synthesis was assessed by immunoprecipitation. Labeled proteins were separated by gel electrophoresis (Lower). The pctrA290 activity was quantitated with a PhosphorImager and is shown as a function of the cell cycle. Flagellin synthesis was assayed as an internal control for cell-cycle progression.
During S phase, the stalked cell elongates and differentiates a new swarmer pole by activating a flagellar transcriptional cascade (5). Then in late S phase, the newly synthesized CcrM DNA methyltransferase brings the replicated chromosomes from the hemimethylated to the fully methylated state before cell division (6, 7). Division of the asymmetric predivisional cell is preceded by the synthesis and assembly of the polar flagellum and the tubulin-like cell division protein, FtsZ (8). All of these cell-cycle events are controlled by a single protein, the essential response regulator CtrA (9–11).
Response regulators are typically activated by receiving a phosphate from a cognate sensor kinase. We have previously shown that CtrA requires phosphorylation for activity (9) and that the CckA histidine kinase is involved in CtrA phosphorylation (12). Phosphorylated CtrA (CtrA≈P) is present in the swarmer cell, rapidly disappears at the swarmer-to-stalked cell transition, and accumulates again in the predivisional cell. After cell division, the protein is maintained in the progeny swarmer cell but is degraded in the progeny stalked cell (13). This cell-cycle pattern of CtrA≈P distribution, and thus cell-cycle progression, is caused by temporally regulated phosphorylation coupled to temporally and spatially restricted proteolysis (13). The transcription of ctrA is also cell-cycle regulated; it is restricted to the predivisional cell (9). We show here that the cell-cycle pattern of ctrA transcription is caused by the differential activity of two temporally regulated promoters that are under CtrA-mediated feedback control. One promoter, P1, is expressed in early predivisional cells, and the other, P2, is expressed in late predivisional cells. We propose that a negative feedback loop at the P1 promoter and a positive feedback loop at the P2 promoter, coupled to the regulated phosphorylation and proteolysis of the CtrA protein, result in the temporally restricted appearance of active CtrA, and thereby contribute to the coordinate expression of multiple cell-cycle events.
MATERIALS AND METHODS
Bacterial Strains, Plasmids, and Growth Conditions.
C. crescentus NA1000 (a synchronizable derivative of the wild-type strain CB15) and derivative strains were grown in PYE complex media or M2G minimal media at 30°C (14). The isogenic temperature-sensitive strain ctrA401 (LS2195) was grown at either 28°C or 37°C (the restrictive temperature) (9). Strain LS2528 contains PxylX-ctrA integrated into the chromosome as the only copy of ctrA in the cell. Antibiotics used include tetracycline (2 μg/ml for Caulobacter and 12.5 μg/ml for Escherichia coli), ampicillin (100 μg/ml), and nalidixic acid (20 μg/ml). Plasmids were mobilized from E. coli strain S17–1 into C. crescentus by bacterial conjugation (14). Transcriptional fusions were constructed by subcloning the different PctrA restriction fragments into pBluescript II SK(+) and then into pRKlac290 (15). Small promoter fragments (<200 bp) were amplified by PCR by using pID1 as a template, ligated into pCR2.1 (Invitrogen), sequenced, and then cloned into pRKlac290. pID1 contains a 470-bp MscI to RsaI fragment of the ctrA promoter region in the SmaI site of pBluescript SKII(+) with PctrA in the same orientation as lacZ. The pctrA290 plasmid was generated by using the dom1 (5′-CGTCAGGCGGATCCGCCGCCAGGG-3′) and the dom2 (5′-GGAGTCCGCTCTAGAAACCCTTCG-3′) oligonucleotides. The Taquence Sequencing kit (United States Biochemical) and the ThermoSequenase kit (Amersham Pharmacia) were used for DNA sequencing.
Site-Directed Mutagenesis of PctrA.
Site-directed mutants in PctrA P1 and P2 were generated by PCR. To create the plasmid pctrA-P1, a mutagenic reverse oligonucleotide (5′-GTGAAACCCTTCGGCCACCCGGCCGGAGAG-3′) that disrupts the −10 region of PctrA P2 was used to amplify the promoter fragment. The altered bases in the −10 region are underlined. In this promoter construct, the AAA sequence at −10 was changed to CGG (see Fig. 2A). In plasmid pctrA-P2, the P1 promoter was mutagenized by inserting 5 bp (GCTTC) at −20, changing the spacing between the −35 region and the TTAAC motif at −10 and their relative orientation on the DNA helix. The mutagenic oligonucleotides used were 5′-GCACCCGATTCGCAAGCTTCATCAGATTAACCATTCC-3′ (forward primer) and 5′-GGAATGGTTAATCTGATGAAGCTTGCGAATCGGGTGC-3′ (reverse primer). The inserted bases are italicized. Both PCR products were sequenced and transcriptional fusions were constructed in pRKlac290, as described above.
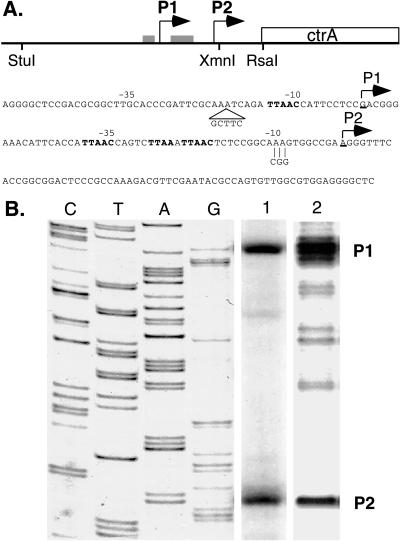
Figure 2.
Identification of ctrA transcription start sites. (A) The diagram shows the location of the P1 and P2 ctrA transcription start sites (bent arrows) and CtrA recognition motifs (gray boxes) relative to the ctrA gene. The −10 and −35 regions of the two promoters are marked above the nucleotide sequence. CtrA recognition motifs and the translation start site are in bold. Individual mutations generated in the P1 promoter (5-bp insert in pctrA-P2) and the P2 promoter (3-bp substitution in pctrA-P1) are indicated. (B) Primer extension and S1 nuclease mapping of the ctrA transcription start sites. The first four lanes show a sequencing ladder generated by using a primer with the same 5′ end as the primer extension primer and the S1 probe. Two transcription start sites were detected. The products common to primer extension (lane 1) and S1 nuclease protection (lane 2) are labeled P1 and P2.
Promoter Activity and ctrA Transcript Analysis.
β-Galactosidase activity of the promoter-lacZ fusions was assayed at 30°C in log-phase cultures as described (16). Assays were done in triplicate with a minimum of three independent cultures for each promoter construct. Transcription during the cell cycle was measured in synchronous cultures obtained by Ludox density centrifugation (17). At 20-min intervals, 1 ml of culture was pulse labeled with 15 μCi [35S]methionine (ICN) for 5 min. Synthesis of β-galactosidase and flagellins was monitored by immunoprecipitation and SDS/PAGE as previously described (18). Radiolabeled proteins were quantitated by using a PhosphorImager (Molecular Dynamics).
Primer extension, S1 nuclease mapping, and RNase protection assays were performed as described (19). Total cellular RNA was isolated from mid-log-phase cultures by hot phenol-SDS lysis (20) or with the RNeasy Midi Kit (Qiagen). Primer extension assays were performed with Superscript II Reverse Transcriptase (GIBCO/BRL) and the oligonucleotide primer 5′-CATCCTCGATCAACAGTACG-3′ located at the ctrA translational start site. The DNA probe for S1 mapping was a 500-bp BamHI to HindIII fragment of pID1. Annealing temperatures of 45°C and 56°C were used for primer extension and S1 mapping, respectively. For RNase protection, a [32P]-labeled cRNA probe (prepared by in vitro transcription by using pID1 as a template and the dom1 primer) was hybridized with Caulobacter RNA at 45°C.
DNase I Footprinting.
DNase I protection experiments were performed with a purified His6-CtrA fusion protein as described (9, 21). Template DNA was a 500-bp BamHI to HindIII fragment of pID1 end labeled with [32P]-γATP.
RESULTS
Transcription of ctrA Is Controlled by Two Cell Cycle-Regulated Promoters.
To define the ctrA promoter region, we constructed a series of plasmids containing fragments of the ctrA 5′ region fused to a promoterless lacZ gene on the low copy number plasmid pRKlac290 (Fig. 1). β-Galactosidase activity (16) was then assayed in wild-type strains carrying these plasmids. The smallest fragment exhibiting full promoter activity (pctrA290; Fig. 1A) is a 190-bp fragment located approximately 65 bp upstream of the ctrA translational start site. The cell-cycle expression of this promoter was assayed by pulse labeling aliquots of synchronized cells with [35S]methionine at 20-min intervals and immunoprecipitating radiolabeled β-galactosidase produced from the transcriptional fusion (Fig. 1B). Transcription from pctrA290 was temporally controlled with transcription peaking in the predivisional cell (Fig. 1B), in agreement with the timing observed for a strain bearing a chromosomal ctrA∷lacZ transcriptional fusion containing the full upstream region (9). These data demonstrate that the 190-bp ctrA promoter region contains all elements essential for the cell cycle-regulated timing of ctrA transcription.
The transcriptional start sites of the ctrA gene were determined by primer extension and S1 nuclease protection assays by using RNA isolated from wild-type cells (Fig. 2B). Both the primer extension and the S1 nuclease protection assays revealed two major transcripts, one initiating at 65 bp (P2) and the other at 122 bp (P1) upstream of the ctrA translational start site (Fig. 2A).
To determine the relative contribution of each of these promoters to the cell-cycle pattern of ctrA transcription, wild-type cells were synchronized, and RNA samples were taken at 20-min intervals during the cell cycle. RNase protection assays performed on these samples confirmed the presence of two transcriptional start sites of the correct size and showed that they were differentially controlled during the cell cycle (Fig. 3A). P1 exhibited peak activity in the early predivisional cell, and P2 was active in the late predivisional cell.
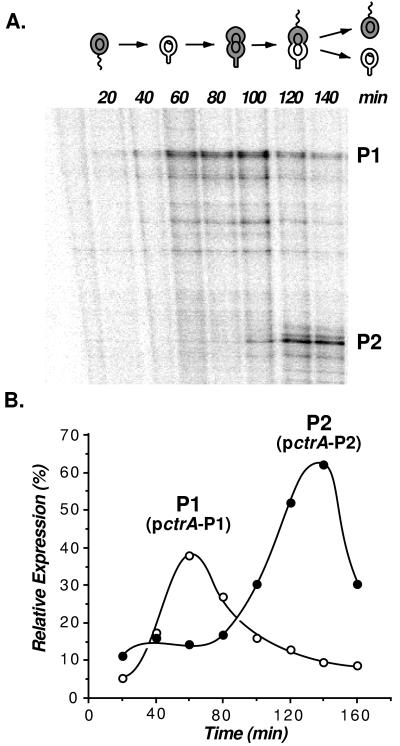
Figure 3.
Temporal expression of the ctrA P1 and P2 promoters. (A) RNase protection analysis. Total cellular RNA was isolated from synchronous wild-type cultures at 20-min intervals. RNase protection assays were performed by using a [32P]dCTP labeled cRNA probe complementary to the ctrA promoter region. Protected RNAs were resolved on a sequencing gel and visualized with a PhosphorImager. The P1 and P2 transcripts are labeled. (B) Expression of pctrA-P1 (o) and pctrA-P2 (●) (diagrammed in Fig. 4A) fused to a promoterless lacZ gene were used to monitor transcription as a function of the cell cycle, as described in the legend to Fig. 1. The relative expression of the two promoters was based on the β-galactosidase activity of pctrA-P1 and pctrA-P2 (3,940 and 6,280 Miller units, respectively).
Separate mutations in the P1 and P2 promoter regions were generated, as shown in Figs. 2A and 4A. The mutation in the P1 promoter was a 5-bp (GCTTC) insert at −20, and the mutation in the P2 promoter was a replacement of AAA in the −10 region with CGG. To confirm that there are two independent and functional ctrA promoters, we used primer extension assays to monitor transcription from the isolated P1 (pctrA-P1) and P2 (pctrA-P2) promoter mutants. With each of the promoter constructs, a single transcript of the predicted size was observed (data not shown). The promoter activity of P1 (pctrA-P1) or P2 (pctrA-P2) was then assayed in synchronized cell populations to determine when each promoter is active (Fig. 3B). The time of lacZ expression from these transcriptional fusions confirmed that the P1 promoter is maximally active in the early predivisional cell and that the stronger P2 promoter is active only in the late predivisional cell.
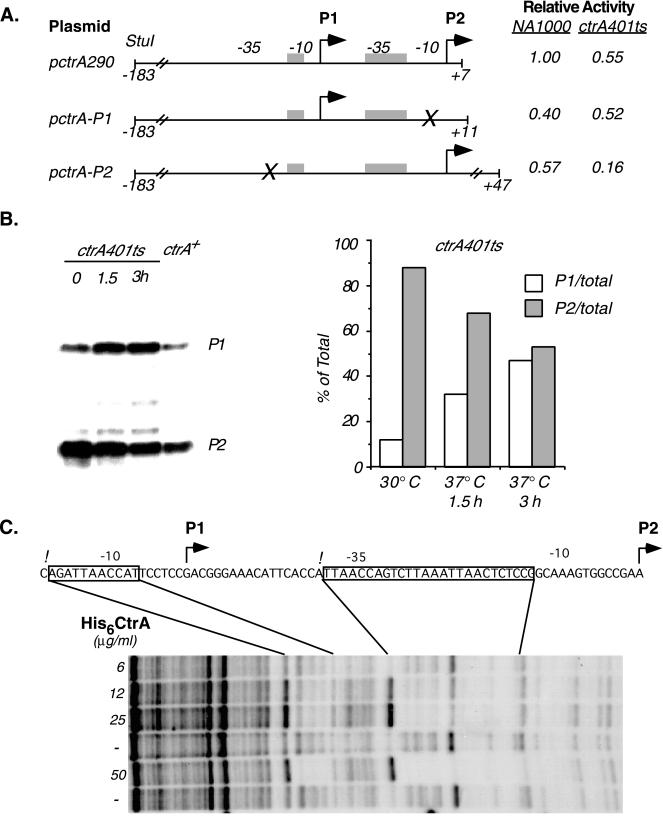
Figure 4.
The CtrA protein controls its own transcription. (A) Promoter activity of wild-type and mutant ctrA promoters. The activity of the wild-type promoter containing both P1 and P2 (plasmid pctrA290) and constructs containing either P1 or P2 activity alone was measured in wild-type cells and in a strain bearing a ctrA401ts allele. Plasmid pctrA-P1 was generated by site-directed mutagenesis of the −10 region of P2 (marked by the X). In pctrA-P2, the P1 promoter was disrupted by inserting 5 bp at −20 (see Fig. 2A). β-Galactosidase activity was measured in wild-type and ctrA401ts cultures after 3 hr at 37°C. Transcription of pctrA-P1 and pctrA-P2 was normalized to pctrA290 activity in the wild-type strain. Values are the average of three determinations. (B) S1 nuclease protection analysis of the P1 and P2 transcripts. Total cellular RNA was isolated from wild-type cultures and cultures containing the ctrA401ts allele grown at 30°C or at 37°C (the restrictive temperature) for 1.5 and 3 h. The P1 and P2 transcripts were resolved on denaturing acrylamide gels. [32P]-labeled DNA standards were used to identify the P1 and P2 transcripts. (Right) Relative levels of the P1 and P2 transcripts, quantitated by using a PhosphorImager, in the mutant background. Data are expressed as the percent of total (P1 + P2) transcripts in each lane. (C) DNase I protection of the ctrA promoter with purified His6-CtrA. His6-CtrA concentrations in each footprinting reaction are indicated. The sequence of the coding strand is shown. Regions protected by His6-CtrA are boxed, the P1 and P2 start sites are marked by bent arrows, and hypersensitive sites are indicated by exclamation marks.
The CtrA Protein Mediates Feedback Control of the ctrA Promoters.
The ctrA promoter region contains sequence motifs that are found in CtrA-regulated promoters (see Figs. 2A and 4C), including the promoters of the class II flagellar genes fliQ and fliL (9), the ccrM gene (9, 11), the ftsZ gene (10), and the promoter in the origin of replication (4). All of these promoters show altered activity in Caulobacter strains carrying the temperature-sensitive ctrA401 allele as the only copy of ctrA. To determine whether the product of the ctrA gene affects its own transcription, we assayed the activity of the ctrA promoter in the ctrA401ts strain (Fig. 4A). The complete promoter region (pctrA290), the P1 promoter (pctrA-P1), and the P2 promoter (pctrA-P2) were assayed in the wild-type background and in the ctrA401 ts strain. At the restrictive temperature (37°C), transcription from the complete ctrA promoter (pctrA290) in the ctrA401 strain was reduced to 55% of the activity in the wild-type background. Because the activity of at least two housekeeping promoters is not affected in the ctrA401ts background and ctrA401ts is a complete loss of function allele at the restrictive temperature (9), this result suggests that CtrA positively regulates the overall transcription of the ctrA gene. However, when the individual ctrA promoters were assayed, we found that transcription from the P1 promoter was somewhat increased in the ctrA401ts strain relative to the wild-type background at the restrictive temperature. In contrast, transcription from the P2 promoter in the ctrA401ts strain at the restrictive temperature dropped to almost one-quarter of the activity found in wild-type cells (Fig. 4A). In a separate series of experiments, we measured P1 activity in a strain in which ctrA is under the control of the xylose-inducible promoter PxylX. When cellular CtrA levels were depleted by growing this strain in the absence of xylose, pctrA-P1 transcription increased 1.7-fold as compared with wild-type cells grown under the same conditions (average values were 4,840 ± 750 and 2,880 ± 260 Miller units for the xylose inducible strain and the wild-type strain, respectively). Taken together, these results argue that CtrA represses transcription from P1 and activates transcription from the P2 promoter.
To confirm these results, we directly measured the relative levels of mRNA produced from the native chromosomal copies of P1 and P2 using S1 nuclease protection assays (19). RNA was isolated from wild-type cells as well as from the ctrA401ts strain grown at the restrictive and permissive temperatures, and S1 nuclease protection assays were performed with these RNA samples (Fig. 4B Left). Relative promoter strength was determined by quantitating the abundance of the P1 and P2 transcripts by using a PhosphorImager (Fig. 4B Right). In the wild-type background, the P2 promoter is the stronger of the two promoters. At the permissive temperature, promoter activity in the ctrA401ts strain was similar to that in the wild-type background. After shifting the cell culture to 37°C for 3 hr, however, the level of the P1 transcript increased approximately 4-fold, whereas the P2 transcript was reduced to about 60% of to the level observed at 30°C (Fig. 4B). These results confirm that CtrA negatively regulates the P1 promoter and positively regulates the P2 promoter.
The presence of CtrA consensus-binding motifs in both the P1 and P2 promoter regions and the genetic evidence for feedback control mediated by CtrA (9) suggest that CtrA might directly bind to its own promoter region. This was confirmed by using a purified His6-CtrA fusion protein (9) to perform DNase I footprinting assays (21) with the ctrA promoter region. Fig. 4C shows that at the lowest concentrations used, the purified His6-CtrA protein specifically protected a 26-bp region from −14 to −39 relative to the P2 transcriptional start site. With increasing concentrations, the purified protein also protected 11 bp in the −10 region of the P1 promoter (from −64 to −74 relative to P2). The 5′ ends of the protected regions were adjacent to CtrA-dependent DNase I-hypersensitive sites. The protected regions contain a consensus TTAAC sequence motif (see also Fig. 2A). This motif was repeated three times in the protected region centered at −26 of the P2 promoter and once in the P1 promoter. Transcriptional repressors often function by competitively binding to RNA polymerase binding sites in the −10 region of the promoter (22). The finding that CtrA binds to the −10 region of the P1 promoter is therefore consistent with the genetic data showing that CtrA represses that promoter.
DISCUSSION
It stands to reason that an essential regulatory factor like CtrA, with global control of temporally and spatially distinct cell-cycle events (refs. 4, 9–11; Fig. 5B) must itself be under strict regulation. We have previously shown that phosphorylation is required for CtrA activity and that cell type-specific proteolysis ensures that CtrA is cleared from the stalked cell and the stalked-cell portion of the late predivisional cell (ref. 13; Fig. 5A). In addition, we have now shown that the transcription of ctrA exhibits complex temporal regulation.
Figure 5.
(A) Schematic of feedback regulation of ctrA transcription during the cell cycle. Shaded areas in the cells indicate the presence of CtrA. The times of CtrA proteolysis and CtrA phosphorylation (13) are indicated. After threshold levels of CtrA≈P accumulate in the predivisional cell, the P1 promoter is repressed, and the P2 promoter is activated by CtrA≈P binding to recognition motifs in these promoters. (B) The regulatory network controlled by CtrA≈P.
The ctrA gene is transcribed from two promoters. Both are cell-cycle regulated, with the activity of the P1 promoter peaking in early predivisional cells and the activity of the stronger P2 promoter peaking in late predivisional cells. Furthermore, we conclude that CtrA directly regulates transcription from its own promoters based on the following evidence: (i) expression of the wild-type ctrA promoter was reduced in the ctrA401 temperature-sensitive strain at the restrictive temperature; (ii) expression assays of the individual P1 and P2 promoters generated by site-directed mutagenesis showed that P1 levels are increased and P2 levels are decreased when CtrA is cleared from the cell; (iii) S1 nuclease protection assays of the individual P1 and P2 promoters revealed that levels of the P1 transcript increased and the P2 transcript decreased in the ctrA401ts background after a shift to the restrictive temperature; and finally, (iv) DNase I footprinting analysis demonstrated specific binding of purified His-tagged CtrA to conserved CtrA-binding motifs in the ctrA promoter region. Because we previously showed that phosphorylation of CtrA is essential for CtrA function and viability (9), we argue that it is CtrA≈P that, in vivo, is responsible for the positive feedback of the P2 promoter and the negative feedback of the P1 promoter (Fig. 5A).
During the cell cycle, CtrA is degraded at the G1 to S transition, which allows the initiation of DNA replication (13). Proteolysis is then turned off after the initiation of DNA replication in the early predivisional cell (Fig. 5A). We propose that the onset of CtrA proteolysis contributes to the control of the timing of ctrA transcription by releasing the P1 promoter from CtrA≈P-mediated repression, allowing transcription of P1 in the early predivisional cell. The cessation of CtrA proteolysis in the early predivisional cell then allows CtrA produced from the early P1 promoter to accumulate. When threshold levels of CtrA≈P are reached, a positive feedback loop activates ctrA transcription from the strong P2 promoter, and transcription from the P1 promoter is repressed (Fig. 5A). The result of this differential regulation of the two ctrA promoters is that transcription of the ctrA gene switches from an early promoter to a stronger late promoter. Thus, the timing of the onset and cessation of CtrA proteolysis would control both the negative feedback loop at P1 and the positive feedback loop regulating ctrA transcription from P2.
The P2 promoter is not active in swarmer cells despite the presence of high levels of phosphorylated CtrA. If threshold levels of phosphorylated CtrA were the only control of the P2 promoter, then it would be active in nascent swarmer cells, as well as in predivisional cells. Thus, it is likely that another factor modulates the P2 promoter: a coactivator present only in predivisional cells, or possibly a repressor present only in nascent swarmer and stalked cells. In a parallel manner, the Class II flagellar promoters are active only in predivisional cells (11), despite the fact that phosphorylated CtrA is present in both swarmer cells and predivisional cells. As is the case of the ctrA promoters, transcription of the CtrA-dependent Class II flagellar promoters may be under the control of additional as yet unidentified factors.
There is precedence for feedback regulation controlling the time of transcription of a global developmental regulator; the Spo0A response regulator that controls the initiation of sporulation in Bacillus subtilis also exhibits feedback transcriptional regulation. Transcription of the spo0A gene occurs from two promoters, a weak vegetative promoter (Pv) and a strong sporulation promoter (Ps) (23, 24). The vegetative promoter constitutively transcribes the spo0A gene at low levels during normal growth. At the initiation of sporulation, the basal level of Spo0A produced from the weak constitutive promoter is activated by phosphorylation. Spo0A≈P binds to sequences within the spo0A promoter region, thereby activating the sporulation Ps promoter and repressing the vegetative Pv promoter. These feedback loops thereby result in a switch from a weak promoter to a strong promoter, yielding a timed pulse of spo0A transcription that initiates the sporulation program. Although Bacillus sporulation and the Caulobacter cell cycle are evolutionarily distant, the logic underlying the transcriptional control of the master regulators of both genetic programs has been conserved.
Acknowledgments
We thank Christine Jacobs for critical reading of the manuscript. This work was supported by National Institutes of Health grants GM32506/5120MZ and GM51426. I.J.D. is a recipient of Medical Scientist Training Program funding from grant GM32506–16.
References
- 1.Shapiro L, Losick R. Science. 1997;276:712–718. doi: 10.1126/science.276.5313.712. [DOI] [PubMed] [Google Scholar]
- 2.Degnen S, Newton A. J Mol Biol. 1972;64:671–680. doi: 10.1016/0022-2836(72)90090-3. [DOI] [PubMed] [Google Scholar]
- 3.Marczynski G T, Lentine K, Shapiro L. Genes Dev. 1995;9:1543–1557. doi: 10.1101/gad.9.12.1543. [DOI] [PubMed] [Google Scholar]
- 4.Quon K C, Yang B, Domian I J, Shapiro L, Marczynski G T. Proc Natl Acad Sci USA. 1998;95:120–125. doi: 10.1073/pnas.95.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts C R, Mohr C D, Shapiro L. Curr Top Dev Biol. 1996;34:207–257. doi: 10.1016/s0070-2153(08)60712-7. [DOI] [PubMed] [Google Scholar]
- 6.Zweiger G, Marczynski G, Shapiro L. J Mol Biol. 1994;235:472–485. doi: 10.1006/jmbi.1994.1007. [DOI] [PubMed] [Google Scholar]
- 7.Stephens C, Reisenauer A, Wright R, Shapiro L. Proc Natl Acad Sci USA. 1996;93:1210–1214. doi: 10.1073/pnas.93.3.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quardokus E, Din N, Brun Y. Proc Natl Acad Sci USA. 1996;93:6314–6319. doi: 10.1073/pnas.93.13.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quon K C, Marczynski G T, Shapiro L. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 10.Kelly A J, Sackett M J, Din N, Quardokus E, Brun Y V. Genes Dev. 1998;12:880–893. doi: 10.1101/gad.12.6.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reisenauer A, Quon K, Shapiro L. J Bacteriol. 1999;181:2430–2439. doi: 10.1128/jb.181.8.2430-2439.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobs C, Domian I J, Maddock J R, Shapiro L. Cell. 1999;97:111–120. doi: 10.1016/s0092-8674(00)80719-9. [DOI] [PubMed] [Google Scholar]
- 13.Domian I J, Quon K C, Shapiro L. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- 14.Ely B. Methods Enzymol. 1991;204:372–384. doi: 10.1016/0076-6879(91)04019-k. [DOI] [PubMed] [Google Scholar]
- 15.Gober J W, Shapiro L. Mol Biol Cell. 1992;3:913–926. doi: 10.1091/mbc.3.8.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 17.Evinger M, Agabian N. J Bacteriol. 1977;132:294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenal U, White J, Shapiro L. J Mol Biol. 1994;243:227–244. doi: 10.1006/jmbi.1994.1650. [DOI] [PubMed] [Google Scholar]
- 19.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1989. [Google Scholar]
- 20.Salser W, Gesteland R, Bolle A. Nature (London) 1967;215:588–590. doi: 10.1038/215588a0. [DOI] [PubMed] [Google Scholar]
- 21.Galas D, Schmitz A. Nucleic Acids Res. 1978;5:3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Record T M, Reznikof W S, Craig M L, McQuade K L, Schlax P J. In: Escherichia coli and Salmonella Cellular and Molecular Biology. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol.; 1996. [Google Scholar]
- 23.Siranosian K J, Grossman A D. J Bacteriol. 1994;176:3812–3815. doi: 10.1128/jb.176.12.3812-3815.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strauch M A, Trach K A, Day J, Hoch J A. Biochimie. 1992;74:619–626. doi: 10.1016/0300-9084(92)90133-y. [DOI] [PubMed] [Google Scholar]