Abstract
Cholesterol 7α-hydroxylase is the first and rate-limiting enzyme in a pathway through which cholesterol is metabolized to bile acids. The gene encoding cholesterol 7α-hydroxylase, CYP7A, is expressed exclusively in the liver. Overexpression of CYP7A in hamsters results in a reduction of serum cholesterol levels, suggesting that the enzyme plays a central role in cholesterol homeostasis. Here, we report the identification of a hepatic-specific transcription factor that binds to the promoter of the human CYP7A gene. We designate this factor CPF, for CYP7A promoter binding factor. Mutation of the CPF binding site within the CYP7A promoter abolished hepatic-specific expression of the gene in transient transfection assays. A cDNA encoding CPF was cloned and identified as a human homolog of the Drosophila orphan nuclear receptor fushi tarazu F1 (Ftz-F1). Cotransfection of a CPF expression plasmid and a CYP7A reporter gene resulted in specific induction of CYP7A-directed transcription. These observations suggest that CPF is a key regulator of human CYP7A gene expression in the liver.
In mammalian cells, cholesterol is an essential membrane component and is required for the synthesis of both sterols and nonsterols necessary for normal cell function. Excess cholesterol causes the formation of toxic precipitates in cells, which may accumulate on arterial walls and eventually lead to atherosclerosis (1). It is therefore crucial that cholesterol levels are maintained under tight control at all times. Three major regulatory pathways are involved in the maintenance of cellular cholesterol homeostasis: (i) uptake of dietary cholesterol via the low density lipoprotein (LDL) receptor, (ii) endogenous cholesterol biosynthesis, and (iii) metabolic conversion of cholesterol to bile acids (2). The link among these regulatory pathways is cholesterol itself. Cholesterol serves as a feedback or feed-forward signal, coordinating the expression of key genes whose products are involved in these pathways (3). When intracellular cholesterol levels are elevated, the transcription of genes encoding the LDL receptor and cholesterol biosynthetic enzymes [including hydroxymethyl glutaryl (HMG)-CoA synthase and HMG-CoA reductase] is suppressed. This negative feedback process is mediated by a family of transcription factors designated sterol regulatory element binding proteins (SREBPs) (4–6). SREBPs contain an N-terminal transcription factor domain, a DNA-binding basic helix–loop–helix–leucine zipper motif, two hydrophobic transmembrane domains that anchor the protein in the endoplasmic reticulum (ER), and a C-terminal regulatory domain (6). When intracellular cholesterol levels are low, a two-step proteolytic cascade releases the N-terminal transcription factor domain of SREBP from the ER membrane (7–9). The transcription factor then enters the nucleus and activates sterol response element-regulated genes (7–9).
Although the sterol regulatory element binding protein pathway, which is responsible for regulating genes involved in cholesterol uptake and biosynthesis, is well characterized, the molecular basis for cholesterol catabolism is largely unknown. The major catabolic pathway for cholesterol removal is the production of bile acids, which occurs exclusively in the liver (10). Cholesterol 7α-hydroxylase (Cyp7a), a member of the cytochrome P450 family, is the first and rate-limiting enzyme in a major bile acid biosynthetic pathway (11). The expression of CYP7A is tightly regulated. The CYP7A gene is expressed exclusively in the liver where it is induced by dietary cholesterol and suppressed by bile acids (11–13). Several independent lines of evidence indicate that cholesterol catabolism plays a central role in cholesterol homeostasis. Treatment of laboratory animals with colestipol or cholestyramine, two bile acid binding resins, decreases serum cholesterol levels (11, 12, 14, 15). In addition, overexpression of the CYP7A gene in hamsters reduces total cholesterol and low density lipoprotein cholesterol levels (16). Thus, regulating the production of Cyp7a represents a potential therapeutic strategy for the discovery of new cholesterol-lowering drugs.
HepG2 cells, a hepatoma-derived cell line, were used as a model system to investigate the molecular mechanisms underlying hepatic-specific expression of the human CYP7A gene (17). In this report, we used DNase I hypersensitivity mapping to characterize the human CYP7A promoter. These studies led to the discovery of a hepatic-specific regulatory element within the CYP7A promoter. We then cloned the gene encoding a CYP7A promoter binding protein and identified it as a human homolog of the orphan nuclear receptor fushi tarazu F1 (Ftz-F1) from Drosophila (18). This transcription factor, designated CYP7A promoter binding factor (CPF), represents a specific transcriptional inducer of human CYP7A gene expression.
MATERIALS AND METHODS
Cells and Plasmids.
HepG2 (a human hepatoma cell line), HEK293 (a transformed human embryonic kidney cell line), and Caco2 (a human colon adenocarcinoma cell line) were purchased from American Type Culture Collection. SV589, a transformed human fibroblast cell line, was a gift from Michael Brown and Joseph Goldstein (University of Texas Southwestern Medical Center, Dallas). Cells were cultured in DMEM/Ham’s F-12 (1:1) supplemented with 10% FCS at 37°C, 5% CO2 in a humidified incubator.
pGL3CYP7Awt was constructed by subcloning the −716/+14 fragment of the human CYP7A gene (a gift from David Russell, University of Texas Southwestern Medical Center) into the pGL3-luciferase reporter plasmid (Promega). pGL3CYP7Am-129/130 and pGL3CYP7Am-61/62 contain mutations at positions −129 and −130 (GG to TT) and −61 and −62 (AA to TC), respectively. The two base-pair substitutions were introduced into pGL3CYP7Awt by using the ExSite mutagenesis kit (Stratagene). pfCPF contains a Flag epitope-tagged sequence at the 5′ end of the CPF gene cloned into pcDNA3 (Invitrogen). Nuclear receptors used in this study were cloned by PCR using QUICK-Clone cDNA purchased from CLONTECH.
DNase I Hypersensitivity Mapping.
HepG2, HEK293, or Caco2 cells (3 × 106) were harvested and lysed in a buffer (1.5 ml) containing 50 mM Tris⋅HCl (pH 7.9), 100 mM KCl, 5 mM MgCl2, 0.05% (vol/vol) saponin, 200 mM 2-mercaptoethanol, and 50% (vol/vol) glycerol. Nuclei were collected by centrifugation and resuspended in a buffer containing 100 mM NaCl, 50 mM Tris⋅HCl (pH 7.9), 3 mM MgCl2, 1 mM DTT, 1× complete protease inhibitor mixture (Boeringer Mannheim), and sequentially diluted DNase I (0.6, 1.7, or 5 units/ml). Nuclei suspensions were incubated at 37°C for 20 min. The reactions were terminated by addition of EDTA to a final concentration of 100 mM. After RNase A and Proteinase K treatment, genomic DNA was prepared and subjected to Southern hybridization (19).
Electrophoretic Mobility-Shift Assay (EMSA).
Nuclear extracts were prepared from cultured cells by the method of Schreiber et al. (20), except that KCl was used instead of NaCl at the indicated concentration. In vitro transcription and translation reactions were performed with the TNT system (Promega). Nuclear extracts (1 μg) or 0.1–1 μl of in vitro-translated product were mixed with 40,000 cpm of 32P-labeled oligonucleotide in a reaction buffer containing 10 mM Hepes (pH 7.6), 1 μg of poly(dI-dC), 100 mM KCl, 7% (vol/vol) glycerol, 1 mM EDTA, 1 mM DTT, 5 mM MgCl2, and 40 pmol of unrelated single-stranded DNA oligonucleotide, and incubated for 20 min at room temperature. The reaction mixtures were separated on a 4% polyacrylamide 0.5× Tris-borate EDTA gel. After electrophoresis, the gels were dried and exposed to x-ray film. In competition experiments, a 30- or 60-fold molar excess of competitor DNA was added to the EMSA reaction mixture. In antibody supershift experiments, an anti-CPF antiserum or preimmune serum was added to the reaction mixtures before addition of the DNA probe.
Transfection and Reporter Gene Analysis.
Cells were plated in 24-well plates (1 × 105/well) 1 day before transfection. In general, 0.5 μg each of luciferase reporter plasmid and expression plasmid along with 50 ng of cytomegalovirus-driven β-galactosidase expression vector were transfected in quadruplicate into cultured cells for 30 h with the use of the calcium phosphate method (21). Cell extracts were prepared and assayed for luciferase activity by using the Luciferase assay system (Promega). Luciferase activity was normalized to the β-galactosidase activity.
Molecular Cloning of CPF.
A human expressed sequence tag clone (GenBank accession no. N59515) that contains the Ftz-F1 DNA-binding domain was used to screen a human liver cDNA library purchased from CLONTECH. The cDNAs in positive clones were recovered by conversion of phage DNA into pTriplEx plasmids and sequenced.
Tissue-Specific Expression of CPF.
Northern (RNA) blots of poly(A)+ RNA extracted from human tissues were purchased from CLONTECH. Hybridization reactions were carried out with the Northern MAX hybridization buffer (Ambion, Austin, TX).
Immunoprecipitation.
A peptide derived from the CPF cDNA sequence (DRMRGGRNFKGPMYKRDR) was used to raise an anti-CPF polyclonal antibody. HepG2 or 293 cells (1 × 107) were cultured in media containing 100 μCi/ml of [35S]methionine for 90 min. Cells were harvested and then lysed by three freeze-thaw cycles in buffer containing 50 mM Tris⋅HCl (pH 7.5), 125 mM NaCl, 5 mM EDTA, and 0.1% (vol/vol) NP-40. Cell lysates then were used for immunoprecipitation with the anti-CPF antibody. Precipitated samples were resolved by 10% SDS/PAGE, and the gels were dried and exposed to x-ray film.
RESULTS
DNase I-Hypersensitive Site Mapping of the Human CYP7A Gene.
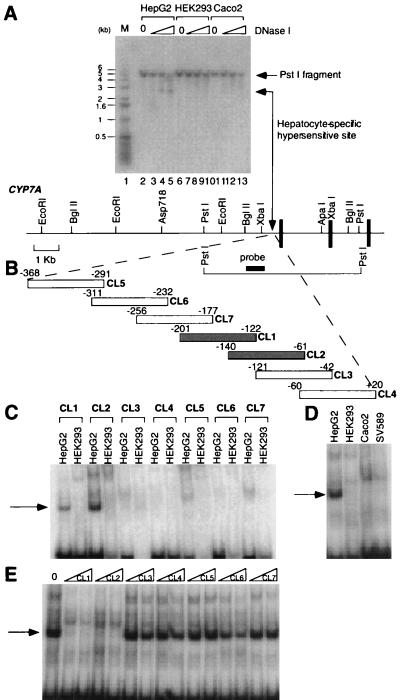
The DNase I-hypersensitive mapping technique was used to identify potential hepatic-specific regulatory regions of the human CYP7A gene. DNase I hypersensitivity is known to be associated with the open chromatin conformations of regulatory regions near transcriptionally active genes (22). Nuclei prepared from HepG2, HEK293, and Caco2 cells were treated with increasing amounts of DNase I. The DNAs then were extracted, digested with PstI, transferred onto nylon, and hybridized with a radio-labeled fragment containing nucleotides −944 to −468 of the CYP7A gene. As indicated in Fig. 1A, this probe hybridized to the predicted 5-kb PstI fragment in DNA isolated from all cell types examined. However, a second 2.8-kb DNA fragment was detected with the same probe and found only in HepG2 cells. As DNase I treatment was extended, the intensity of the 2.8-kb DNA band increased, while the intensity of the parental 5-kb DNA band diminished concomitantly. These observations revealed the existence of a DNase I-hypersensitive site located roughly 200 bp upstream from the CYP7A transcription start site. The 2.8-kb band was observed only in HepG2 cells and not in HEK293 or Caco2 cells, suggesting the existence of a hepatic-specific regulatory element.
Figure 1.
A hepatic-specific DNase I-hypersensitive site in the CYP7A promoter region. (A) DNAs prepared from DNase I-treated nuclei (0, 0.6, 1.7, or 5.0 units/ml) from HepG2, HEK293, and Caco2 cells were separated by 1.0% agarose gel electrophoresis and subjected to Southern hybridization. A radio-labeled fragment corresponding to base pairs −944 to −468 of the CYP7A gene was used as a probe. A 5.0-kb PstI fragment was observed in each lane. A 2.8-kb hepatic-specific fragment (lanes 3–5) is indicated by the double arrow. The lane that displays molecular size markers is indicated by M and contains the 1-kb ladder (GIBCO/BRL). The sizes (in kb) of bands of the 1-kb ladder are indicated at the left of the gel. Below the gel is diagrammed a partial restriction map of the promoter region. The vertical bars represent exons 1–3 of the CYP7A gene. The arrow between the XbaI site and the first exon represents the location of the hepatic-specific DNase I-hypersensitive site. The heavy horizontal bar denotes the location of the probe used in this experiment. The 5.0-kb PstI fragment is also diagrammed. (B) A schematic diagram indicating the location of double-stranded oligonucleotides CL1–CL7 within the CYP7A promoter region that were used as radio-labeled probes. +1 denotes the transcription start site. (C–E) Formation of hepatic-specific DNA-protein complexes. EMSAs were performed with the indicated nuclear extracts and radio-labeled, double-stranded CL1–CL7 oligonucleotide probes diagrammed in B. (C) EMSAs were performed by using nuclear extracts prepared from HepG2 and 293 cells. The arrow indicates the hepatic-specific DNA-protein complex. (D) Nuclear extracts from the indicated cell lines were used in EMSAs with CL1 as the radio-labeled probe. The arrow indicates the hepatic-specific DNA-protein complex. (E) Competition EMSAs using unlabeled CL1–CL7 fragments as competitors. An EMSA was performed by using HepG2 nuclear extracts and radio-labeled CL1 with the addition of either a 30- or 60-fold molar excess of unlabeled CL1–CL7 (indicated by triangles at the top of the gel lanes). The DNA-protein complexes are indicated by the arrows. 0, no competitor.
Identification of a Hepatic-Specific CYP7A Promoter Element.
To elucidate further the hepatic-specific element of the CYP7A gene, seven overlapping oligonucleotides (CL1–CL7) that spanned the region between nucleotides −368 and +20 were synthesized, radio-labeled, and used in EMSAs with nuclear extracts (Fig. 1B). As shown in Fig. 1C, hepatic-specific DNA-protein complexes were observed only when CL1 and CL2 were used as radio-labeled probes. The relative mobilities of these two DNA-protein complexes were nearly identical, suggesting that the same nuclear protein bound to each probe. Importantly, the DNA-protein complex formation was specific to HepG2 nuclear extracts. No equivalent complexes were observed with HEK293, Caco2, or SV589 nuclear extracts (Fig. 1D). To decipher the DNA-binding specificity of this DNA-protein complex, an EMSA was performed by using CL1 as the radio-labeled probe and fragments CL1–CL7 as unlabeled competitors. As shown in Fig. 1E, only the addition of excess CL1 and CL2 fragments prevented complex formation. These observations suggested that the hepatic-specific expressed factor bound to the overlapping sequences of probes CL1 and CL2. These sequences are located within the region that contains the hepatic-specific DNase I-hypersensitive site.
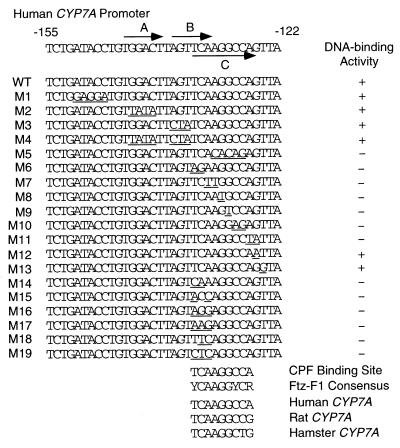
Several 6-bp elements that are known binding sites for nuclear hormone receptors (23) were discovered by visual inspection of the two overlapping sequences. To determine the exact sequence responsible for hepatic-specific binding, we synthesized a series of oligonucleotides with mutations in the putative nuclear receptor response elements (Fig. 2, M1–M19) and tested their abilities to compete with complex formation in EMSAs. As summarized in Fig. 2, oligonucleotides containing mutations in the direct repeats spaced by 1 nt (DR1, repeats A and B) were effective competitors of complex formation. In contrast, oligonucleotides containing mutations in repeat C failed to compete effectively, suggesting that the DNA sequence of repeat C is essential for hepatic-specific binding. To further dissect the sequence required for complex formation, we synthesized oligonucleotides containing point mutations in repeat C and adjacent sequences and tested them in EMSAs. The results indicated that a consensus element containing 9 nt is required for complex formation. The 9-bp element, conserved in CYP7A human and rodent promoters, is identical to the consensus binding site for the Ftz-F1 family of nuclear hormone receptors (18). We designated this element a CPF binding site.
Figure 2.
Sequences (+ strand only) of oligonucleotides containing the overlapping region of CL1 and CL2, and mapping of the binding site of CPF. The DR1 site is indicated by the tandem overlining arrows (labeled repeats A and B). The 9-bp CPF binding site is indicated by the underlining arrow (labeled repeat C). The wild-type (WT) sequence is shown above the mutant oligonucleotide sequences (M1–M19). Bases that are underlined are those that were mutated. + denotes the formation of a DNA-protein complex, and − the absence of such a complex. The CPF-binding site within the CYP7A gene promoter and the Ftz-F1 consensus binding site are listed at the bottom, along with a sequence comparison of the binding site from human, rat, and hamster CYP7A promoters.
The CPF Binding Site Is Essential for Hepatic-Specific Expression of the Human CYP7A Gene.
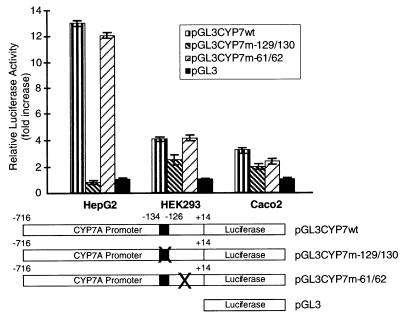
To determine the role of the CPF binding site in human CYP7A gene expression, two G to T nucleotide substitutions were introduced into the 9-bp recognition sequence TCAAGGCCA (−126 to −134). As a control, mutations were introduced into a region of the promoter outside the CPF binding site (two nucleotide substitutions at positions −61 and −62). Promoter fragments (−716 to +14) that contained either the wild-type sequence, the mutated CPF binding site, or the adjacent mutation sequences were cloned into the luciferase reporter plasmid pGL3. These constructs were transfected into HepG2, HEK293, and Caco2 cells, and promoter activity was measured by determination of luciferase activity levels. Mutations in the CPF binding site completely abolished promoter activity in HepG2 cells, while showing little or no effect in HEK293 and Caco2 cells (Fig. 3). Mutations in the region outside of the CPF binding site had no effect on promoter activity irrespective of cell type.
Figure 3.
Effect of mutations in the CPF-binding site on CYP7A gene expression. The indicated cell lines were transfected with reporter constructs by using the calcium phosphate method (19). Luciferase assays were performed as described in Materials and Methods. Relative luciferase units were normalized to β-galactosidase activity to control for variations in transfection efficiency. The cross-out refers to the presence of two point mutations (as described in Materials and Methods). The pGL3 reporter is a vector-only control.
Cloning of the Hepatic-Specific CYP7A Promoter-Binding Protein.
Having identified the CPF binding element, we next sought to clone the gene encoding the binding protein. Ftz-F1 is a Drosophila melanogaster DNA-binding protein (18) that belongs to the nuclear receptor superfamily (23, 24). Analogous to other nuclear receptors, Ftz-F1 contains a zinc finger DNA-binding domain and a putative ligand-binding domain. The DNA-binding domain of the Ftz-F1 family members contains a unique 26-aa extension (called the Ftz-F1 domain) that resides C-terminal to its two zinc finger domains (25). The sequence of the Ftz-F1 domain has been conserved from D. melanogaster to rodent and is largely responsible for the DNA-binding specificity of the Ftz-F1 family of nuclear receptors (25). Identification of the Ftz-F1 binding site in the human CYP7A promoter raised the possibility that a human Ftz-F1-like protein might bind to this element. To clone the human homolog of Drosophila Ftz-F1, we searched an expressed sequence tag (EST) database by using the DNA sequence derived from the conserved Ftz-F1 domain. A human EST clone (GenBank accession no. N59515) was found to share more than 80% DNA sequence similarity with the mouse liver receptor homolog (mLRH-1) (26). This sequence then was used as a probe to screen a human liver cDNA library. Several distinct cDNA clones were isolated, and sequence analysis revealed that these clones could be classified into three closely related groups, represented by clones 113, 105, and 36.
Characterization of CPF.
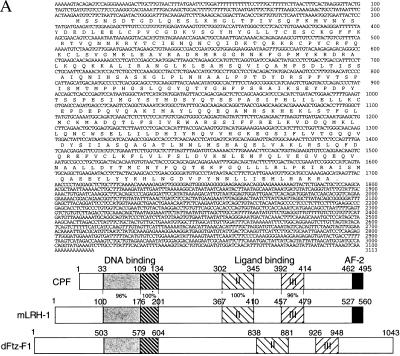
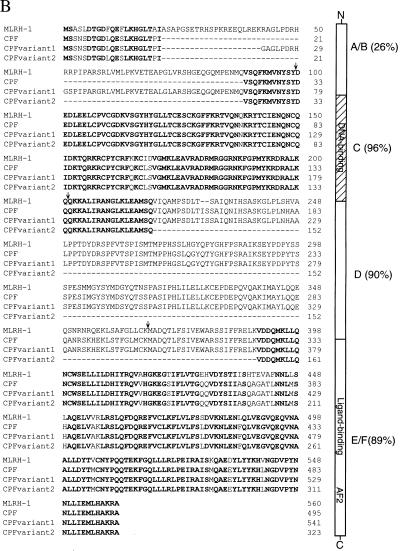
Clone 113 encodes a polypeptide 495 aa in length. The sequence contained an in-frame translational stop codon 30 nt upstream of the first ATG (Fig. 4A). The protein encoded by the ORF of clone 113 is designated CPF. Sequence analysis revealed that CPF represents an additional member of the Ftz-F1 family, most closely related to mouse LRH-1 (Fig. 4B). Clone 105, designated CPF variant 1 to distinguish it from CPF, encodes a polypeptide identical to CPF except for a 46-aa insertion at amino acid 21 of the A/B domain. The 46-aa insertion is also present in mLRH-1, sharing 87% homology with CPF variant 1 in this region. Clone 36, or CPF variant 2, contains a 172-aa deletion within the D and E domains. Both CPF variants probably are derived from alternative splicing. Our study focused on CPF as it is apparently the major form expressed in HepG2 cells (see below).
Figure 4.
(A) cDNA and predicted amino acid sequence of CPF. A human liver cDNA library was screened with a probe based on the conserved DNA-binding domain of the Drosophila Ftz-F1 nuclear receptor. Nucleotide positions are shown on the right. The predicted amino acid sequence is written in single-letter abbreviations below the nucleotide sequence. A schematic representation of CPF, mLRH-1, and dFtz-F1 is shown below the CPF nucleotide and protein sequence. We have noted that during preparation of our manuscript, a similar but shorter, cDNA sequence with an identical coding region and slightly different 5′ and 3′ untranslated regions was reported by Li et al. (41). (B) Amino acid sequence alignment of mLRH-1 and human CPF, along with CPF variants 1 and 2. The amino acids conserved among all four clones are highlighted in bold. The structural/functional domains of CPF are illustrated to the right of the alignment with percent identity to mLRH-1 in parenthesis. A/B: N-terminal variable region; C: DNA-binding domain; D: Variable hinge region; E/F: Ligand-binding domain. Arrows above the sequence alignment indicate boundaries of the domains.
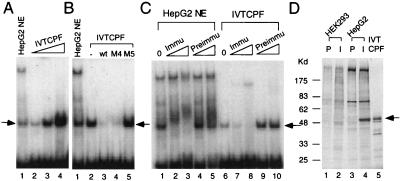
To test whether the cloned CPF gene encoded the factor responsible for the CYP7A promoter binding activity observed in HepG2 cells, in vitro-synthesized CPF and HepG2 nuclear extracts were tested in parallel in EMSAs using CL1 as the radio-labeled DNA probe. As shown in Fig. 5 A and B, the binding specificity of the in vitro-synthesized CPF was identical to that of the endogenous protein present in the HepG2 extracts, and the DNA-protein complexes formed by both exhibited identical mobilities in the EMSAs. To further test whether the binding activity in the HepG2 nuclear extract was caused by the presence of endogenous CPF, EMSAs were performed with a peptide antibody raised against the DNA-binding domain of CPF. As shown in Fig. 5C, the anti-CPF antibody, but not preimmune serum, prevented DNA-protein complex formation by both HepG2 nuclear extracts and in vitro-translated CPF. Moreover, 35S-methionine-labeled extracts from HepG2 and HEK293 were immunoprecipitated with the CPF-specific antibody, and the immuno-complexes were analyzed by 10% SDS/PAGE. As shown in Fig. 5D, the antibody recognized a hepatic-specific protein that comigrated with in vitro-translated CPF.
Figure 5.
Functional similarity of in vitro-synthesized and endogenous CPF. (A) EMSAs with in vitro-synthesized CPF-1 (IVTCPF) and CL1 as the radio-labeled probe. Lanes 2–4, 0.1, 0.5 and 1.0 μl of in vitro-synthesized CPF-1 (IVTCPF), respectively, and lane 1, 1 μg of HepG2 nuclear extract (NE). The arrow indicates the CPF binding complex. (B) Competition EMSAs. EMSAs were performed as described above with the addition of unlabeled competitor oligonucleotides, including wild type (wt) and those with mutations in either the CPF binding site (M5) or a region outside of the CPF binding site (M4). A 60-fold molar excess of each competitor was used. The arrow denotes the location of the bound DNA-protein complex. NE, HepG2 nuclear extract; −, no competitor, IVTCPF, in vitro-translated CPF. (C) EMSA in the presence of CPF-specific antibodies. EMSAs were performed with HepG2 nuclear extracts and in vitro-synthesized CPF with the addition of either preimmune serum (Preimmu) or rabbit anti-CPF antibodies (Immu). Lanes 1 and 6, no antibody (0); lanes 2, 4, 7, and 9, 0.1 μl of serum; lanes 3, 5, 8, and 10, 1 μl of serum. The arrow denotes the location of the bound DNA-protein complex. (D) Immunoprecipitation of CPF in HepG2 cells. HEK293 and HepG2 cells were grown in the presence of [35S]methionine. Preimmune (P) and rabbit anti-CPF antibodies (I) then were used in immunoprecipitation experiments with HEK293 (lanes 1 and 2) and HepG2 (lanes 3 and 4) extracts. 35S-methionine-labeled in vitro-synthesized CPF (IVTCPF) was loaded into lane 5. The precipitated samples were separated by 10% SDS/PAGE, and gel was dried and exposed to x-ray film. The arrow denotes the immunoprecipitated and in vitro-synthesized CPF. Molecular size markers (in kd) are shown on the left.
Transcriptional Inducing Activity of CPF.
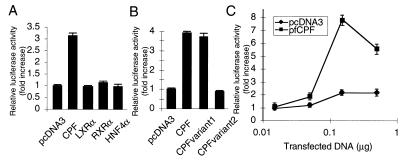
To examine its activity as a transcription factor, CPF was assayed for its ability to induce CYP7A expression in HEK293 cells, which lack endogenous CPF expression (Fig. 5D). We first transfected HEK293 cells with expression plasmids encoding either full-length CPF, liver X receptor α, retinoid X receptor α, hepatic nuclear factor 4α, or control plasmid pcDNA3, along with luciferase reporter plasmid containing the human CYP7A promoter sequence (−716 to +14). As shown in Fig. 6A, among the nuclear receptors tested, only overexpression of CPF stimulates human CYP7A promoter activity. Transcriptional activity of the CPF variants we isolated also was examined. As shown in Fig. 6B, both CPF and CPF variant 1 induced CYP7A promoter activity whereas CPF variant 2 failed to induce, suggesting that the hinge region and ligand-binding domain of CPF is important for its function. We further show in Fig. 6C that cotransfection of a flag-tagged CPF (pfCPF) resulted in a dose-dependent up-regulation of human CYP7A promoter activity. These results support the notion that CPF functions as a transcription regulator for the human CYP7A gene.
Figure 6.
Activation of CYP7A gene expression by CPF. Various expression plasmids and control plasmid pcDNA3 (0.5 μg) were cotransfected into HEK293 cells with 0.5 μg of luciferase reporter plasmid containing the human CYP7A promoter sequence (−716 to +14). (A) Full-length CPF, liver X receptor α (LXRα), retinoid X receptor α (RXRα), and hepatic nuclear factor 4α (HNF4α). (B) Full-length CPF, and its variants. (C) Control plasmid pcDNA3 or an expression plasmid encoding a flag-tagged full-length CPF (0, 0.015, 0.05, 0.15, or 0.5 μg) was cotransfected into 293 cells with the human CYP7A promoter luciferase reporter plasmid. Relative luciferase activity normalized to β-galactosidase activity is shown for each transfection.
Tissue-Specific Expression of CPF.
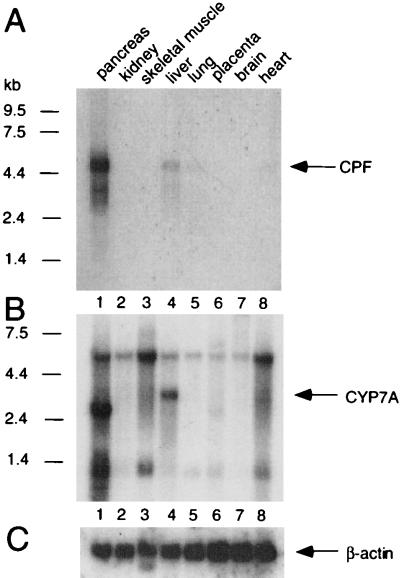
It has been reported that the CYP7A gene in rodents is expressed exclusively in the liver (11). If the human CYP7A gene behaves like its counterpart in rodents, CPF would be expected to be expressed in the liver. To determine the tissue expression profile of the CPF gene, RNA tissue blots were hybridized with either labeled CPF cDNA or CYP7A cDNA. As shown in Fig. 7A, the expression of the CPF gene apparently was enriched in the pancreas and liver, with a low level of expression in the heart and lung. The human CYP7A gene apparently was expressed only in the liver (Fig. 7B), consistent with the liver-specific expression observed in rodent.
Figure 7.
Tissue blot analyses of CPF and CYP7A mRNA expression. (A) The tissue blot was hybridized with radio-labeled CPF cDNA. (B) The same blot was stripped and rehybridized either with a riboprobe derived from CYP7A cDNA or with (C) radio-labeled β-actin cDNA as a control. Molecular size markers (in kb) are shown at the left of A and B.
DISCUSSION
A number of transcription factors including HNF3, C/EBP, DBP, and the nuclear hormone receptors hepatic nuclear factor 4α, ARP-1, retinoic acid receptor α, COUP, and glucocorticoid receptor have been implicated in the regulation of CYP7A promoter activity (27–30). Of these putative regulators, however, few or none have been shown to directly regulate CYP7A expression. In this report we identified a hepatic-specific element within the CYP7A promoter by DNase I hypersensitivity mapping of the human CYP7A gene, and then cloned the gene encoding a hepatic-expressed orphan nuclear receptor CPF that binds to this promoter element. Four lines of evidence support the interpretation that CPF is a bona fide activator of human CYP7A gene expression. First, in vitro-translated CPF binds to the same sequence within the CYP7A promoter as does the endogenous factor. Second, DNA-protein complexes formed with in vitro-translated CPF and the endogenous factor are indistinguishable, and a CPF-specific antibody perturbs the formation of both complexes. Third, mutations in the CPF binding site within the CYP7A promoter abolish hepatic-specific expression of the CYP7A gene. Last, transient transfection assays show that CPF activates expression of the reporter gene under control of the human CYP7A promoter sequence.
CPF is a unique member of the Ftz-F1 family of class IV orphan nuclear receptor superfamily (23), which bind DNA as a monomer and have intrinsic transcriptional activity (25, 26, 31). Ftz-F1 family members have been conserved throughout evolution and been identified in many species ranging from zebrafish (32), to Xenopus (33), to rodents (26), and now to human. Although our cotransfection experiments indicate that CPF has intrinsic transcriptional activity, the identification of nuclear receptor CPF as a regulator of CYP7A expression raises a very intriguing question of whether the activity of CPF, like many other nuclear receptors, is regulated by a small molecule ligand. Because CPF regulates expression of an enzyme that plays a key role in cholesterol and bile acid homeostasis, it seems plausible that certain metabolites may further regulate CPF activity. Most known endogenous ligands for orphan nuclear receptors are lipophilic molecules involved in specific metabolic pathways: e.g., retinoids for the retinoic acid receptors (34, 35), 9-cis-retinoic acid for the retinoid X receptors (36, 37), vitamin D3 for the vitamin D receptor (38), and oxysterols for the liver X receptors (39, 40). Identification of the ligand for CPF may help clarify the biological actions of CPF and further define key steps in the regulation of cholesterol homeostasis.
Acknowledgments
We thank David Russell for invaluable advice and providing human CYP7A promoter and CYP7A cDNA. We thank Marc Learned for CYP7A luciferase reporter construct and Marc Learned, Tim Hoey, Jin-long Chen, Josh Schultz, Kevin Lustig, and Terry Rosen for their critical comments.
ABBREVIATIONS
- CYP7A
cholesterol 7α-hydroxylase
- CPF
CYP7A promoter binding factor
- EMSA
electrophoretic mobility-shift assay
- mLRH-1
mouse liver receptor homolog
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF146343).
References
- 1.Brown M S, Goldstein J L. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 2.Turley S D, Dietschy J M. In: The Liver: Biology and Pathology. Arias I, Popper H, Schachter D, Shafritz D A, editors. New York: Raven; 1982. pp. 467–492. [Google Scholar]
- 3.Goldstein J L, Brown M S. Nature (London) 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 4.Brown M S, Goldstein J L. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 5.Hua X, Yokoyama C, Wu J, Briggs M R, Brown M S, Goldstein J L, Wang X. Proc Natl Acad Sci USA. 1993;90:11603–11607. doi: 10.1073/pnas.90.24.11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yokoyama C, Wang X, Briggs M R, Admon A, Wu J, Hua X, Goldstein J L, Brown M S. Cell. 1993;75:187–197. [PubMed] [Google Scholar]
- 7.Wang X, Sato R, Brown M S, Hua X, Goldstein J L. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 8.Sakai J, Duncan E A, Rawson R B, Hua X, Brown M S, Goldstein J L. Cell. 1996;85:1037–1046. doi: 10.1016/s0092-8674(00)81304-5. [DOI] [PubMed] [Google Scholar]
- 9.Hua X, Sakai J, Brown M S, Goldstein J L. J Biol Chem. 1996;271:10379–10384. doi: 10.1074/jbc.271.17.10379. [DOI] [PubMed] [Google Scholar]
- 10.Russell D W, Setchell K D R. Biochemistry. 1992;31:4737–4749. doi: 10.1021/bi00135a001. [DOI] [PubMed] [Google Scholar]
- 11.Jelinek D F, Andersson S, Slaughter C A, Russell D W. J Biol Chem. 1990;265:8190–8197. [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y C, Wang D P, Chiang J Y. J Biol Chem. 1990;265:12012–12019. [PubMed] [Google Scholar]
- 13.Stravitz R T, Hylemon P B, Heuman D M, Hagey L R, Schteingart C D, Ton-Nu H T, Hofmann A F, Vlahcevic Z R. J Biol Chem. 1993;268:13987–13993. [PubMed] [Google Scholar]
- 14.Noshiro M, Nishimoto M, Okuda K. J Biol Chem. 1990;265:10036–10041. [PubMed] [Google Scholar]
- 15.Sundseth S S, Waxman D J. J Biol Chem. 1990;265:15090–15095. [PubMed] [Google Scholar]
- 16.Spady D K, Cuthbert J A, Willard M N, Meidell R S. J Clin Invest. 1995;96:700–709. doi: 10.1172/JCI118113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pandak W M, Stravitz R T, Lucas V, Heuman D M, Chiang J Y L. Am J Physiol. 1996;270:G401–G410. doi: 10.1152/ajpgi.1996.270.3.G401. [DOI] [PubMed] [Google Scholar]
- 18.Lavorgna G, Ueda H, Clos J, Wu C. Science. 1991;252:848–851. doi: 10.1126/science.1709303. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 20.Schreiber E, Matthias P, Muller M M, Schaffner W. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wigler M, Silverstein S, Lee L S, Pellicer A, Cheng Y C, Axel R. Cell. 1977;11:223–232. doi: 10.1016/0092-8674(77)90333-6. [DOI] [PubMed] [Google Scholar]
- 22.Weintraub H, Groudine M. Science. 1976;193:848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- 23.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R M. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mangelsdorf D J, Evans R M. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 25.Ueda H, Sun G C, Murata T, Hirose S. Mol Cell Biol. 1992;12:5667–5672. doi: 10.1128/mcb.12.12.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galarneau L, Pare J F, Allard D, Hamel D, Levesque L, Tugwood L D, Green S, Belanger L. Mol Cell Biol. 1996;16:3853–3865. doi: 10.1128/mcb.16.7.3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavery D J, Schibler U. Genes Dev. 1993;7:1871–1884. doi: 10.1101/gad.7.10.1871. [DOI] [PubMed] [Google Scholar]
- 28.Wang D, Stroup D, Marrapodi M, Crestani M, Galli G, Chiang J Y L. J Lipid Res. 1996;37:1831–1841. [PubMed] [Google Scholar]
- 29.Crestani M, Sadeghpour A, Stroup D, Galli G, Chiang J Y. J Lipid Res. 1998;39:2192–2200. [PubMed] [Google Scholar]
- 30.Cooper A D, Chen J, Botelho-Yetkinler M J, Cao Y, Taniguchi T, Levy-Wilson B. J Biol Chem. 1997;272:3444–3452. [PubMed] [Google Scholar]
- 31.Lala D S, Rice D A, Parker K L. Mol Endocrinol. 1992;6:1249–1258. doi: 10.1210/mend.6.8.1406703. [DOI] [PubMed] [Google Scholar]
- 32.Liu D, Le Drean Y, Ekker M, Xiong F, Hew C L. Mol Endocrinol. 1997;11:877–890. doi: 10.1210/mend.11.7.9947. [DOI] [PubMed] [Google Scholar]
- 33.Ellinger-Ziegelbauer H, Hihi A K, Laudet V, Keller H, Wahli W, Dreyer C. Mol Cell Biol. 1994;14:2786–2797. doi: 10.1128/mcb.14.4.2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giguere V, Ong E S, Segui P, Evans R M. Nature (London) 1987;330:624–629. doi: 10.1038/330624a0. [DOI] [PubMed] [Google Scholar]
- 35.Petkovich M, Brand N J, Krust A, Chambon P. Nature (London) 1987;330:444–450. doi: 10.1038/330444a0. [DOI] [PubMed] [Google Scholar]
- 36.Heyman R A, Mangelsdorf D J, Dyck J A, Stein R B, Eichele G, Evans R M, Thaller C. Cell. 1992;68:397–406. doi: 10.1016/0092-8674(92)90479-v. [DOI] [PubMed] [Google Scholar]
- 37.Levin A A, Sturzenbecker L J, Kazmer S, Bosakowski T, Huselton C, Allenby G, Speck J, Kratzeisen C, Rosenberger M, Lovey A, Grippo J F. Nature (London) 1992;355:359–361. doi: 10.1038/355359a0. [DOI] [PubMed] [Google Scholar]
- 38.McDonnell D P, Mangelsdorf D J, Pike J W, Haussler M R, O’Malley B W. Science. 1987;235:1214–1217. doi: 10.1126/science.3029866. [DOI] [PubMed] [Google Scholar]
- 39.Lehmann J M, Kliewer S A, Moore L B, Smith-Oliver T A, Oliver B B, Su J L, Sundseth S S, Winegar D A, Blanchard D E, Spencer T A, Willson T M. J Biol Chem. 1997;272:3137–3140. doi: 10.1074/jbc.272.6.3137. [DOI] [PubMed] [Google Scholar]
- 40.Janowski B A, Willy P J, Devi T R, Falck J R, Mangelsdorf D J. Nature (London) 1996;383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- 41.Li M, Xie Y-H, Kong Y-Y, Wu X, Zhu L, Wang Y. J Biol Chem. 1998;273:29022–29031. doi: 10.1074/jbc.273.44.29022. [DOI] [PubMed] [Google Scholar]