Summary
The anaphase promoting complex/cyclosome (APC/C) is a conserved multi-subunit E3 ubiquitin ligase required to signal the degradation of key cell cycle regulators. Using single particle cryo-electron microscopy (cryo-EM) we have determined a three-dimensional (3D) structure of the core APC/C from Schizosaccharomyces pombe bound to the APC/C activator Slp1/Cdc20. At the 27 Å resolution of our density map, the APC/C is a triangular-shaped structure, ~19 × 17 × 15 nm in size, with a deep internal cavity and a prominent horn-like protrusion emanating from a lip of the cavity. Using antibody labeling and mutant analysis we have localized twelve of thirteen APC/C components, as well as the position of the activator Slp1, enabling us to propose a structural model of APC/C organization. Comparison of the APC/C with another multi-protein E3 ligase, the SCF complex, uncovers remarkable structural similarities.
Keywords: APC/C, Cdc20, Slp1, E3 ubiquitin ligase, cryo-electron microscopy
Introduction
Cell cycle progression is regulated by the transient activation and inactivation of cyclin-dependent protein kinases. It is also regulated by proteasome-mediated degradation of key proteins following their polyubiquitination. Ubiquitin (Ub) becomes covalently attached to substrate proteins via an enzyme cascade consisting of activating (E1), conjugating (E2), and ligating (E3) enzymes. E3 ubiquitin ligases vary widely in size, composition, and enzymology, reflecting their regulatory role in substrate recognition (Pickart, 2001). The anaphase-promoting complex or cyclosome (APC/C) is a RING-finger type E3 ubiquitin ligase that facilitates the transfer of Ub from E2s to specific substrates involved in cell cycle events.
The APC/C becomes active at the metaphase/anaphase transition and remains active during G1 (reviewed in Harper et al., 2002; Peters, 2002; Thornton and Toczyski, 2006). During these stages it is responsible for directing the degradation of central cell cycle regulatory proteins, particularly mitotic cyclins whose destruction allows mitotic exit, and securin whose destruction allows chromosome segregation. The activation of the APC/C is mediated in part by the binding of transiently available activating subunits typified by the founding member of the family, Cdc20 (reviewed in Harper et al., 2002; Peters, 2002; Thornton and Toczyski, 2006). Cdc20 binds the core APC/C in mitosis and is critical for anaphase initiation. A second family member, Cdh1, plays distinct roles in mitotic exit and G1. Cdc20 and Cdh1 not only bind the APC/C, but also bind substrates directly through their WD40 motifs and therefore act as substrate specific adaptors essential for APC/C-mediated ubiquitination in cells (Burton and Solomon, 2001; Burton et al., 2005; Hilioti et al., 2001; Kraft et al., 2005; Passmore et al., 2003; Pfleger and Kirschner, 2000; Pfleger et al., 2001). Cdc20 also serves as the target of the spindle assembly checkpoint that prevents anaphase onset until chromosomes are correctly attached to the mitotic spindle (reviewed in Cleveland et al., 2003; Kadura and Sazer, 2005; Musacchio and Hardwick, 2002; Taylor et al., 2004). In Schizosaccharomyces pombe, Slp1 is the Cdc20 ortholog important for mitotic progression (Matsumoto, 1997) and Ste9 is the Cdh1 homolog important for G1 arrest in preparation for mating (Blanco et al., 2000; Kitamura et al., 1998).
In S. pombe and Saccharomyces cerevisiae the core APC/C is composed of 13 subunits (Hall et al., 2003; Passmore et al., 2003; Schwickart et al., 2004; Yoon et al., 2002) and most of these are conserved in higher eukaryotes (reviewed in Thornton and Toczyski, 2006). Interestingly, only Apc2 and Apc11, a cullin-repeat and RING-finger containing protein respectively are required for substrate dependent catalytic activity in vitro (Gmachl et al., 2000; Tang et al., 2001), leaving the roles of many subunits unclear. One of the TPR (Tetratrico Peptide Repeat) containing subunits, Cdc27 (S. pombe Nuc2), is able to bind activator subunits directly and is therefore implicated in mediating interactions with substrates (Burton et al., 2005; Kraft et al., 2005; Vodermaier et al., 2003). Other core APC/C subunits, particularly Apc10/Doc1, have been implicated in substrate binding also and processivity of substrate ubiquitination (Carroll et al., 2005; Meyn et al., 2002; Nourry et al., 2004; Passmore et al., 2003; Yamano et al., 2004). Thus, these and other core components might act as molecular scaffolds to position the substrate, the RING-finger domain, and the E2 enzyme into the correct orientation to facilitate the transfer of ubiquitin from the E2 to the substrate. An additional complication is that there are two E2 enzymes necessary for different aspects of cyclin B ubiquitination in S. pombe, Ubc4 and Ubc11 (Seino et al., 2003) and core subunits other than Apc11 and Apc2 could be involved in regulating their specific action. In other words, proteins not directly involved in the transfer of Ub are likely to be important for creating the correct structural environment for this transfer to occur or for localizing the APC/C correctly within the cell.
To understand how APC/C components organize to form a tightly regulated, multi-protein E3 ubiquitin ligase it is necessary to determine the structure formed when they come together. Thus far, cell cycle-specific changes in composition and post-translational modification of the APC/C, as well as its low cellular abundance, have thwarted efforts to structurally characterize this complex using X-ray crystallography. Electron microscopy (EM), however, is well suited for dealing with multi-protein complexes at relatively low concentrations, and EM structures of the mammalian, Xenopus, and S. cerevisiae APC/C have been presented (Dube et al., 2005; Gieffers et al., 2001; Passmore et al., 2005). Given the conservation of APC/C function and composition, the four reported structures are surprisingly dissimilar. Moreover, the positions of only three core APC/C components have been determined in a subset of these structures, leaving the question of overall structural organization unresolved.
We present the 27 Å 3D structure of an active form of the S. pombe APC/C purified from cells blocked in mitosis and bound to the mitotic activator, Slp1, that we obtained by cryo-EM of vitrified samples. The density map reveals an asymmetric particle with a prominent central cavity and a horn shaped structure protruding from its lip. Using antibody labeling and mutant analysis, we have mapped the positions of 12 of the 13 APC/C core components, as well as the activator Slp1, within this structure to provide the most comprehensive structural analysis of APC/C organization to date. Our model of APC/C architecture reveals striking similarity with that of another RING-type E3, the SCF complex, that leads to postulated implications for APC/C function and regulation.
Results
Purification and characterization of the S. pombe APC/C
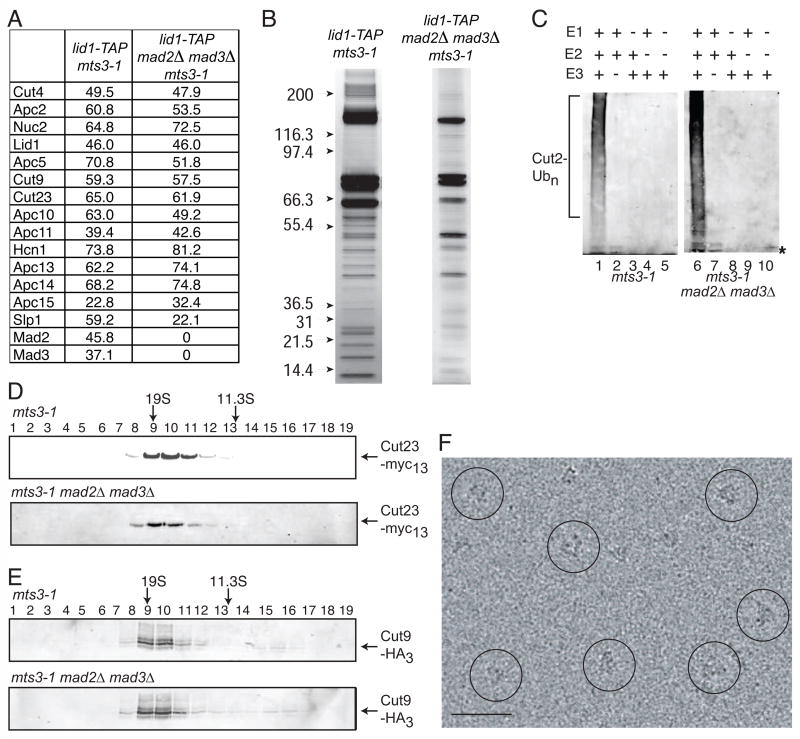
The S. pombe APC/C was first purified from mts3-1 arrested cells using a tandem affinity purification (TAP) strategy targeting S. pombe Lid1 (Yoon et al., 2002). S. pombe Mts3, a subunit of the proteasome, is essential for proteasome-mediated degradation of ubiquitin conjugates and mts3 mutants arrest at the metaphase-to-anaphase transition (Gordon et al., 1996; Seeger et al., 1996). Following the two affinity purification steps, the protein content of the TAP complex was analyzed by MudPIT mass spectrometry and viewed by silver staining (Fig. 1A and 1B). As from asynchronously growing cells, all 13 core APC/C components (Yoon et al., 2002) were present in this purification (Fig. 1A). In addition to core APC/C components, the APC/C activator Slp1/Cdc20 and also the spindle checkpoint proteins Mad2 and Mad3 co-purified with Lid1-TAP (Fig. 1A). Since some fraction of the APC/C is active at this arrest, as measured by the ubiquitination of target proteins in vivo (Berry et al., 1999) and in vitro (Yoon et al., 2002), and Mad2 and Mad3 are inhibitors of APC/C function, we reasoned that Mad2 and Mad3 are present in only a subset of the purified APC/C complexes. Thus, to eliminate a source of heterogeneity in our purifications, Lid1-TAP complexes were purified from mts3-1 mad2Δ mad3Δ arrested cells. Once again, all 13 core APC/C components and the Slp1/Cdc20 activator were present as determined by mass spectrometry (Fig. 1A) and the composition appeared similar by silver staining as well (Fig. 1B). Semi-quantitative analysis of the mass spectrometric results by spectral counting (Liu et al., 2004) indicated that the core APC/C was greatly enriched over background contaminants in both strains (data not shown). The purified APC/C-TAP complexes were active as they could mediate the ubiquitination of purified maltose-binding protein-Cut2/securin in an E1-, E2-, and ATP-specific manner (Fig. 1C). Lid1-TAP complexes purified from mad2Δ mad3Δ strains, like the ones purified from the mts3-1 strain, sedimented in a discrete peak at ~20S by sucrose gradient analysis indicating that the purified APC/C complexes were intact and might be suitable for structural analysis (Fig. 1D and E).
Figure 1. Purification and characterization of the S. pombe APC/C.
(A) TAP/mass spectrometry results from APC/C particles purified from mts3-1 and mts3-1 mad2Δ mad3Δ arrested cells. Numbers represent the percent sequence coverage for each protein. (B) Silver stained gels of a portion of the Lid1-TAP complexes purified from mts3-1 and mts3-1 mad2Δ mad3Δ arrested cells. Note that several components have similar molecular masses and are present in more than one copy (see Fig. 2H). (C) The APC/C purified from mts3-1 (left panel) or mts3-1 mad2Δ mad3Δ (right panel) arrested cells was subjected to ubiquitination assays in the presence (+) or absence (−) of the indicated components. Ubiquitin (Ub)-conjugated derivatives were detected by immunoblotting with anti-Ub antibody. “*” indicates unbound Ub. (D and E) Protein lysates from the Lid1-TAP complex following its purification from either mts3-1 cut23-Myc2 or mts3-1 cut9-HA3 (upper panels) or mts3-1 cut23-Myc13 mad2Δ mad3Δ or mts3-1 cut9-HA3 mad2Δ mad3Δ (lower panels) arrested cells were resolved on a 10–30% sucrose gradient. Fractions were collected from the bottom (fraction 1) and immunoblotted with anti-myc antibodies to detect Cut23-Myc or anti-HA antibodies to detect Cut9-HA3. The different mobility forms of Cut9 are due to phosphorylation (Yamada et al., 1997). The peaks of thyroglobulin (19S) and catalase (11.3S) collected from parallel gradients are indicated. (F) A typical electron micrograph area of APC/C particles in vitrified ice. A few of the particles are circled in black. Scale bar represents 50 nm.
To examine the structural homogeneity of our preparation, Lid1-TAP particles purified from mad2Δ mad3Δ mts3-1 arrested cells were negatively stained with uranyl formate and examined by EM. The particles were mono-disperse and homogenous in size (Fig. S1). Classification of approximately three thousand particles selected from images of untilted specimens into twenty groups revealed that a majority of the class averages contained particles with very similar structural features (see Fig. S1, inset), and, importantly, structures calculated from the tilted particles corresponding to these class averages looked almost indistinguishable (data not shown). Additionally, structural analysis of the APC/C particles using the random conical tilt approach (RCT) (Radermacher et al., 1987) in cryo-negative stain demonstrated that the purified complexes are structurally homogenous and amenable to further analysis in vitrified ice (Fig. S3). Thus, Lid1-TAP particles purified from mts3-1 mad2Δ mad3Δ arrested cells were vitrified and examined by cryo-EM. The particles were mono-disperse and homogenous in size (Fig. 1F). We did not detect dimerization of the S. pombe APC/C particles by negative stain or cryo-EM (Fig. S1, S3A, 1F) as was observed for the S. cerevisiae APC/C (Passmore et al., 2005).
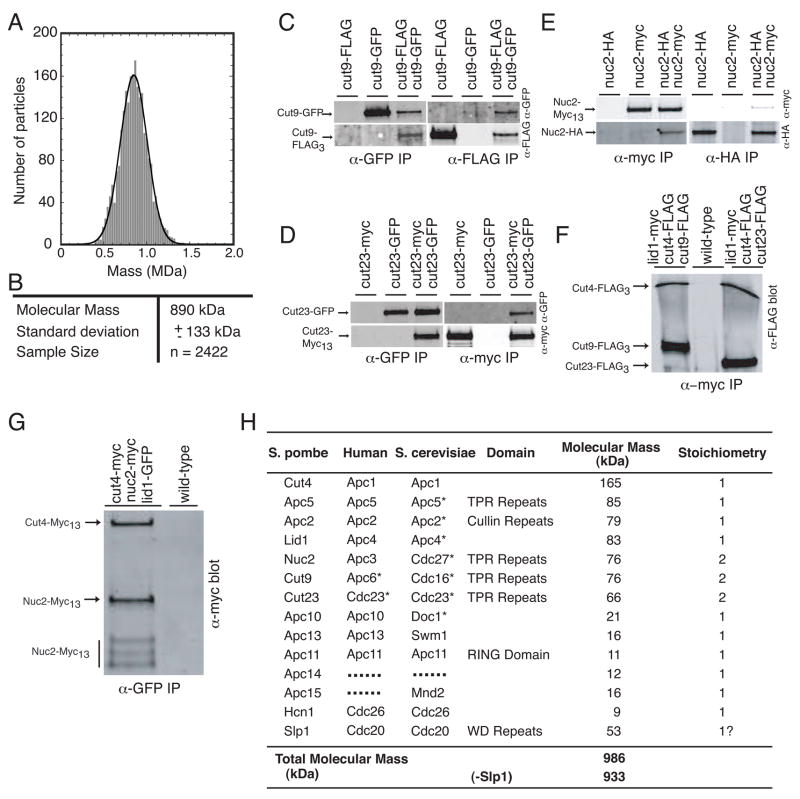
To obtain an accurate molecular mass of the APC/C complex we analyzed freeze-dried, unstained particles by scanning transmission electron microscopy (STEM). The 2,422 selected particles segregated into one Gaussian shaped curve, indicating a complex with a mass of 890 kDa (+/− 133 kDa) (Fig. 2A and B). Summation of the mass of the APC/C components, as identified by mass spectrometry (Fig. 1A), yields a molecular weight of 768 kDa; however, it seemed likely that the S. pombe APC/C, such as the mammalian and S. cerevisiae APC/C complexes (Dube et al., 2005; Passmore et al., 2005), contains subunits that are present in more than one copy per particle.
Figure 2. Mass measurement of the S. pombe APC/C by STEM and determination of APC/C subunit stoichiometry.
(A) Histogram showing the mass distribution of S. pombe APC/C particles. (B) Summary of the STEM analysis of S. pombe APC/C. (C) S. pombe Cut9-GFP and Cut9-FLAG associate in vivo. An anti-GFP (upper panel) and an anti-FLAG (lower panel) immunoblot of immunoprecipitates from cut9-GFP, cut9- flag3, and cut9-GFP cut9-flag3 strains. Immunoprecipitations were performed with anti-GFP antibodies or anti-flag antibodies. (D) S. pombe Cut23-GFP and Cut23-Myc13 associate in vivo. An anti-GFP (upper panel) and an anti-myc (lower panel) immunoblot of immunoprecipitates from cut23-GFP, cut23-Myc13, and cut23-GFP cut23-Myc13 strains. Immunoprecipitations were performed with anti-GFP antibodies or anti-Myc antibodies. (E) S. pombe Nuc2-HA3 and Nuc2-Myc13 associate in vivo. An anti-Myc (upper panel) and an anti-HA (lower panel) immunoblot of immunoprecipitates from nuc2-HA3, nuc2-Myc13, and nuc2-HA3 nuc2-Myc13 strains. Immunoprecipitations were performed with anti-HA antibodies or anti-Myc antibodies. (F) S. pombe Cut9 and Cut23 are present within the APC/C in a 2.1:1 ratio with Cut4. An anti-FLAG immunoblot of an anti-Myc immunoprecipitate of the APC/C-containing fractions from a sucrose gradient from either wild-type, lid1-Myc13 cut4- flag3 cut9- flag3, or lid1-Myc13 cut4- flag3 cut23- flag3strains. Bands were quantified on an Odyssey instrument. (G) S. pombe Nuc2 is present within the APC/C in a 2.2:1 ratio with Cut4. An anti-Myc immunoblot of an anti-GFP immunoprecipitate from the APC/C-containing sucrose gradient fractions from either wild-type or lid1-GFP cut4-Myc13 nuc2- Myc13 cells. Nuc2-Myc13 was also present in three degradation bands indicated by the bar that were considered in the quantitation. All bands were quantified on an Odyssey instrument. (H) Summary of S. pombe APC/C molecular weight calculations using subunit stoichiometries. Human and S. cerevisiae APC/C subunits reported to be present in more than one copy (Dube et al., 2005; Passmore and Barford, 2005) are marked with an asterisk.
To establish the number of each component within a single APC/C particle, a series of strains were constructed in which one of the thirteen APC/C subunits was produced with either a Myc13, HA3, GFP, or FLAG3 C-terminal epitope tag. These tags did not affect APC/C function significantly as all epitope-tagged strains grew well (data not shown). Thirteen diploid strains were then constructed that each produced two different epitope-tagged versions of a single APC/C component and co-immunoprecipitation analyses were performed on the diploid strains to determine if one epitope-tagged component could co-immunoprecipitate the other epitope-tagged version of itself. For example, in a diploid strain producing both Cut9-FLAG and Cut9-GFP, anti-GFP antibody pulled down both Cut9-GFP and Cut9-FLAG (Fig. 2C), while Cut9-FLAG was not immunoprecipitated from the strain producing only Cut9-GFP, indicating the specificity of the antibody (Fig. 2C). In the reciprocal experiment, anti-FLAG immunoprecipitates from the diploid but not the single GFP-tagged strain contained Cut9-GFP in addition to Cut9-FLAG (Fig. 2C). Through similar analyses of all 13 double-tagged diploid strains, we found that only three proteins, Cut9, Cut23 and Nuc2, self-associated in vivo (Fig. 2C–E), indicating that these proteins are present in more than one copy per APC/C particle. Conversely, one epitope-tagged variant of Cut4, Apc5, Apc2, Lid1, Apc10, Apc13, Apc11, Apc14, Apc15, and Hcn1 did not co-immunoprecipitate a second epitope-tagged variant of itself (Fig. S2), indicating that these proteins are present in only one copy per APC/C particle (summarized in Fig. 2H).
To determine the stoichiometry of Cut9, Cut23, and Nuc2 within the APC/C we compared the abundance of these proteins relative to Cut4, a subunit found only once in each APC/C particle (Fig. S2A). For this analysis, lid1-Myc13 cut4-flag3 cut9-flag3, lid1-Myc13 cut4-flag3 cut23- flag3, and lid1-GFP cut4-Myc13 nuc2-Myc13 strains were generated. The APC/C was first isolated through anti-Myc (for the Lid1-Myc13 strains) or anti-GFP (for the Lid1-GFP strain) immunoprecipitations. These immunoprecipitations were then immunoblotted with anti-FLAG or anti-Myc antibodies. By quantitating the intensities of the anti-FLAG and anti-Myc signals on immunoblots, we determined that Cut9-FLAG and Cut23-FLAG were each 2.1-fold more abundant than Cut4-FLAG, and Nuc2-Myc13 was 2.2 times more abundant than Cut4-Myc13 (Fig. 2F–G). These data provide compelling evidence that the APC/C contains two copies of each of these three TPR proteins (summarized in Fig. 2H). Taking this subunit stoichiometry into account, the calculated mass of the S. pombe APC/C becomes 986 kDa (933 kDa minus Slp1) (Fig. 2H). This value is consistent with the mass determined by STEM of 890 kDa (+/− 133 kDa) (Fig. 2A and B), especially when considering that Slp1 may not remain bound to all of the APC/C particles examined in the STEM analysis.
Cryo-electron microscopy and three-dimensional reconstruction
Using cryo-EM we collected images of S. pombe APC/C particles purified from mts3-1 mad2Δ mad3Δ cells in vitrified ice. The individual APC/C particles were clearly visible in the raw images (Fig. 1F). 28,540 particles were manually selected from micrographs, band-pass filtered, and subjected to multi-reference alignment, multivariate statistical analysis, and classification to obtain averages with improved signal-to-noise ratio (Fig. S4A, bottom rows). Using the program FREALIGN (Grigorieff, 2007), the class averages were aligned directly to the low-resolution cryo-negative stain structure determined using the RCT approach (Fig. S3D), and the structure was refined until stable at a resolution of 40 Å. After five refinement cycles, individual particle images were aligned to the reference map obtained with the class averages. The density map was refined and corrected for the contrast transfer function (CTF) using FREALIGN (Grigorieff, 2007). The resolution of our final density map was estimated by the Fourier shell correlation (FSC) method, which yielded 27 Å based on the FSC = 0.5 cut-off (Bottcher et al., 1997) (Fig. S4B). Although particles adopt preferred orientations on the carbon support film, all projections are represented in the final 3D map (Fig. S4C). Reprojections of the 27 Å density map (Fig. S4A, middle panel) correspond very well to the raw images (Fig. S4A, upper panel) and the class averages (Fig. S4A, lower panel) demonstrating the consistency of the 3D map with the projection data. The contour level of the refined density map was selected to encompass a volume corresponding to the molecular mass of 980 kDa, as predicted by the subunit stoichiometry analysis of the complex and confirmed by STEM (Fig. 2 and S2).
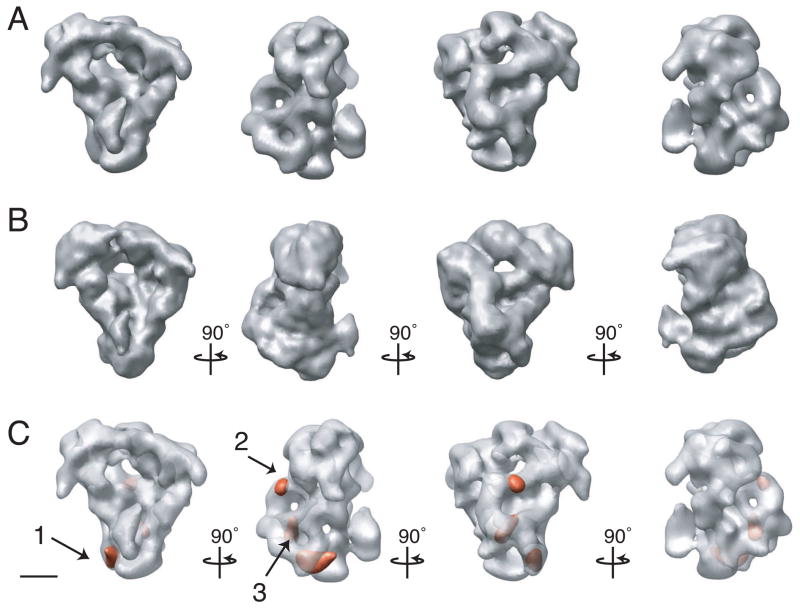
The S. pombe APC/C adopts an asymmetric, tricorn-shaped structure, ~19 × 17 × 15 nm in size, with a deep internal cavity and a prominent horn-like protrusion emanating from near the bottom of the cavity lip (Fig. 3). The size of the central cavity is ~11.5 × 9.5 × 6.5 nm and it contains one small channel that pierces the back wall of the complex. This channel, at a diameter of ~3.0 nm, is not large enough to allow an ubiquitin molecule (2.1 × 3.0 × 3.3 nm) to pass. The cavity is large enough to hold both an E2-Ub conjugated complex and an APC/C substrate, such as the Cdk (cyclin dependent kinase)-cyclin complex (data not shown). The horn-like structure protrudes from the bottom lip of the central cavity at an angle of ~15° from the main cavity and is ~4.5 × 2.5 × 4.5 nm in size. At its most extended point the horn is ~3.5 nm away from the opening of the cavity.
Figure 3. Three-dimensional reconstruction of the S. pombe APC/C.
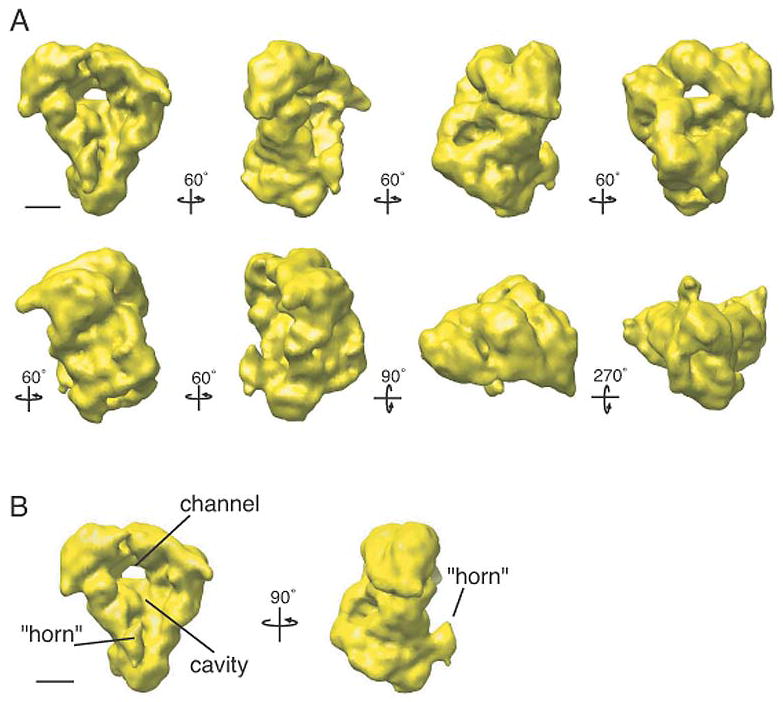
(A) Views of the APC/C tilted stepwise about the vertical axis in 60 increments or about the horizontal axis by 90° or 270° steps (in reference to the top left structure) as indicated by arrows. The density map was contoured to a mass of 980 kDa. (B) The APC/C is shown in two orientations tilted about its vertical axis by 90°. The complex has a tricorn shape with a large central cavity and a prominent horn. The handedness of the structure was not experimentally determined. Scale bars represent 5 nm.
To confirm the accuracy of our density map, we calculated an additional, independent structure using an ab initio structure determination method (Penczek et al., 1996). In this method, orientation parameters of the particles are assigned to the class averages obtained from the vitrified APC/C data yielding a low-resolution model of the structure. After subsequent refinement using the whole cryo-dataset, both this and the structure calculated using the RCT density map as an initial model looked very similar to each other within the resolution range claimed (compare Fig. 4A and 4B), validating our APC/C structure.
Figure 4. Ab initio structural characterization and variance mapping of the APC/C.
(A) 3D reconstruction of the APC/C in vitrified ice determined using an ab initio approach without using an initial model. (B) 3D reconstruction of the APC/C in vitrified ice determined using the structure shown in Figure S3 as an initial model. (C) 3D variance of the APC/C density map shown in panel A. Variance peaks are shown in red. All density maps are tilted stepwise about the vertical axis by 90° steps as indicated by arrows. Scale bars represent 5 nm.
Analysis of the 3D variance in the APC/C density map
The fact that the refinement of the structure of the APC/C yielded a map only at 27 Å resolution indicated possible structural heterogeneity in the data set as a limiting factor. However, images and class averages of negatively stained APC/C appeared homogenous (Fig. S1), suggesting that any variation in the sample was subtle. To further test the homogeneity of the sample, we analyzed the 3D variance of the cryo-EM density map. The analysis was carried out using the resampling technique described in Penczek et. al. (Penczek et al., 2006). We were able to detected three small but well-defined regions of high variance (Fig. 4C). The largest of these areas (Fig. 4C, peak 1) most likely results from the loss of the activator Slp1 from a portion of the APC/C population, as determined by a combination of difference mapping and immuno-labeling (see below); however, the source of the other two variance peaks is unclear.
Localization of APC/C core subunits
To gain insight into how the APC/C components are organized within this structure, we performed antibody labeling experiments to localize the C-termini of all core subunits. To do this, 13 strains were constructed in which each APC/C subunit was individually engineered to have a Myc2 epitope at its C-terminus. These alleles were then combined with the mts3-1 mutation and either the lid1-TAP or in the case of Lid1-Myc2, apc13-TAP so that the APC/C could again be purified from mitotically arrested cells. To confirm that intact APC/C particles had been purified in each case, the TAP eluates were analyzed by silver staining and in most cases, also by mass spectrometry (Fig. S5). Antibody against the Myc epitope was added after the second affinity purification step in each of the 13 purifications and the particles were analyzed by negative stain EM. In all but one purification, antibody could be observed associated with the APC/C particles at specific positions (Fig. 5A–L). We suspect that the Myc2 epitope on Apc14 was buried within the APC/C and inaccessible to the antibody. Between 579 and 1,369 antibody decorated particles were selected for each labeling experiment to ensure that the location of the antibody bound to the APC/C was correctly determined for each of the Myc-tagged strains (Fig. 5A–L, left gallery of panels). Confirming our co-immunoprecipitation results (Fig. 2C–G), a majority of Cut9-Myc2, Nuc2-Myc2 and Cut23-Myc2 containing APC/C particles were decorated with two antibodies (Fig. 5J–L). Class averages were calculated for each labeling experiment to more clearly show the location of each bound antibody (Fig. 5A–J, right panels). This worked well for all the subunits with the exception of Cut9 and Cut23, where the position of the two bound antibodies could not be clearly resolved into one class average. The relative visibility of the antibodies in the averaged images likely reflects the relative flexibility of the different C-termini. Through these analyses we were able to localize the approximate position of the C-termini of 12 of the 13 APC/C core subunits within the EM density map (Fig. 5M).
Figure 5. Localization of APC/C subunits using antibody labeling.

(A–L) Antibody (9E10) labeling of the APC/C that contains a C-terminal Myc2 epitope-tagged version of either Apc13 (A), Apc10 (B), Apc2 (C), Apc5 (D), Hcn1 (E), Lid1 (F), Apc11 (G), Cut4 (H), Apc15 (I), Nuc2 (J), Cut9 (K), or Cut23 (L). (A and L) Galleries (left panels) and a representative class average (right panel) of antibody labeled APC/C particles. Numbers of labeled particles picked for each experiment are given after the subunit and the number of particles in each projection average is shown underneath the average. Schematic representations are shown below the EM images. Positions of antibodies are shown in red. Side lengths of panels are 57.3 nm. (M) Two-dimensional view of antibody positions (marked with “*”) on the 3D model of the APC/C. Scale bar represents 5 nm.
Localization of the APC/C activator Slp1
Cdc20 family members, with their ability to bind both the APC/C and specific substrates, play an essential role in APC/C-mediated ubiquitination (reviewed in Harper et al., 2002; Peters, 2002; Thornton and Toczyski, 2006). To identify the position of Slp1 within the APC/C particle, we used two complementary approaches. First, we purified the APC/C from a strain containing the slp1-362 temperature sensitive allele. Because Slp1-362 can no longer associate with the APC/C when this strain is shifted to its non-permissive temperature (Yamada et al., 2000), APC/C particles purified from slp1-362 arrested cells do not contain bound Slp1, Mad2, or Mad3, while the composition of the core APC/C is unchanged (Fig. S6). Therefore, a comparison of 3D maps generated from APC/C particles purified from mts3-1 mad2Δ mad3Δ cells with those from slp1-362 cells were expected to reveal the location of Slp1. We collected 8,893 APC/C particles in vitrified ice purified from slp1-362 arrested cells and calculated a 3D structure using FREALIGN. Only one significant difference was detected when the 3D structure of the APC/C lacking Slp1 was compared with the structure of the APC/C determined from mts3-1 mad2Δ mad3Δ cells (Fig. 6A). This difference peak, representing the location of Slp1, is located at the lower corner of the structure in close proximity to both the “horn-like” protrusion and the position of the C-terminus of the RING protein, Apc11 (Fig. 5G and M). Interestingly, this difference peak (Fig. 6A) matches the position of one of the main variance peaks mapped in the APC/C complex (Fig. 4C, peak 1), suggesting that the variance found in this part of the structure most likely arises from the absence of Slp1 in some of the APC/C particles purified from mts3-1 mad2Δ mad3Δ arrested cells. Also of note, the presence or absence of Slp1 does not seem to cause any obvious changes in the overall structure of the S. pombe APC/C.
Figure 6. Localization of the activator Slp1 on the APC/C.
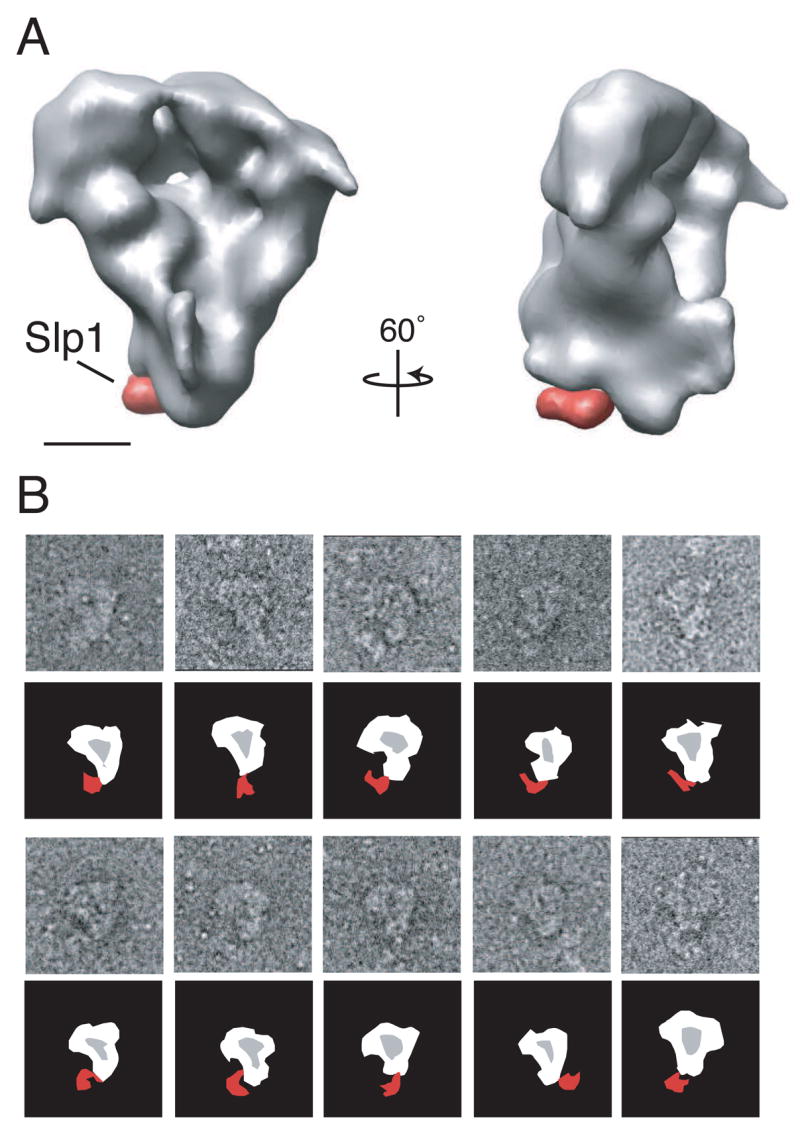
(A) The difference in density found between APC/C structures that either contain or do not contain Slp1. The APC/C purified from a slp1-362 genetic background (lacking Slp1-362) is shown in grey. The missing density in the APC/C slp1-362 when compared with the APC/C structure purified from an mts3-1 mad2Δ mad3Δ genetic background (containing Slp1) is shown in red. The complex is shown in two orientations tilted about its vertical axis by 60°. Scale bar represents 5 nm. (B) HA antibody labeling of APC/C particles containing Slp1-HA3. Schematic representations are shown below the raw particle images. Position of antibody is shown in red. Side lengths of panels are 57.3 nm.
The second approach we took to identify the position of Slp1 was antibody labeling. The slp1+ locus was engineered to express a C-terminal HA3 epitope and this allele was combined with lid1-TAP mts3-1. The protein composition of the purification was verified by mass spectrometry and silver staining (Fig. S6). Other tagged alleles of Slp1 were temperature-sensitive on their own (for example, the Myc2 tag) and could not be used for this analysis. Antibody against the HA epitope was added to the purification after the second step of purification to allow labeling of the particles. When analyzed by negative stain EM, antibody could be seen associated with the APC/C particles at the lower corner of the complex (Fig. 6B) in an identical position to the difference observed in particles purified from the slp1-362 strain (Fig. 6A). Thus, antibody labeling confirms the position of Slp1 determined by the difference map.
Discussion
The APC/C is a conserved RING-finger ubiquitin ligase essential for cell cycle regulation. There are many unsolved questions concerning how the APC/C carries out specific ubiquitination events and how its activity is regulated throughout the cell cycle (reviewed in Peters, 2006; Thornton and Toczyski, 2006). One feature of the APC/C complicating its analysis is its large mass and subunit complexity. Comprised of at least 13 core subunits and additional activators, it has not been clear how these components cooperate to carry out polyubiquitination of target proteins. To advance our understanding of APC/C structural organization, we have characterized the size of the S. pombe APC/C by STEM analysis and determined the subunit stoichiometry using a simple but comprehensive epitope tagging and co-immunoprecipitation approach. We have also determined the 3D structure of the S. pombe APC/C bound to the activator Slp1 by cryo-EM at a resolution of 27 Å. Antibody labeling and mutant analysis, together with EM analysis, allowed us to map the positions of the C-termini of 12 of 13 APC/C core components as well as the location of the mitotic activator Slp1. Our analysis provides the first comprehensive structural study of the molecular organization of the mitotic form of the APC/C.
APC/C complexes purified from different organisms are expected to be similar in structure considering that they are comprised of conserved subunits and are functional orthologs (reviewed in Thornton et al., 2006). Indeed, our conclusions regarding S. pombe core APC/C particle size and composition are similar to those reached in studies of the S. cerevisiae and vertebrate APC/Cs although our estimate of its mass is somewhat smaller at ~900 kDa to 1 MDa compared with 1.4–1.7 MDa for the others (Dube et al., 2005; Passmore et al., 2005). There are several possibilities to account for the differences in reported sizes including the lack of a dimer form in our preparations, more stringent and therefore more homogenous purification from S. pombe, and/or species-specific differences in subunit composition. We also determined that, similar to the human APC/C (Dube et al., 2005), the S. pombe APC/C contains two copies of three TPR components (Nuc2, Cut9 and Cut23) and one copy of all other subunits (summarized in Fig. 2H). In addition, individual particles of the S. pombe APC/C purified from mts3-1 mad2Δ mad3Δ arrested cells appear similar by EM to images of mammalian, Xenopus, and S. cerevisiae APC/C particles (Dube et al., 2005; Gieffers et al., 2001; Passmore et al., 2005).
However, although all reported APC/C 3D structures are generally “triangular” in shape, the structures vary in many details, especially in the presence and/or prominence of a central cavity. The first cryo-negative stain structure reported for the mammalian APC/C featured a slightly triangular shape with a prominent central cavity that the authors noted was large enough to be involved in sequestering and/or positioning the E2 and substrate for ubiquitin transfer (Gieffers et al., 2001). A second cryo-negative stain APC/C structure of the mammalian and frog APC/C determined by the same group does not include this large, open cavity (Dube et al., 2005), even though similar purification and structural approaches were used to determine the maps (Dube et al., 2005; Gieffers et al., 2001). The S. cerevisiae APC/C structure, calculated from particles in vitrified ice, has an overall triangular shape; however, rather than having a prominent central cavity, the S. cerevisiae map contains a tunnel-like feature that is not large enough to hold an E2, ubiquitin and substrate simultaneously (Passmore et al., 2005). In the latter studies, the RCT approach was used to calculate an additional APC/C structure in order to help confirm the validity of the respective cryo-EM structure (Dube et al., 2005; Passmore et al., 2005). We note, however, that the RCT maps were not used as initial models for aligning the cryo-EM data (Dube et al., 2005; Passmore et al., 2005), even though this approach has been shown to be a good way to generate a reliable cryo-EM structure (Cheng et al., 2006). Interestingly, our S. pombe vitrified ice structure (Fig. 3) looks very similar to the negative stain EM structure of the S. cerevisiae APC/C calculated using the RCT approach (Passmore et al., 2005) which, similarly as our S. pombe cryo-EM structure, contains a large central cavity and a protruding “horn-like” feature emanating from the lip of the cavity.
Our initial reconstruction, done by aligning ice images using the cryo-negative stain structure as an initial model, appeared to be significantly different in major structural features, such as the presence of a large central cavity and a prominent “horn-like” structure, from the three published cryo-EM structures (Dube et al., 2005; Gieffers et al., 2001; Passmore et al., 2005), which as discussed above are also different from each other. Concerned by these differences, we calculated a 3D reconstruction of the S. pombe APC/C in an entirely different manner, namely assigning orientation parameters ab initio to the class averages from the vitrified particles and then calculating a 3D map (Fig. 4A). The advantage of this approach is that no initial model is used to align the images, eliminating the possibility of model bias. The resulting density map looked very similar in almost all structural features to the one obtained using the cryo-negative stain structure as the reference model (Fig. 4A and B). That two unrelated structure determination methods yielded such similar structural results is strong evidence that our 3D map is a faithful representation of the S. pombe APC/C bound to Slp1.
The differences between the S. pombe structure and the vertebrate and S. cerevisiae APC/C structures might lie in the structural approaches used to determine the structures, as discussed above. However, another potential explanation is that the S. pombe APC/C particles were purified from cells arrested in mitosis while the vertebrate and S. cerevisiae APC/C particles were collected from lysates or cells that were primarily in interphase cell cycle stages (Dube et al., 2005; Gieffers et al., 2001; Passmore et al., 2005). It is possible that the core APC/C adopts different conformations at different points in the cell cycle to favor either Cdc20/Slp1 or Ste9/Cdh1 binding, or to inhibit activator binding. Indeed, antibody labeling experiments show that Cdh1 and Slp1/Cdc20 apparently bind to different regions of the APC/C (Fig. 6 and Dube et al., 2005), making it likely that different regulatory signals are involved in supporting or inhibiting their binding. Differences in core particle structure during mitosis favoring Cdc20 binding could conceivably be induced by the plethora of mitotic-specific phosphorylation events that have been linked to tighter Cdc20/Slp1 interactions (Kraft et al., 2003; Rudner and Murray, 2000). Such a scenario would be consistent with our finding that the core APC/C structure does not appear to change significantly depending on Slp1 binding whereas it was reported that a flexible arc domain within the Xenopus APC/C shifts position when Cdh1 binds (Dube et al., 2005). Potential structural variation in the APC/C due to cell cycle regulation is unlikely, however, to account for the reported differences between the vertebrate and S. cerevisiae structures (Dube et al., 2005; Gieffers et al., 2001; Passmore et al., 2005).
Only two APC/C components are essential for ubiquitination activity in vitro, but many more are required for in vivo function. Our 3D map shows that the APC/C forms a structure with a large central pocket, providing an attractive explanation for why additional APC/C components are required for its function in vivo. The size and central location of the cavity creates a large number of potential protein-protein interaction surfaces that could all spatially orient a substrate near the RING-finger domain and hence, the E2-Ub complex. These potential interaction surfaces could easily be modified by phosphorylation such that they would become available only during distinct times of the cell cycle. A growing body of evidence has accumulated showing that while many substrates require the activators Slp1/Cdc20 or Ste9/Cdh1 to initially interact with the APC/C (Burton and Solomon, 2001; Burton et al., 2005; Hilioti et al., 2001; Kraft et al., 2005; Passmore et al., 2003; Pfleger and Kirschner, 2000; Pfleger et al., 2001), additional core APC/C components, such as Apc10, are also important in mediating APC/C-substrate interactions (Meyn et al., 2002; Nourry et al., 2004; Passmore et al., 2003; Yamano et al., 2004). The presence of a central cavity makes it tempting to propose a unifying model in which APC/C activators could initially recruit substrates to the cavity; however, how tightly the substrate is bound and how processive the ubiquitination reaction becomes would be regulated by core components. In this model substrates associated within the APC/C pocket could interact with activators and core components simultaneously. In addition, the position of the horn-like protrusion extending from the bottom lip at an angle from the cavity could also provide another level of APC/C specificity by creating an accessible scaffold located near the central pocket and the Slp1 binding site that could facilitate APC/C interactions with substrates, activators, inhibitors and/or E2-Ub conjugates. Localizing the position of a bound substrate would be an important next step in gaining insight into APC/C function.
Gratifyingly, our localization results for core subunits support and extend previous studies aimed at understanding APC/C subunit associations and structural organization. Our map indicates that three TPR proteins (Cut23, Cut9, and Nuc2), Hcn1, and Apc13 cluster around the top lip of the complex, the C-termini of Cut4, Lid1, and Apc5 are positioned near the lower corner of the structure, and the catalytic subunits (Apc2, Apc11, and Apc10) are localized along the right side of the structure (Fig. 5M). This overall organization is very consistent with a recently proposed model dividing the S. cerevisiae APC/C into three sub-complexes: a scaffold complex consisting of Apc1, Apc4, and Apc5, a catalytic complex comprised of Apc11, Apc2 and Apc10, and a TPR complex comprising the remaining subunits (Thornton et al., 2006). The relative positions of the components are also consistent with other previous studies that provided evidence for various subunit interactions and stable APC/C sub-complexes (Schwickart et al., 2004; Vodermaier et al., 2003; Wendt et al., 2001; Yoon et al., 2006). The one apparent variation with previous reports involves the positioning of the Slp1 C-terminus near the Apc11 and Apc2 C-termini since it has previously been determined that the APC/C activator Cdh1 associates through a C-terminal IR motif (also present in Slp1) with the TPR repeat protein Cdc27 (S. pombe Nuc2) (Vodermaier et al., 2003; Kraft et al., 2005). It should be noted, however, that the relative placements of the Slp1 and Nuc2 C-termini do not preclude the possibility of this association within the APC/C given the resolution of our map. It should also be emphasized that Cdc20 lacking its C-terminal IR motif remains capable of APC/C interaction indicating that other subunits are involved in docking Cdc20 family activators to the APC/C (Thornton and Toczyski, 2006).
It has been postulated that the structural organization of the multi-protein E3 ligases is essential for positioning the substrate in the correct orientation to optimize the transfer of ubiquitin from the E2 by the RING-finger component (Zheng et al., 2002). Interestingly, the localization of the Apc11 C-terminus at the lip of the central cavity and near the protruding density indicates that the E2-Ub complexes could be positioned either in or near the cavity. Additionally, difference mapping and antibody localization show that Slp1 is found in close proximity to the C-terminus of Apc11. Thus this APC/C activator is located close enough to the RING-finger domain to help position substrates correctly for ubiquitin transfer from the E2-Ub complex.
The atomic structure of another multi-protein RING-type E3 ubiquitin ligase, the SCF (Skp1-Cullin-F-box) complex has been determined (Zheng et al., 2002). This crystal structure is composed of hCul1 (a cullin-repeat containing protein and human homolog of S. pombe Cdc53), hRbx1, hSkp1, and the conserved F-box domain of protein Skp2. hCul1 is composed of three cullin repeats and a C-terminal globular domain (CTD) that binds to the RING-finger containing protein hRbx1. hCul1 acts as a rigid scaffold separating hRbx1 from the S. cerevisiae Skp2 F-box, the substrate interacting domain, by ~110 Å (Zheng et al., 2002) (Fig. 7). Like the SCF complex, the APC/C contains both a cullin-repeat component (Apc2) and a RING-finger containing protein (Apc11). In addition, the crystal structure of the S. cerevisiae Apc2 CTD can be superimposed directly onto the structure of the hCul1 CTD, even though the domains share no significant sequence homology (Zheng et al., 2002). Thus, although the APC/C is composed of more protein components than the SCF, these two E3 ligases are likely to share some structural similarity and, when the structure of the quaternary SCF complex is compared with the APC/C structure (Fig. 7), three notable similarities become apparent. First, the distance between the bottom and the top lip of the APC/C cavity is approximately the same distance (~115 Å) as between hRbx1 and the F-box in the SCF complex. Second, the curvature of the APC/C cavity is very similar to the arc created by hCul1 in SCF. Third, just as the C-terminal domain of hCul1 interacts with hRbx1 (Zheng et al., 2002), the C-termini of S. pombe Apc2 and Apc11 are both found near the lower corner of the complex (Fig. 5C, G and M). These structural similarities make it tempting to speculate that the cullin repeats of Apc2 form part of the back wall of the central cavity. Therefore, our data support the idea that the SCF and the APC/C likely adopt similar structural backbones that facilitate the transfer of ubiquitin from an E2 to a substrate.
Figure 7. Structural comparison of the APC/C and SCF complex.
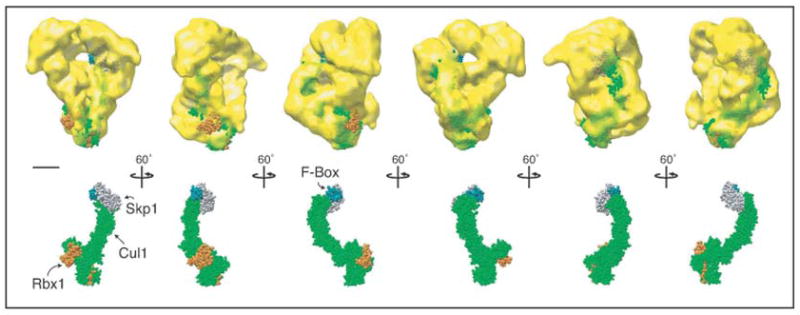
The SCF structure is composed of four proteins: hCul1 (green), hRbx1 (the RING-finger protein, orange), hSkp1 (grey), and the conserved F-box of S. cerevisiae Skp2 (dark green) (Zheng et al., 2002). The SCF complex has been placed into the APC/C density map with its RING-finger domain positioned near the bottom corner of the APC/C structure, the approximate location of S. pombe Apc11-Myc2, (see Fig. 5C and M). Scale bar represents 5 nm.
Experimental Procedures
Strains and media
S. pombe strains used in this study (Supplemental Table 1) were grown in yeast extract medium or EMM minimal medium with appropriate supplements (Moreno et al., 1991). Transformations were performed by the lithium acetate method (Gietz et al., 1995; Keeney and Boeke, 1994). Strains expressing epitope-tagged versions of proteins were constructed as described previously (Bahler et al., 1998; Longtine et al., 1998; Tasto et al., 2001). Each ORF was tagged endogenously at its 3′ end with either a myc2-Kanr, myc13-Kanr, HA3- Kanr, GFP-Kanr, FLAG3-Kanr, or TAP-Kanr cassette. Appropriate tagging was confirmed by PCR and immunoblotting. All tagged strains were viable at temperatures ranging from 25 °C to 36 °C. S. pombe genes were deleted and replaced by the ura4+ marker by a one-step gene disruption method described previously (Bahler et al., 1998).
Immunoblotting and sucrose gradients
200 μl corresponding to ~200 μg of protein lysate was layered onto a 10–30% sucrose gradient and centrifuged at 30,000 rpm for 21 h in a Beckman SW50.1 rotor. Sedimentation markers were fractionated on gradients prepared and spun in parallel. Fractions were collected from the bottom of the gradient and resolved on SDS 6–20% gradient gels. For immunoblotting, samples were transferred by electroblotting to PVDF and proteins were detected using anti-HA, anti-Myc, anti-GFP (Roche, Indianapolis, IN), anti-FLAG (Sigma, St. Louis, MO) or anti-Ub (Sigma, St. Louis, MO) antibodies. Immunoblots were developed and quantified with an Odyssey instrument (Licor Corp., Lincoln, NE) using the protocol and reagents supplied by the manufacturer. Quantitation was performed using Odyssey version 1.2.
In vitro ubiquitination assay
In vitro ubiquitination assays were carried out as previously described (Leverson et al., 2000; Yoon et al., 2002) using human E1 (Biochem), bacterially produced His6-Ubc5, and bacterially produced MBP-Cut2 as a substrate (Yoon et al., 2002).
APC/C purifications
APC/C-TAP purifications were performed as described previously (Yoon et al., 2002).
Antibody labeling of the APC/C
Labeling was performed by incubating APC/C particles containing either Apc11-Myc2, Apc10-Myc2, Apc2-Myc2, Apc13-Myc2, Hcn1-Myc2, Apc15-Myc2, Cut9-Myc2, Nuc2-Myc2, Cut23-Myc2, Apc5-Myc2, Cut4-Myc2, Lid1-Myc2, Apc14-Myc2, or Slp1-HA3 with anti-myc (9E10) or anti-HA (12CA5) antibodies overnight at 4°C in 10 mM Tris, pH 8.0, 150 mM NaCl, 0.02% NP-40, 1 mM Mg2+ acetate, 1 mM imidazole, 2 mM EGTA.
Scanning transmission EM (STEM)
Specimen preparation and electron microscopy
Image processing of particles in negative stain and vitrified ice
De novo structure calculation and variance mapping
Supplementary Material
Acknowledgments
We are grateful to Martha Simon for her assistance with STEM imaging, Joshua Rosenberg for constructing the Myc2 epitope tagging cassette, Dawn Clifford Hart, Laura Lee, Janel McLean, Ryoma Ohi, Claudia Petit, Rachel Roberts, Brenda Schulman, and Jianqui Wang for comments on the manuscript, and Lidiya Pudnor, and Jianqui Wang for assistance in yeast strain construction. This work was supported by the Giovanni Armenise-Harvard Foundation (T.W.), NIH Grants P01 GM064692 and GM 60635 (P.A.P.) and the Howard Hughes Medical Institute (K.L.G). M.D.O. was an Agouron Institute Fellow of the Jane Coffin-Childs Foundation and a fellow of the Charles A. King Medical Foundation. The BNL STEM is a NIH Supported Resource Center, NIH 5 P41 EB2181, with additional support provided by DOE, OBER. The molecular EM facility at Harvard Medical School is maintained by funds from NIH Grant GM62580 (T.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Berry LD, Feoktistova A, Wright MD, Gould KL. The schizosaccharomyces pombe dim1(+) gene interacts with the anaphase-promoting complex or cyclosome (APC/C) component lid1(+) and is required for APC/C function. Mol Cell Biol. 1999;19:2535–2546. doi: 10.1128/mcb.19.4.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco MA, Sanchez-Diaz A, de Prada JM, Moreno S. APC(ste9/srw1) promotes degradation of mitotic cyclins in G(1) and is inhibited by cdc2 phosphorylation. Embo J. 2000;19:3945–3955. doi: 10.1093/emboj/19.15.3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottcher B, Wynne SA, Crowther RA. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- Burton JL, Solomon MJ. D box and KEN box motifs in budding yeast Hsl1p are required for APC-mediated degradation and direct binding to Cdc20p and Cdh1p. Genes Dev. 2001;15:2381–2395. doi: 10.1101/gad.917901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton JL, Tsakraklides V, Solomon MJ. Assembly of an APC-Cdh1-substrate complex is stimulated by engagement of a destruction box. Mol Cell. 2005;18:533–542. doi: 10.1016/j.molcel.2005.04.022. [DOI] [PubMed] [Google Scholar]
- Carroll CW, Enquist-Newman M, Morgan DO. The APC subunit Doc1 promotes recognition of the substrate destruction box. Curr Biol. 2005;15:11–18. doi: 10.1016/j.cub.2004.12.066. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Wolf E, Larvie M, Zak O, Aisen P, Grigorieff N, Harrison SC, Walz T. Single particle reconstructions of the transferrin-transferrin receptor complex obtained with different specimen preparation techniques. J Mol Biol. 2006;355:1048–1065. doi: 10.1016/j.jmb.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- Dube P, Herzog F, Gieffers C, Sander B, Riedel D, Muller SA, Engel A, Peters JM, Stark H. Localization of the coactivator Cdh1 and the cullin subunit Apc2 in a cryo-electron microscopy model of vertebrate APC/C. Mol Cell. 2005;20:867–879. doi: 10.1016/j.molcel.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Gieffers C, Dube P, Harris JR, Stark H, Peters JM. Three-dimensional structure of the anaphase-promoting complex. Mol Cell. 2001;7:907–913. doi: 10.1016/s1097-2765(01)00234-9. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Gmachl M, Gieffers C, Podtelejnikov AV, Mann M, Peters JM. The RING-H2 finger protein APC11 and the E2 enzyme UBC4 are sufficient to ubiquitinate substrates of the anaphase-promoting complex. Proc Natl Acad Sci U S A. 2000;97:8973–8978. doi: 10.1073/pnas.97.16.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon C, McGurk G, Wallace M, Hastie ND. A conditional lethal mutant in the fission yeast 26 S protease subunit mts3+ is defective in metaphase to anaphase transition. J Biol Chem. 1996;271:5704–5711. doi: 10.1074/jbc.271.10.5704. [DOI] [PubMed] [Google Scholar]
- Grigorieff N. FREALIGN: high-resolution refinement of single particle structures. J Struct Biol. 2007;157:117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Hall MC, Torres MP, Schroeder GK, Borchers CH. Mnd2 and Swm1 are core subunits of the Saccharomyces cerevisiae anaphase-promoting complex. J Biol Chem. 2003;278:16698–16705. doi: 10.1074/jbc.M213109200. [DOI] [PubMed] [Google Scholar]
- Harper JW, Burton JL, Solomon MJ. The anaphase-promoting complex: it's not just for mitosis any more. Genes Dev. 2002;16:2179–2206. doi: 10.1101/gad.1013102. [DOI] [PubMed] [Google Scholar]
- Hilioti Z, Chung YS, Mochizuki Y, Hardy CF, Cohen-Fix O. The anaphase inhibitor Pds1 binds to the APC/C-associated protein Cdc20 in a destruction box-dependent manner. Curr Biol. 2001;11:1347–1352. doi: 10.1016/s0960-9822(01)00399-2. [DOI] [PubMed] [Google Scholar]
- Kadura S, Sazer S. SAC-ing mitotic errors: how the spindle assembly checkpoint (SAC) plays defense against chromosome mis-segregation. Cell Motil Cytoskeleton. 2005;61:145–160. doi: 10.1002/cm.20072. [DOI] [PubMed] [Google Scholar]
- Keeney JB, Boeke JD. Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics. 1994;136:849–856. doi: 10.1093/genetics/136.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura K, Maekawa H, Shimoda C. Fission yeast Ste9, a homolog of Hct1/Cdh1 and Fizzy-related, is a novel negative regulator of cell cycle progression during G1-phase. Mol Biol Cell. 1998;9:1065–1080. doi: 10.1091/mbc.9.5.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C, Herzog F, Gieffers C, Mechtler K, Hagting A, Pines J, Peters JM. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. Embo J. 2003;22:6598–6609. doi: 10.1093/emboj/cdg627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C, Vodermaier HC, Maurer-Stroh S, Eisenhaber F, Peters JM. The WD40 propeller domain of Cdh1 functions as a destruction box receptor for APC/C substrates. Mol Cell. 2005;18:543–553. doi: 10.1016/j.molcel.2005.04.023. [DOI] [PubMed] [Google Scholar]
- Leverson JD, Joazeiro CA, Page AM, Huang H, Hieter P, Hunter T. The APC11 RING-H2 finger mediates E2-dependent ubiquitination. Mol Biol Cell. 2000;11:2315–2325. doi: 10.1091/mbc.11.7.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- Matsumoto T. A fission yeast homolog of CDC20/p55CDC/Fizzy is required for recovery from DNA damage and genetically interacts with p34cdc2. Mol Cell Biol. 1997;17:742–750. doi: 10.1128/mcb.17.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyn MA, 3rd, Melloy PG, Li J, Holloway SL. The destruction box of the cyclin Clb2 binds the anaphase-promoting complex/cyclosome subunit Cdc23. Arch Biochem Biophys. 2002;407:189–195. doi: 10.1016/s0003-9861(02)00467-8. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Musacchio A, Hardwick KG. The spindle checkpoint: structural insights into dynamic signalling. Nat Rev Mol Cell Biol. 2002;3:731–741. doi: 10.1038/nrm929. [DOI] [PubMed] [Google Scholar]
- Nourry C, Maksumova L, Pang M, Liu X, Wang T. Direct interaction between Smad3, APC10, CDH1 and HEF1 in proteasomal degradation of HEF1. BMC Cell Biol. 2004;5:20. doi: 10.1186/1471-2121-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore LA, Barford D. Coactivator functions in a stoichiometric complex with anaphase-promoting complex/cyclosome to mediate substrate recognition. EMBO Rep. 2005;6:873–878. doi: 10.1038/sj.embor.7400482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore LA, Booth CR, Venien-Bryan C, Ludtke SJ, Fioretto C, Johnson LN, Chiu W, Barford D. Structural analysis of the anaphase-promoting complex reveals multiple active sites and insights into polyubiquitylation. Mol Cell. 2005;20:855–866. doi: 10.1016/j.molcel.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Passmore LA, McCormack EA, Au SW, Paul A, Willison KR, Harper JW, Barford D. Doc1 mediates the activity of the anaphase-promoting complex by contributing to substrate recognition. Embo J. 2003;22:786–796. doi: 10.1093/emboj/cdg084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penczek PA, Chao Y, Frank J, Spahn CMT. Estimation of variance in single particle reconstruction using the bootstrap technique. J Struct Biol. 2006;154:168–183. doi: 10.1016/j.jsb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Penczek PA, Zhu J, Frank J. A common-lines based method for determining orientations for N > 3 particle projections simultaneously. Ultramicroscopy. 1996;63:205–218. doi: 10.1016/0304-3991(96)00037-x. [DOI] [PubMed] [Google Scholar]
- Peters JM. The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol Cell. 2002;9:931–943. doi: 10.1016/s1097-2765(02)00540-3. [DOI] [PubMed] [Google Scholar]
- Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Pfleger CM, Kirschner MW. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- Pfleger CM, Lee E, Kirschner MW. Substrate recognition by the Cdc20 and Cdh1 components of the anaphase-promoting complex. Genes Dev. 2001;15:2396–2407. doi: 10.1101/gad.918201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Radermacher M, Wagenknecht T, Verschoor A, Frank J. Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J Microsc. 1987;146(Pt 2):113–136. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- Rudner AD, Murray AW. Phosphorylation by Cdc28 activates the Cdc20-dependent activity of the anaphase-promoting complex. J Cell Biol. 2000;149:1377–1390. doi: 10.1083/jcb.149.7.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwickart M, Havlis J, Habermann B, Bogdanova A, Camasses A, Oelschlaegel T, Shevchenko A, Zachariae W. Swm1/Apc13 is an evolutionarily conserved subunit of the anaphase-promoting complex stabilizing the association of Cdc16 and Cdc27. Mol Cell Biol. 2004;24:3562–3576. doi: 10.1128/MCB.24.8.3562-3576.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M, Gordon C, Ferrell K, Dubiel W. Characteristics of 26 S proteases from fission yeast mutants, which arrest in mitosis. J Mol Biol. 1996;263:423–431. doi: 10.1006/jmbi.1996.0586. [DOI] [PubMed] [Google Scholar]
- Seino H, Kishi T, Nishitani H, Yamao F. Two ubiquitin-conjugating enzymes, UbcP1/Ubc4 and UbcP4/Ubc11, have distinct functions for ubiquitination of mitotic cyclin. Mol Cell Biol. 2003;23:3497–3505. doi: 10.1128/MCB.23.10.3497-3505.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Li B, Bharadwaj R, Zhu H, Ozkan E, Hakala K, Deisenhofer J, Yu H. APC2 Cullin protein and APC11 RING protein comprise the minimal ubiquitin ligase module of the anaphase-promoting complex. Mol Biol Cell. 2001;12:3839–3851. doi: 10.1091/mbc.12.12.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasto JJ, Carnahan RH, McDonald WH, Gould KL. Vectors and gene targeting modules for tandem affinity purification in Schizosaccharomyces pombe. Yeast. 2001;18:657–662. doi: 10.1002/yea.713. [DOI] [PubMed] [Google Scholar]
- Taylor SS, Scott MI, Holland AJ. The spindle checkpoint: a quality control mechanism which ensures accurate chromosome segregation. Chromosome Res. 2004;12:599–616. doi: 10.1023/B:CHRO.0000036610.78380.51. [DOI] [PubMed] [Google Scholar]
- Thornton BR, Ng TM, Matyskiela ME, Carroll CW, Morgan DO, Toczyski DP. An architectural map of the anaphase-promoting complex. Genes Dev. 2006;20:449–460. doi: 10.1101/gad.1396906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton BR, Toczyski DP. Precise destruction: an emerging picture of the APC. Genes Dev. 2006;20:3069–3078. doi: 10.1101/gad.1478306. [DOI] [PubMed] [Google Scholar]
- Vodermaier HC, Gieffers C, Maurer-Stroh S, Eisenhaber F, Peters JM. TPR subunits of the anaphase-promoting complex mediate binding to the activator protein CDH1. Curr Biol. 2003;13:1459–1468. doi: 10.1016/s0960-9822(03)00581-5. [DOI] [PubMed] [Google Scholar]
- Wendt KS, Vodermaier HC, Jacob U, Gieffers C, Gmachl M, Peters JM, Huber R, Sondermann P. Crystal structure of the APC10/DOC1 subunit of the human anaphase-promoting complex. Nat Struct Biol. 2001;8:784–788. doi: 10.1038/nsb0901-784. [DOI] [PubMed] [Google Scholar]
- Yamada HY, Matsumoto S, Matsumoto T. High dosage expression of a zinc finger protein, Grt1, suppresses a mutant of fission yeast slp1(+), a homolog of CDC20/p55CDC/Fizzy. J Cell Sci. 2000;113(Pt 22):3989–3999. doi: 10.1242/jcs.113.22.3989. [DOI] [PubMed] [Google Scholar]
- Yamada H, Kumada K, Yanagida M. Distinct subunit functions and cell cycle regulated phosphorylation of 20S APC/cyclosome required for anaphase in fission yeast. J Cell Sci. 1997;110(Pt 15):1793–1804. doi: 10.1242/jcs.110.15.1793. [DOI] [PubMed] [Google Scholar]
- Yamano H, Gannon J, Mahbubani H, Hunt T. Cell cycle-regulated recognition of the destruction box of cyclin B by the APC/C in Xenopus egg extracts. Mol Cell. 2004;13:137–147. doi: 10.1016/s1097-2765(03)00480-5. [DOI] [PubMed] [Google Scholar]
- Yoon HJ, Feoktistova A, Chen JS, Jennings JL, Link AJ, Gould KL. Role of Hcn1 and its phosphorylation in fission yeast anaphase-promoting complex/cyclosome function. J Biol Chem. 2006;281:32284–32293. doi: 10.1074/jbc.M603867200. [DOI] [PubMed] [Google Scholar]
- Yoon HJ, Feoktistova A, Wolfe BA, Jennings JL, Link AJ, Gould KL. Proteomics analysis identifies new components of the fission and budding yeast anaphase-promoting complexes. Curr Biol. 2002;12:2048–2054. doi: 10.1016/s0960-9822(02)01331-3. [DOI] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.