Human ADP-ribosylhydrolase 3 (ARH3), which has been identified as an ARH by a sequence-similarity search and which cleaves the glycosidic bond of ADP-ribose attached to a protein, has been cloned, expressed, purified and crystallized in two different space groups.
Keywords: ADP-ribosylhydrolase, ADP-ribose, post-translational modification
Abstract
ADP-ribosylhydrolases catalyze the release of ADP-ribose from ADP-ribosylated proteins via hydrolysis of the glycosidic bond between ADP-ribose and a specific amino-acid residue in a target protein. Human ADP-ribosylhydrolase 3, consisting of 347 amino-acid residues, has been cloned and heterologously expressed in Escherichia coli, purified and crystallized in two different space groups. Preliminary X-ray diffraction studies yielded excellent diffraction data to a resolution of 1.6 Å.
1. Introduction
ADP-ribosylation is one of the enzyme-catalyzed post-translational modifications that regulate protein function (Aktories & Just, 2000 ▶; Haag & Koch-Nolte, 1997 ▶). Opposing reactions are catalyzed by ADP-ribosyltransferases (ARTs) and protein-ADP-ribosylhydrolases (ARHs). ADP-ribosylation results in either inhibition or activation of the function of the target protein. For example, in phototrophic bacteria the key enzyme of nitrogen fixation, dinitrogenase reductase (DR), is completely inhibited by ADP-ribosylation, whereas de-ADP-ribosylation restores its enzyme activity (Ludden, 1994 ▶). ADP-ribosylation has also been described as the pathogenic principle of many bacterial toxins with significant impact on human health (Aktories & Just, 2000 ▶). For example, ADP-ribosylation of G-proteins by cholera and pertussis toxins interferes with signal transduction, ADP-ribosylation of actin and rho by salmonella and clostridial toxins disturbs the cell cytoskeleton and ADP-ribosylation of elongation factor 2 by diphtheria and pseudomonas toxins shuts off protein synthesis.
Endogenous ADP-ribosylating and de-ADP-ribosylating enzymes have also been discovered in mammalian cells (Haag & Koch-Nolte, 1997 ▶). For example, T-cell surface ADP-ribosyltransferase ART2 ADP-ribosylates the P2X7 purinoceptor and induces the formation of calcium-permeable membrane pores (Seman et al., 2003 ▶). In liver mitochondria, a reversible ADP-ribosylation cycle controls the enzymatic activity of glutamate dehydrogenase, a key enzyme in amino-acid metabolism (Herrero-Yraola et al., 2001 ▶).
While the three-dimensional structures of several bacterial ADP-ribosylating toxins (Bell & Eisenberg, 1996 ▶; Choe et al., 1992 ▶; Han et al., 1999 ▶, 2001 ▶; Li et al., 1996 ▶; Sixma et al., 1993 ▶; Stein et al., 1994 ▶; Weiss et al., 1995 ▶; Zang et al., 1995 ▶) as well as that of a mammalian ART (Mueller-Dieckmann et al., 2002 ▶) have been determined, no structure is yet available for a de-ADP-ribosylating enzyme. Two de-ADP-ribosylating enzymes have been purified and cloned by traditional biochemical techniques: dinitrogenase reductase-activating glycohydrolase (DRAG) from Rhodospirillum rubrum (Fitzmaurice et al., 1989 ▶) and an ADP-ribosyl-arginine-hydrolase (ARH1) from rat brain (Moss et al., 1992 ▶). In silico analyses revealed that these proteins show significant amino-acid similarity over the entire length of the protein and thus are possibly derived from a common ancestor. Database searches of the recently completed human genome revealed the presence of three DRAG/ARH1-related genes, designated ARH1–ARH3 (Glowacki et al., 2002 ▶; gene IDs 141, 113622 and 54936). Here, we report the cloning, recombinant production, purification and crystallization of human ADP-ribosylhydrolase 3 (hARH3) consisting of 347 amino-acid residues.
2. Experimental methods
2.1. Cloning
A cDNA clone encompassing the coding region for hARH3 was obtained from the IMAGE consortium (IMAGE Consortium Clone ID 4803538; Lennon et al., 1996 ▶). The ARH coding region excluding a putative N-terminal signal sequence (residues 2–17) was PCR amplified with appropriate primers (5′-CGATGACCATATGAGATCTCTCTCGCGCTTCCGA-3′ and 5′-CAATTCTCGAGGCTCTTCTGGAAGACACGGTGC-3′) and cloned into the NdeI and XhoI sites of the prokaryotic expression plasmid pET26b (Novagen). This cloning strategy resulted in the removal of the pelB-leader sequence from pET26b, the deletion of residues 2–17 of hARH3 and a C-terminal extension of two amino acids encoded by the XhoI recognition sequence (Leu-Glu) followed by a hexahistidine tag. The correct cloning of the gene was verified by DNA sequencing.
2.2. Expression and purification
The plasmid pET26b-hARH3 was transformed into Escherichia coli BL21-CodonPlus(DE3)-RIL cells (Stratagene). 5 ml of an overnight preculture grown at 310 K in LB broth medium containing kanamycin (50 µg ml−1) and chloramphenicol (30 µg ml−1) was used to inoculate a 1 l culture. After the cells reached an optical density at 600 nm of 0.7, the temperature was decreased to 293 K and the culture was induced with isopropyl β-d-thiogalactopyranoside (IPTG) at a final concentration of 1 mM. 15 h later, cells were harvested and kept frozen at 253 K until further processing.
1 g of cells was dissolved in 4 ml lysis buffer [50 mM Tris–HCl pH 8.0, 150 mM NaCl, 10 mM MgCl2, 3 mM β-mercaptoethanol (β-ME) and 0.1%(v/v) Triton X-100]. Lysis was performed by sonication for 2 × 4 min in 15 s pulses at 277 K. Cell debris was removed by centrifugation at 20 000 rev min−1 for 30 min at 277 K. The crude lysate was then directly applied onto an Ni–NTA Superflow column (Qiagen) pre-equilibrated in lysis buffer. To remove contaminating protein, the column was washed with at least five times the column volume of wash buffer (50 mM Tris–HCl pH 8.0, 300 mM NaCl, 10 mM MgCl2 and 3 mM β-ME). Elution of the protein was performed with 1.5 column volumes of elution buffer (50 mM Tris–HCl pH 8.0, 300 mM NaCl, 10 mM MgCl2, 3 mM β-ME and 500 mM imidazole). For further purification, the eluate was concentrated to about 50 mg ml−1 and subsequently applied onto a Superdex S75 16/60 gel-filtration column (Pharmacia-Amersham) pre-equilibrated with filtration buffer [50 mM Tris–HCl pH 8.0, 150 mM NaCl, 10 mM MgCl2 and 3 mM dithiothreitol (DTT)]. The protein eluted with an approximate molecular weight of 40 kDa in accordance with the presence of a monomeric protein in solution. Pure protein fractions were concentrated to 10 mg ml−1 and extensively dialyzed against crystallization buffer (20 mM Tris–HCl pH 8.0, 100 mM NaCl and 3 mM DTT).
2.3. Crystallization
Crystals of the orthorhombic crystal form were obtained by the hanging-drop method on mixing equal volumes of protein solution (10 mg ml−1 protein in 20 mM Tris–HCl pH 8.0, 100 mM NaCl and 3 mM DTT) and reservoir solution [100 mM 2-morpholinoethanesulfonic acid (MES) pH 6.0, 10%(w/v) PEG 3350] at 293 K. Crystals appeared as plates from a heavy precipitate after about two weeks and typically grew to dimensions of 300 × 200 × 50 µm. They diffracted X-rays to 1.6 Å resolution.
Crystals of the monoclinic crystal form were obtained by mixing protein solution (10 mg ml−1 protein in 20 mM Tris–HCl pH 8.0, 100 mM NaCl, 3 mM DTT and 2 mM ADP) with twice the volume of reservoir solution [100 mM 4-(2-hydroxyethyl)-1-piperazineethane sulfonic acid (HEPES) pH 7.0, 200 mM calcium acetate and 18%(w/v) PEG 8000]. Needle-shaped crystals (∼75 × 75 × 300 µm) appeared within a few days at 293 K and diffracted X-rays to about 2.0 Å resolution.
2.4. Diffraction data collection and processing
Diffraction data collection was performed at 100 K in an N2 cryostream. For data collection, crystals of the orthorhombic crystal form were cryoprotected for about 30 s in a solution consisting of reservoir solution with 20%(v/v) glycerol. This was performed stepwise starting with a reservoir solution containing 5%(v/v) glycerol; the glycerol concentration was then increased in 5% steps until it reached a final concentration of 20%. After successful flash-cooling, diffraction data were collected at the BW7A beamline at EMBL Outstation Hamburg, Germany at a wavelength of 1.127 Å using a 165 mm MAR CCD detector.
Crystals of the monoclinic crystal form were cryoprotected in dry paraffin oil. After flash-cooling to 100 K, diffraction data were collected at a wavelength of 1.50 Å at the X12 beamline (EMBL Outstation Hamburg, Germany) using a MAR mosaic CCD detector.
All data were indexed and integrated using DENZO (Otwinowski & Minor, 1997 ▶) and scaled using SCALEPACK (Otwinowski & Minor, 1997 ▶). The redundancy-independent merging R factor (R r.i.m.) and the precision-indicating merging R factor (R p.i.m.) (Weiss, 2001 ▶) were calculated using the program RMERGE (available from MSW upon request or from http://www.embl-hamburg.de/~msweiss/projects/msw_qual.html). Intensities were converted to structure-factor amplitudes using the program TRUNCATE (French & Wilson, 1978 ▶; Collaborative Computational Project, Number 4, 1994 ▶). The optical resolution of the data sets was calculated using the program SFCHECK (Vaguine et al., 1999 ▶). Calculation of the self-rotation function was carried out using the program MOLREP (Collaborative Computational Project, Number 4, 1994 ▶) and structure-factor amplitudes to a maximum resolution of 4.0 Å.
3. Results and discussion
Human ARH3 has been cloned and overexpressed with a C-terminal His6 tag in E. coli. 1 l of bacterial culture yielded about 10 mg pure protein. About 50% of the overexpressed hARH3 appeared to be in the soluble fraction. After affinity chromatography and subsequent gel filtration, where the protein eluted as a monomer, the protein was about 99% pure as assessed by gel electrophoresis. Single crystals of hARH3 (Fig. 1 ▶) were obtained using the conditions described above in two different space groups. In the absence of ADP crystals were obtained in the orthorhombic space group P212121, whereas in the presence of ADP crystals grew in the monoclinic space group P21. For data collection, one crystal of each crystal form was used. The orthorhombic crystal form diffracted X-rays to better than 1.6 Å resolution and a high-quality highly redundant data set was collected using a wavelength of 1.13 Å (Table 1 ▶). In contrast, the monoclinic crystal form only diffracted X-rays to about 2.05 Å resolution. All relevant data-collection and processing parameters are given in Table 1 ▶.
Figure 1.
Images of crystals of hARH3. (a) shows the orthorhombic crystal form in a heavy precipitate and (b) the monoclinic crystal form.
Table 1. Data-collection statistics.
| Crystal form | Orthorhombic | Monoclinic |
|---|---|---|
| Data-collection wavelength (Å) | 1.1271 | 1.50 |
| Space group | P212121 | P21 |
| Unit-cell parameters | ||
| a (Å) | 56.34 | 57.33 |
| b (Å) | 60.35 | 60.63 |
| c (Å) | 97.17 | 102.88 |
| β (°) | 96.43 | |
| Resolution limits (Å) | 99–1.60 (1.63–1.60) | 99–2.05 (2.09–2.05) |
| Mosaicity (°) | 0.38 | 0.97 |
| Total reflections | 1394837 | 301539 |
| Unique reflections | 44141 | 44248 |
| Redundancy | 31.6 | 6.8 |
| 〈I/σ(I)〉 | 73.5 (15.7) | 17.2 (3.3) |
| Completeness (%) | 99.4 (99.9) | 99.9 (100) |
| Rmerge† (%) | 4.7 (36.5) | 9.0 (64.4) |
| Rr.i.m.‡ (%) | 4.8 (37.6) | 9.8 (70.3) |
| Rp.i.m.§ (%) | 0.8 (6.7) | 3.7 (27.9) |
| Optical resolution (Å) | 1.34 | 1.71 |
| Overall B factor from Wilson plot (Å2) | 19.9 | 39.0 |
R
merge(I) = 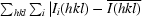
 .
.
R
r.i.m. = 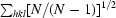
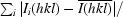
 .
.
R
p.i.m. = 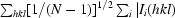 −
− 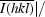
 .
.
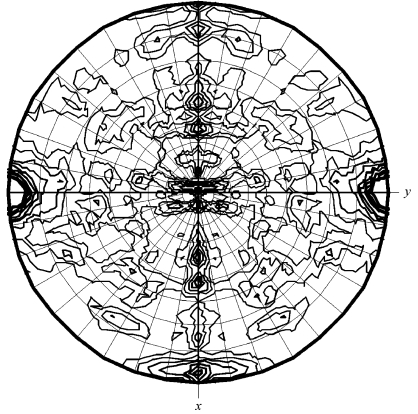
Based on the Matthews parameter (Matthews, 1968 ▶), the orthorhombic crystal form contains one monomer per asymmetric unit (V M = 2.06 Å3 Da−1 and 40.4% solvent content) and the monoclinic crystal form two monomers per asymmetric unit (V M = 2.22 Å3 Da−1 and 44.6% solvent content). Based on this and on the unit-cell parameters of the two crystal forms, it is clearly obvious that the two forms are related. This notion is corroborated by the calculation of a self-rotation function using the data collected from the monoclinic crystal form, which clearly shows the presence of two twofold axes approximately parallel to the x and z axes. Thus, the monoclinic crystal form can be described as pseudo-orthorhombic (Fig. 2 ▶).
Figure 2.
Self-rotation function, κ = 180° section. This figure was produced using the program MOLREP (Collaborative Computational Project, Number 4, 1994 ▶).
The most likely reason for the observation of the two crystal forms is the absence of ADP in crystallization condition 1 and the presence of ADP in condition 2, although the reservoir composition and the pH also differ slightly between the two conditions. Determination of whether ADP is present in the structure of hARH3 in the monoclinic crystal form will have to await the determination of the three-dimensional structure. The structure solution by isomorphous replacement methods is currently under way.
In summary, hARH3 from Homo sapiens is the first eukaryotic protein of the family of ADP-ribosylhydrolases crystallized to date. Both the apo structure and the potential binary ADP-containing complex will lead to a deeper understanding of its enzymatic mechanism. Together with the already known structure of the ADP-ribosyltransferase 2 from Rattus norvegicus (Mueller-Dieckmann et al., 2002 ▶), the structure of hARH3 will provide structural prototypes for both branches of ADP-ribosylation cycles in eukaryotes.
Acknowledgments
Parts of the work described in this study represent the partial fulfilment of the requirements of the doctoral thesis of SK at the University of Hamburg. SK is the recipient of a stipend from the German National Academic Foundation. This work was supported by grants from the Deutsche Forschungsgemeinschaft (to FK-N and MSW) and the Studienstiftung des Deutschen Volkes (to SK).
References
- Aktories, K. & Just, I. (2000). Bacterial Protein Toxins. Berlin: Springer–Verlag.
- Bell, C. E. & Eisenberg, D. (1996). Biochemistry, 35, 1137–1149. [DOI] [PubMed] [Google Scholar]
- Choe, S., Bennett, M. J., Fujii, G., Curmi, P. M., Kantardjieff, K. A., Collier, R. J. & Eisenberg, D. (1992). Nature (London), 357, 216–222. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Fitzmaurice, W. P., Saari, L. L., Lowery, R. G., Ludden, P. W. & Roberts, G. P. (1989). Mol. Gen. Genet.218, 340–347. [DOI] [PubMed] [Google Scholar]
- French, G. S. & Wilson, K. S. (1978). Acta Cryst. A34, 517–525. [Google Scholar]
- Glowacki, G., Braren, R., Firner, K., Nissen, M., Kuhl, M., Reche, P., Bazan, F., Cetkovic-Cvrlje, M., Leiter, E., Haag, F. & Koch-Nolte, F. (2002). Protein Sci.11, 1657–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag, F. & Koch-Nolte, F. (1997). Adv. Exp. Med. Biol.419, 459–462. [PubMed] [Google Scholar]
- Han, S., Arvai, A. S., Clancy, S. B. & Tainer, J. A. (2001). J. Mol. Biol.305, 95–107. [DOI] [PubMed] [Google Scholar]
- Han, S., Craig, J. A., Putnam, C. D., Carozzi, N. B. & Tainer, J. A. (1999). Nature Struct. Biol.6, 932–936. [DOI] [PubMed] [Google Scholar]
- Herrero-Yraola, A., Bakhit, S. M. A., Franke, P., Weize, C., Schweiger, M., Jorcke, D. & Ziegler, M. (2001). EMBO J.20, 2404–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon, G., Auffray, C., Polymeropoulos, M. & Soares, M. B. (1996). Genomics, 33, 151–152. [DOI] [PubMed] [Google Scholar]
- Li, M., Dyda, F., Benhar, I., Pastan, I. & Davies, D. R. (1996). Proc. Natl Acad. Sci. USA, 93, 6902–6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludden, P. W. (1994). Mol. Cell. Biochem.138, 123–129. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Moss, J., Stanley, S. J., Nightingale, M. S., Murtagh, J. J. Jr, Monaco, L., Mishima, K., Chen, H.-C., Williamson, K. C. & Tsai, S.-C. (1992). J. Mol. Biol.267, 10481–10488. [PubMed]
- Mueller-Dieckmann, C., Ritter, H., Koch-Nolte, F. & Schulz, G. E. (2002). J. Mol. Biol.322, 687–696. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Seman, M., Adriouch, S., Scheuplein, F., Krebs, C., Freese, D., Glowacki, G., Deterre, P., Haag, F. & Koch-Nolte, F. (2003). Immunity, 19, 571–582. [DOI] [PubMed] [Google Scholar]
- Sixma, T. K., Kalk, K. H., van Zanten, B. A. M., Dauter, Z., Kingma, J., Witholt, B. & Hol, W. G. J. (1993). J. Mol. Biol.230, 890–913. [DOI] [PubMed] [Google Scholar]
- Stein, P. E., Boodhoo, A., Armstrong, G. D., Cockle, S. A., Klein, M. H. & Read, R. J. (1994). Structure, 2, 45–57. [DOI] [PubMed] [Google Scholar]
- Vaguine, A. A., Richelle, J. & Wodak, S. J. (1999). Acta Cryst. D55, 191–205. [DOI] [PubMed] [Google Scholar]
- Weiss, M. S., Blanke, S. R., Collier, R. J. & Eisenberg, D. (1995). Biochemistry, 34, 773–781. [DOI] [PubMed] [Google Scholar]
- Weiss, M. S. (2001). J. Appl. Cryst.34, 130–135. [Google Scholar]
- Zhang, R.-G., Scott, D. L., Westbrook, M. L., Nance, S., Spangler, B. D., Shipley, G. G. & Westbrook, E. M. (1995). J. Mol. Biol.251, 563–573. [DOI] [PubMed] [Google Scholar]