A C-terminal fragment of the Epstein–Barr virus lytic switch protein ZEBRA has been crystallized in complex with DNA.
Keywords: Epstein–Barr virus, ZEBRA, BZLF1, Zta
Abstract
A C-terminal fragment of the Epstein–Barr virus immediate-early transcription factor ZEBRA has been expressed as a recombinant protein in Escherichia coli and purified to homogeneity. The fragment behaves as a dimer in solution, consistent with the presence of a basic region leucine-zipper (bZIP) domain. Crystals of the fragment in complex with a DNA duplex were grown by the hanging-drop vapour-diffusion technique using polyethylene glycol 4000 and magnesium acetate as crystallization agents. Crystals diffract to better than 2.5 Å resolution using synchrotron radiation (λ = 0.976 Å). Crystals belong to space group C2, with unit-cell parameters a = 94.2, b = 26.5, c = 98.1 Å, β = 103.9°.
1. Introduction
Epstein–Barr virus (EBV) is a human γ-herpesvirus which causes infectious mononucleosis and is associated with numerous malignanicies (Rickinson & Kieff, 2001 ▶; Young & Rickinson, 2004 ▶). EBV has a biphasic life cycle consisting of a latent and a lytic replicative phase. A key regulator in the switch from latency to the lytic state is the immediate-early transcription factor ZEBRA (BZLF1, Zta, Z or EB1; Chevallier-Greco et al., 1986 ▶; Countryman & Miller, 1985 ▶; Takada et al., 1986 ▶). ZEBRA activates the promoters of EBV lytic genes by binding to target sites termed ZEBRA response elements (ZREs; Farrell et al., 1989 ▶; Lieberman & Berk, 1990 ▶; Taylor et al., 1991 ▶; Urier et al., 1989 ▶). ZEBRA activates its own expression and that of the only other immediate-early protein, Rta (Flemington & Speck, 1990a ▶; Sinclair et al., 1991 ▶). Together, ZEBRA and Rta induce the entire complement of early and late lytic EBV genes (Feederle et al., 2000 ▶).
ZEBRA is a 245-residue protein with sequence similarity to basic region leucine-zipper (bZIP) proteins such as Fos and Jun (Farrell et al., 1989 ▶; Kouzarides et al., 1991 ▶). These proteins bind DNA through their ∼60-residue bZIP domain, comprising an N-terminal basic region that contacts the DNA and a C-terminal leucine zipper that dimerizes by forming a coiled coil. The bZIP domain of ZEBRA (approximately residues 175–220) is unusual because it lacks the canonical leucine-zipper motif (Chang et al., 1990 ▶; Flemington & Speck, 1990b ▶; Kouzarides et al., 1991 ▶). Moreover, a peptide spanning the putative zipper region fails to form a stable coiled coil at physiological temperature (Hicks et al., 2001 ▶) and the deletion of residues C-terminal to this region compromises DNA binding (Hicks et al., 2003 ▶), suggesting that ZEBRA dimerizes and binds DNA differently from prototypical bZIP proteins.
Structures have been determined for the bZIP domains of a number of transcription factors. These include GCN4 (Ellenberger et al., 1992 ▶; Konig & Richmond, 1993 ▶; Keller et al., 1995 ▶), c-Fos/c-Jun (Glover & Harrison, 1995 ▶), Pap-1 (Fujii et al., 2000 ▶), c/EBPβ (Podust et al., 2001 ▶; Tahirov et al., 2001 ▶) and ATF-2/c-Jun (Panne et al., 2004 ▶). Here, we report the cloning, overexpression and purification of a C-terminal fragment of ZEBRA that encompasses the bZIP domain and its crystallization in complex with a DNA duplex.
2. Materials and methods
2.1. Cloning of ZEBRA and C-terminal fragments
Polyadenylated mRNA was purified from an EBV-infected B lymphocyte cell line, B95-8, using the mRNA capture kit (Roche). The cDNA encoding the full-length ZEBRA protein (residues 1–245; GenBank accession No. CAD53423) was obtained by RT-PCR using the Titan One Tube RT-PCR System (Roche). Primers used were 5′-GGAATTCCATATGATGGACCCAAACTC-3′ and 5′-CGGGATCCTTAGAAATTTAAGAGATCCTCG-3′, which contain NdeI and BamHI restriction sites (bold), respectively. PCR products were ligated into the cloning vector pGEM-Teasy (Promega) and transformed into Escherichia coli strain XL1-blue. Plasmid DNA was amplified and digested with NdeI and BamHI. The resulting 735 bp fragment was ligated into the NdeI and BamHI sites of the expression vector pET28a (Novagen) and the ligation product was transformed into E. coli strain BL21(DE3). The ZEBRA175–245 construct was generated by deletion PCR using the ExSite kit (Stratagene) and the primers 5′-GGTATATCTCCTTCTTAAAGTTAAAC-3′ and 5′-ATGCTAGAAATAAAGCGATACAAGAATCGGG-3′. A construct encoding residues 175–236 was generated by the introduction of a stop codon at position 237 using the Stratagene QuikChange kit. This construct was used to generate the construct ZEBRA175–236/mut by introducing point mutations S186A and C189S using the Stratagene QuikChange kit.
2.2. Overexpression and purification of ZEBRA constructs
E. coli BL21(DE3) cells expressing full-length ZEBRA bearing an N-terminal hexahistidine tag were grown in LB medium containing 25 µg ml−1 kanamycin to an OD600 of 0.6 and were induced with 0.5 mM IPTG for 16 h at 296 K. Cells were lysed by sonication in buffer A (20 mM sodium phosphate pH 7, 300 mM NaCl and 1 mM PMSF) and the cleared lysate was applied onto a Talon cobalt-affinity resin (Clontech). The resin was extensively washed with buffer A containing 1 M NaCl to eliminate nucleic acids and the bound protein was eluted with buffer A plus 0.5 M imidazole. The protein was further purified by Superdex-75 (Pharmacia) chromatography in buffer A.
The C-terminal fragments ZEBRA175–245 and ZEBRA175–236/mut were expressed as untagged proteins in E. coli BL21 (DE3) cells using the same protocol as for the full-length protein. Cells were lysed in 20 mM Tris–HCl pH 6.8, 10 mM NaCl, 5 mM β-mercaptoethanol and 1 mM PMSF. Nucleic acids were removed from the cleared lysate by polyethyleneimine precipitation [0.3%(v/v)]. The protein was purified by SP Sepharose chromatography (Amersham) using a 0.01–1 M NaCl gradient. Fractions containing the ZEBRA fragment were pooled and subjected to ammonium sulfate precipitation [30%(w/v)]. The protein was resuspended in 20 mM Tris pH 7.5, 150 mM NaCl, 150 mM ammonium acetate, 5 mM DTT and 0.2 mM PMSF and purified by Superdex-75 (Pharmacia) chromatography.
2.3. Preparation of DNA duplex
DNA oligonucleotides were designed based on the promoter sequence of the EBV early lytic gene BSLF2/BMLF1, which is a natural target site of ZEBRA (Farrell et al., 1989 ▶). The oligonucleotides yielding the best crystals were 5′-AAGCACTGACTCATGAAGT-3′ and 5′-TACTTCATGAGTCAGTGCT-3′ (ZEBRA binds directly to the sequence in bold), which upon annealing generate a 19-mer duplex with a single-base overhang. Oligonucleotides chemically synthesized by BioSpring were dissolved in 10 mM NaOH and 100 mM NaCl and purified by MonoQ HR10/10 (Pharmacia) chromatography using a 0–1 M NaCl gradient (in 10 mM NaOH). Fractions were pooled and extensively dialysed against deionized water. Samples were further desalted by applying them onto a Econo-Pac 10DG (Biorad) column pre-equilibrated in deionized water. Samples were lyophilized to dryness and resuspended in H2O to a final concentration of 20 mg ml−1. Complementary oligonucleotides were mixed in a 1:1 molar ratio, heated to 368 K and annealed by slow cooling.
2.4. Crystallization
The protein–DNA complex was prepared by mixing the ZEBRA175–236/mut homodimer and DNA duplex in an equimolar ratio and incubating at room temperature for 15 min. Crystallization trials were carried out by the hanging-drop vapour-diffusion technique at 293 K by mixing equal volumes of reservoir solution and protein–DNA complex. Crystals grew from 200 mM ammonium acetate, 150 mM magnesium acetate, 50 mM HEPES pH 7 and 5% PEG 4K. Crystals grew as fine needles and appeared within 1–2 weeks.
3. Results
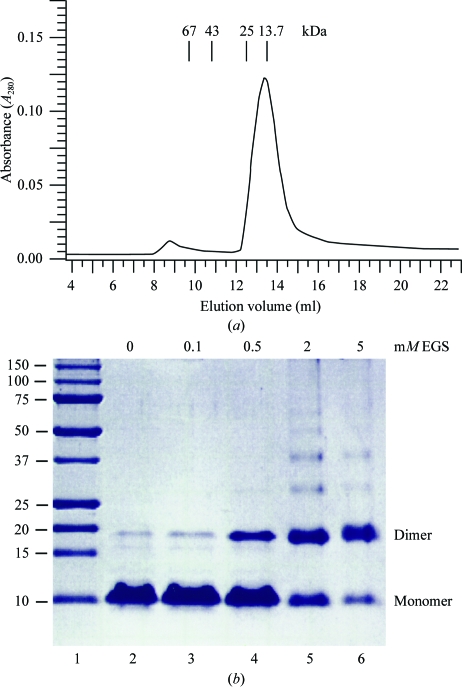
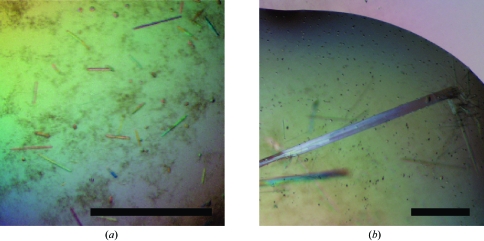
Initial attempts to purify recombinant full-length ZEBRA indicated that the protein was readily degraded by endogenous E. coli proteases. Limited proteolysis with trypsin yielded a partially stable 8 kDa fragment, which was determined by N-terminal sequencing and mass spectrometry to correspond to the C-terminal 71 residues of ZEBRA (data not shown). Accordingly, we made a construct spanning this region, ZEBRA175–245, which includes the bZIP domain. This fragment was expressed in E. coli BL21(DE3) cells, yielding ∼0.5 mg of highly purified protein per litre of bacterial culture (Fig. 1 ▶, lane 2). The fragment elutes from a Superdex-75 HR10/30 gel-filtration column as a single peak corresponding to an apparent molecular weight of 15 kDa (Fig. 2 ▶ a) or approximately twice the calculated molecular weight of monomeric ZEBRA175–245 (8380 Da). This indicates that ZEBRA175–245 is homodimeric in solution, consistent with the presence of the bZIP domain. The existence of a homodimer was confirmed by a cross-linking study using ethylene glycol bis(succinimidyl succinate) (EGS; Fig. 2 ▶ b). Purified ZEBRA175–245 has a tendency to aggregate in solution and we were unable to concentrate it to concentrations higher than ∼3 mg ml−1. Attempts to crystallize ZEBRA175–245 in complex with oligonucleotide duplexes of various lengths resulted in several crystal forms (e.g. Fig. 3 ▶ a); however, none of these were suitable for structure determination.
Figure 1.
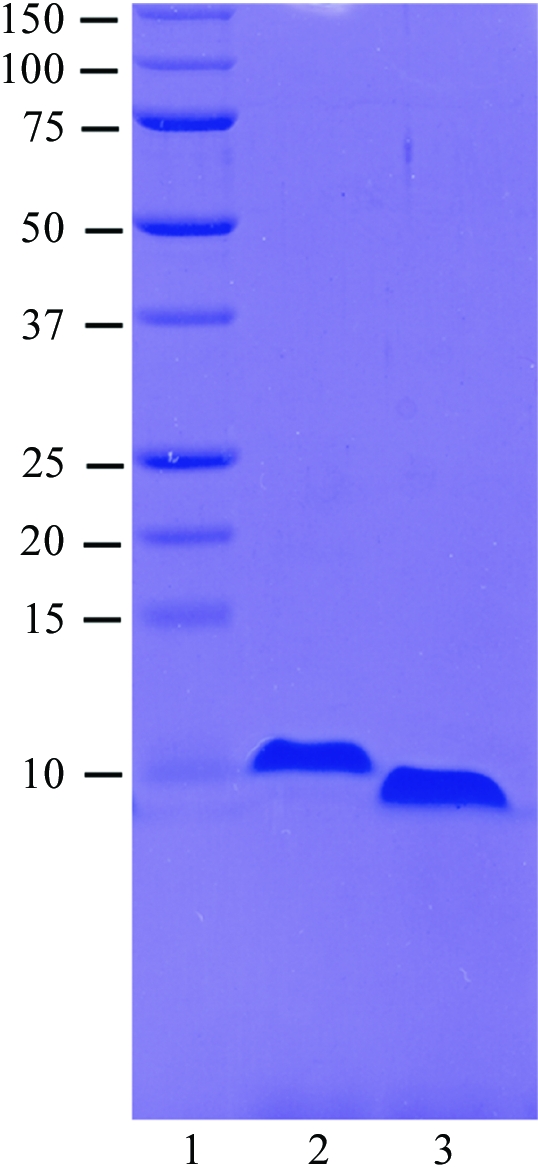
Purification of ZEBRA constructs. 12% SDS–PAGE showing molecular-weight markers (kDa) in lane 1, ZEBRA175–245 in lane 2 and ZEBRA175–236/mut in lane 3. Approximately 5 µg of protein are loaded in lanes 2 and 3.
Figure 2.
Evidence for dimer formation. (a) Analytical gel-filtration profile of ZEBRA175–245 on a Superdex-75 HR10/30 column. Elution volumes of molecular-weight standards are indicated. (b) Cross-linking study with EGS. ZEBRA175–245 (2.5 µg) was incubated with increasing concentrations of EGS in a final reaction volume of 10 µl at 293 K for 30 min. The reaction was quenched by the addition of 40 mM Tris pH 7.5 buffer and analyzed by 12% SDS–PAGE.
Figure 3.
Crystals of ZEBRA C-terminal constructs in complex with DNA. (a) ZEBRA175–245 (b) ZEBRA175–236/mut. In both cases cocrystallization was with the same 19-mer DNA duplex (see §2). The scale bar corresponds to 0.2 mm.
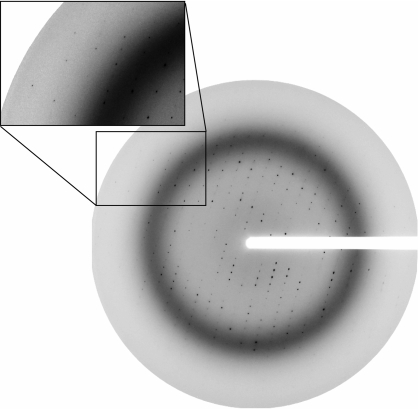
We therefore began crystallization trials with a modified construct, ZEBRA175–236/mut, obtained by deleting nine C-terminal residues (VLHEDLLNF) and introducing two point mutations, S186A and C189S. The deletion eliminates five hydrophobic residues and considerably increases protein solubility, allowing us to concentrate the purified protein to >10 mg ml−1. The deletion does not appreciably alter the DNA-binding affinity, as measured in an electrophoretic mobility shift assay (not shown). The C189S mutation stabilizes the protein against oxidation and corresponds to the mutation which yielded improved crystals of Fos-Jun-DNA (Glover & Harrison, 1995 ▶), while the S186A mutation renders the ZEBRA sequence more similar to that of Fos and Jun. The typical degree of chemical purity obtained is illustrated in Fig. 1 ▶ (lane 3). The homogeneity of the protein and of protein–DNA complexes was confirmed by mass spectrometry (data not shown). Crystallization trials with ZEBRA175–236/mut and DNA duplexes of various lengths yielded several promising crystal forms. One crystal form, obtained in complex with a 19-mer DNA duplex and characterized by a needle-like morphology (Fig. 3 ▶ b), reproducibly diffracted synchrotron radiation to better than 3 Å resolution (Fig. 4 ▶).
Figure 4.
Diffraction pattern of a ZEBRA175–236/mut–DNA crystal. The crystal was exposed at 100 K after soaking in 25% PEG 400 for cryoprotection. The rotation angle used was 1°, the crystal-to-detector distance was 150 mm and the wavelength was 0.9755 Å. The inset shows a detail of the diffraction pattern with spots visible near the edge of the detector (2.5 Å).
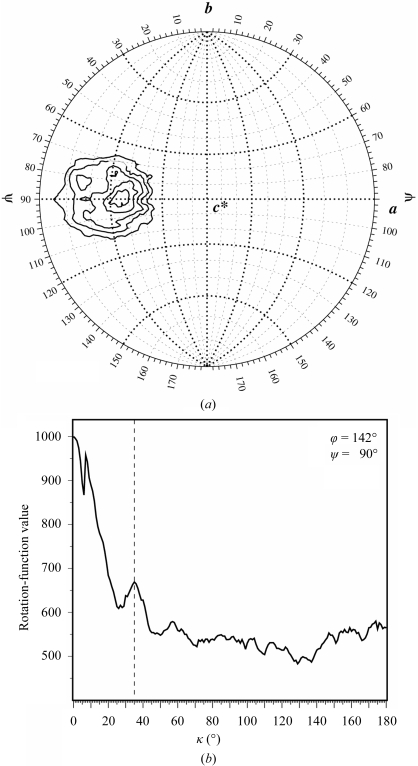
Using the microfocus beamline ID13 at the ESRF, a complete 2.5 Å data set could be collected from a single frozen crystal by successively exposing different crystal volumes to X-radiation. Data processing was carried out in space group C2 using XDS (Kabsch, 1993 ▶). Data-collection statistics are summarized in Table 1 ▶. Packing-parameter calculations suggest that the asymmetric unit contains one ZEBRA175–236/mut homodimer bound to one DNA duplex. This corresponds to a Matthews coefficient (Matthews, 1968 ▶) of 2.3 Å3 Da−1, with a solvent content of 45%. Self-rotation functions show the consistent presence of a strong peak (>50% of the origin peak) at polar angles ϕ = 142, ψ = 90° for all values of the polar angle κ between 0 and 180°, corresponding to an axis of symmetry parallel to the ac plane (Fig. 5 ▶ a). A scan along the κ angle displays a local maximum at κ = 35° (Fig. 5 ▶ b). This is close to the mean helical twist of 36° between successive base pairs for B-DNA, suggesting that the DNA axis may be oriented in this direction.
Table 1. Data-collection and processing statistics.
Values in parentheses are for the highest resolution shell (2.5–2.6 Å).
| X-ray source | ID13, ESRF |
| Wavelength (Å) | 0.9755 |
| Data-collection temperature (K) | 100 |
| Detector | MAR CCD 165 mm |
| Space group | C2 |
| Unit-cell parameters (Å, °) | a = 94.2, b = 26.5, c = 98.1, β = 103.9 |
| Resolution (Å) | 30–2.5 |
| Observations | 45256 (5251) |
| Unique reflections (F > 0) | 8542 (954) |
| Mosaicity (°) | 0.38 |
| Wilson B factor (Å2) | 43 |
| I/σ(I) | 12.9 (4.3) |
| Rmerge (%) | 8.0 (38.3) |
| Completeness (%) | 99.8 (99.9) |
| Redundancy | 5.3 (5.5) |
Figure 5.
Self-rotation function calculated using GLRF (Tong & Rossmann, 1997 ▶) with an integration radius of 15 Å and data in the resolution range 30–2.5 Å. (a) The κ = 36° section. Contour lines are drawn at intervals of 2σ starting from 16σ. The peak at (ϕ, ψ) = (142, 90°) is observed in all κ sections between 0 and 180° and is consistently present in self-rotation functions calculated using various high-resolution cutoffs (2.5–5 Å) and integration radii (10–25 Å). (b) Scan along κ showing the local maximum at 35°.
Acknowledgments
We thank M. Moulin, J. B. Artero and F. Baudin for important technical contributions, the EMBL/ESRF Joint Structural Biology Group for access and support at the ESRF beamlines and D. Flot for assistance at ESRF beamline ID13.
References
- Chang, Y. N., Dong, D. L., Hayward, G. S. & Hayward, S. D. (1990). J. Virol.64, 3358–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevallier-Greco, A., Manet, E., Chavrier, P., Mosnier, C., Daillie, J. & Sergeant, A. (1986). EMBO J.5, 3243–3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Countryman, J. & Miller, G. (1985). Proc. Natl Acad. Sci. USA, 82, 4085–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberger, T. E., Brandl, C. J., Struhl, K. & Harrison, S. C. (1992). Cell, 71, 1223–1237. [DOI] [PubMed] [Google Scholar]
- Farrell, P. J., Rowe, D. T., Rooney, C. M. & Kouzarides, T. (1989). EMBO J.8, 127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feederle, R., Kost, M., Baumann, M., Janz, A., Drouet, E., Hammerschmidt, W. & Delecluse, H. J. (2000). EMBO J.19, 3080–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemington, E. & Speck, S. H. (1990a). J. Virol.64, 1227–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemington, E. & Speck, S. H. (1990b). Proc. Natl Acad. Sci. USA, 87, 9459–9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, Y., Shimizu, T., Toda, T., Yanagida, M. & Hakoshima, T. (2000). Nature Struct. Biol.7, 889–893. [DOI] [PubMed] [Google Scholar]
- Glover, J. N. & Harrison, S. C. (1995). Nature (London), 373, 257–261. [DOI] [PubMed] [Google Scholar]
- Hicks, M. R., Al-Mehairi, S. S. & Sinclair, A. J. (2003). J. Virol.77, 8173–8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks, M. R., Balesaria, S., Medina-Palazon, C., Pandya, M. J., Woolfson, D. N. & Sinclair, A. J. (2001). J. Virol.75, 5381–5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch, W. (1993). J. Appl. Cryst.26, 795–800. [Google Scholar]
- Keller, W., Konig, P. & Richmond, T. J. (1995). J. Mol. Biol.254, 657–667. [DOI] [PubMed] [Google Scholar]
- Konig, P. & Richmond, T. J. (1993). J. Mol. Biol.233, 139–154. [DOI] [PubMed] [Google Scholar]
- Kouzarides, T., Packham, G., Cook, A. & Farrell, P. J. (1991). Oncogene, 6, 195–204. [PubMed] [Google Scholar]
- Lieberman, P. M. & Berk, A. J. (1990). J. Virol.64, 2560–2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Panne, D., Maniatis, T. & Harrison, S. C. (2004). EMBO J.23, 4384–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podust, L. M., Krezel, A. M. & Kim, Y. (2001). J. Biol. Chem.276, 505–513. [DOI] [PubMed] [Google Scholar]
- Rickinson, A. B. & Kieff, E. (2001). Fields Virology, edited by D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman & S. E. Straus, pp. 2575–2672. Philadelphia, USA: Lippincott, Williams & Wilkins.
- Sinclair, A. J., Brimmell, M., Shanahan, F. & Farrell, P. J. (1991). J. Virol.65, 2237–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahirov, T. H., Inoue-Bungo, T., Morii, H., Fujikawa, A., Sasaki, M., Kimura, K., Shiina, M., Sato, K., Kumasaka, T., Yamamoto, M., Ishii, S. & Ogata, K. (2001). Cell, 104, 755–767. [DOI] [PubMed] [Google Scholar]
- Takada, K., Shimizu, N., Sakuma, S. & Ono, Y. (1986). J. Virol.57, 1016–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, N., Flemington, E., Kolman, J. L., Baumann, R. P., Speck, S. H. & Miller, G. (1991). J. Virol.65, 4033–4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, L. & Rossmann, M. G. (1997). Methods Enzymol.276, 594–611. [PubMed] [Google Scholar]
- Urier, G., Buisson, M., Chambard, P. & Sergeant, A. (1989). EMBO J.8, 1447–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, L. S. & Rickinson, A. B. (2004). Nature Rev. Cancer, 4, 757–768. [DOI] [PubMed]