A novel cytochrome c nitrite reductase (TvNiR) has been isolated from the haloalkalophilic bacterium T. nitratireducens. TvNiR crystals were grown by the hanging-drop vapour-diffusion technique.
Keywords: cytochrome c nitrite reductase
Abstract
A novel cytochrome c nitrite reductase (TvNiR) was isolated from the haloalkalophilic bacterium Thioalkalivibrio nitratireducens. The enzyme catalyses nitrite and hydroxylamine reduction, with ammonia as the only product of both reactions. It consists of 525 amino-acid residues and contains eight haems c. TvNiR crystals were grown by the hanging-drop vapour-diffusion technique. The crystals display cubic symmetry and belong to space group P213, with unit-cell parameter a = 194 Å. A native data set was obtained to 1.5 Å resolution. The structure was solved by the SAD technique using the data collected at the Fe absorption peak wavelength.
1. Introduction
Nitrite is a component of the biological nitrogen cycle which is produced by the reduction of nitrate or the oxidation of ammonia in biological habitats. It widely used by bacteria as an electron acceptor under anaerobic conditions. Reduction of nitrite can be regarded as an assimilatory, respiratory or dissimilatory process. Assimilatory nitrite reduction serves in the production of ammonia, which is incorporated into cell material. This process is catalyzed by a cytoplasmic sirohaem-containing nitrite reductase that uses either NAD(P)H or ferredoxin as an electron donor (Richardson & Watmough, 1999 ▶; Simon, 2002 ▶). Respiratory nitrite reduction is catalyzed by three different classes of enzymes. Cytochrome cd 1 nitrite reductases and copper-containing nitrite reductases catalyze one-electron nitrite reduction to nitric oxide (NO). Multihaem cytochrome c nitrite reductases catalyze six-electron reduction of nitrite to ammonia (Richardson & Watmough, 1999 ▶; Simon, 2002 ▶). Bacterial cytochrome c nitrite reductases form a well studied group of enzymes with similar molecular and catalytic properties (Simon, 2002 ▶). High-resolution X-ray structures are known for four examples (Cunha et al., 2003 ▶; Bamford et al., 2002 ▶; Einsle et al., 1999 ▶, 2000 ▶). These enzymes are single-domain proteins with a unique secondary structure containing five haems c as prosthetic groups and occur as dimers in the crystals. Four haems of the enzymes have standard bis-histidine coordination, while the active-site haem is coordinated by lysine. Such coordination only occurs in cytochrome c nitrite reductases.
A novel cytochrome c nitrite reductase (TvNiR) was isolated from the soluble fraction of the denitrifying sulfur-oxidizing bacteria Thioalkalivibrio nitratireducens strain ALEN 2 (Sorokin, Antipov et al., 2003 ▶; Sorokin, Touruva et al., 2003 ▶).
TvNiR catalyzes the reduction of nitrite and hydroxylamine to ammonium ions without the release of any intermediates (Tikhonova et al., 2006 ▶),
According to gel-filtration data, TvNiR exists in solution mainly as a hexamer (76–93% of the total protein for a freshly prepared enzyme sample).
The TvNiR gene was sequenced (EMBL Nucleotide Sequence Database accession No. AJ880678). The translated amino-acid sequence consists of 525 residues and includes seven regions containing the CXXCH haem-binding motif and one region containing the CXXCK haem-binding motif characteristic of binding of haems c.
Multiple alignment of the TvNiR sequence with sequences from the NCBI database, carried out using ClustalW (http://www.ebi.ac.uk/clustalw/index.html), demonstrated that the TvNiR sequence has about 20% identity to cytochrome c nitrite reductases (ccNiRs). Nevertheless, all the catalytically important active-site residues as well as all five haem-binding motifs of ccNiRs sequences successfully coincide with those of the TvNiR sequence.
Here, we describe the crystallization, preliminary X-ray analysis and solution of the TvNiR structure.
2. Crystallization
The protein was isolated and purified to homogeneity as described in Tikhonova et al. (2006 ▶).
Crystals of TvNiR were grown by the hanging-drop vapour-diffusion technique at 278 K (Fig. 1 ▶). Hampton Research Crystal Screen Cryo was used to search for the initial crystallization conditions. Crystals were obtained using condition Nos. 13, 26 and 38. Crystals suitable for high-resolution X-ray data collection were grown using the following reservoir solutions. Condition 1: 26.3% MPD, 0.09 M sodium citrate buffer (pH 5.6), 0.175 M ammonium acetate; condition 2: 27% PEG 400, 0.09 M Tris buffer (pH 8.5), 0.18 M sodium citrate. The drops contained 5 µl of the purified enzyme in 0.05 M Tris–borate buffer pH 8.7 at a concentration of 14.5 mg ml−1 and 5 µl reservoir solution. Crystals of an appropriate size (up to 300 µm) grew within two months.
Figure 1.
Crystals of TvNiR obtained by hanging-drop vapour diffusion at 278 K.
3. X-ray data collection
A complete data set at 1.5 Å resolution was collected from a native TvNiR crystal grown under condition 1 at beamline BW7A of EMBL/DESY (Hambug, Germany) using a MAR CCD detector (MAR Research, Germany). A complete anomalous difference data set at 2.36 Å resolution was collected at the Fe absorption peak from a crystal grown under condition 2. The crystals were directly flash-cooled in a stream of cold nitrogen gas at 100 K using an Oxford Cryosystems cooling device (Oxford Cryosystems Ltd, England). There was no need for prior crystal transfer to a cryoprotecting solution. X-ray diffraction images were indexed, integrated and subsequently scaled using the programs DENZO and SCALEPACK (Otwinowski & Minor, 1997 ▶); the CCP4 package (Collaborative Computational Project, Number 4, 1994 ▶) was used for data reduction. Crystal and data-collection statistics are summarized in Table 1 ▶
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Data set | Fe peak SAD | Native |
|---|---|---|
| Conditions (reservoir solution) | 26.3% MPD, 0.09 M sodiumcitrate buffer pH 5.6, 0.175 M ammonium acetate | 27% PEG 400, 0.09 M Tris buffer pH 8.5, 0.18 M sodium citrate |
| Wavelength (Å) | 1.7375 | 0.9600 |
| Resolution (Å) | 2.36 (2.39–2.36) | 1.50 (1.52–1.50) |
| Rsym | 0.063 (0.273) | 0.036 (0.476) |
| Completeness (%) | 99.8 | 97.4 |
| I/σ(I) | 29.20 (7.76) | 29.0 (3.37) |
| Space group | P213 | P213 |
| Unit-cell parameters (Å) | a = b = c = 193.7 | a = b = c = 193.9 |
| No. of reflections measured | 4616298 | 7027135 |
| No. of unique reflections | 97669 | 383704 |
4. Data analysis
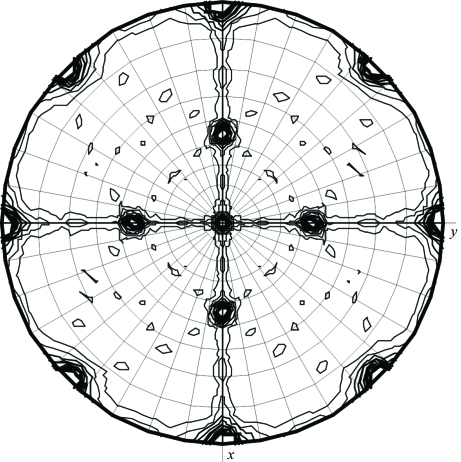
Matthews coefficient calculations showed that the asymmetric unit may contain between two and six TvNiR molecules. Self-rotation function calculations (using MOLREP) showed the presence of a non-crystallographic (NCS) twofold symmetry axis parallel to the face diagonal of the unit cell. The κ = 180° section of the self-rotation function is shown in Fig. 2 ▶.
Figure 2.
Self-rotation function for twofold axes (3 Å resolution; the integration radius was 35 Å).
The structure of TvNiR was solved by the SAD technique. The positions of 16 Fe atoms were revealed using the SHELXD program package (Schneider & Sheldrick, 2002 ▶). The solution suggested the presence of two molecules in the asymmetric unit with a high solvent content (75%). This hypothesis was followed in a subsequent structure solution and was found to be correct. The phases were calculated using the MLPHARE program implemented in the CCP4 package (Collaborative Computational Project, Number 4, 1994 ▶) and improved by solvent flattening/histogram matching in DM. Protein model building was carried out automatically using ARP/wARP (Perrakis et al., 1999 ▶) with only four gaps for the two molecules in the asymmetric unit. The high quality of the electron-density maps allowed us to construct the complete atomic model of the protein without gaps and include eight haems c for each molecule. The positions of five haems (the fourth to eighth haems; the haems are numbered in accordance with the position of the haem-binding motif in the primary structure) were determined by superimposing the preliminary TvNiR model onto the structure of cytochrome c nitrite reductase from Wolinella succinogenes (Einsle et al., 2000 ▶) using the MOLREP program (Vagin & Teplyakov, 1997 ▶). The other three haems were inserted into electron-density maps manually.
The refinement of the model containing 520 amino-acid residues and eight haems c per subunit is now in progress.
References
- Bamford, V. A., Angove, H. C., Seward, H. E., Thomson, A. J., Cole, J. A., Butt, J. N., Hemmings, A. M. & Richardson, D. J. (2002). Biochemistry, 41, 2921–2931. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Cunha, C. A., Macieira, S., Dias, J. M., Almeida, G., Goncalves, L. L., Costa, C., Lampreia, J., Huber, R., Moura, J. J., Moura, I. & Romao, M. J. (2003). J. Biol. Chem.278, 17455–17465. [DOI] [PubMed] [Google Scholar]
- Einsle, O., Messerschmidt, A., Stach, P., Bourenkov, G. P., Bartunik, H. D., Huber, R. & Kroneck, P. M. H. (1999). Nature (London), 400, 476–480. [DOI] [PubMed] [Google Scholar]
- Einsle, O., Stach, P., Messerschmidt, A., Simon, J., Kroger, A., Huber, R. & Kroneck, P. M. H. (2000). J. Biol. Chem.275, 39608–39616. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Perrakis, A., Morris, R. M. & Lamzin, V. S. (1999). Nature Struct. Biol.6, 458–463. [DOI] [PubMed] [Google Scholar]
- Richardson, D. J. & Watmough, N. J. (1999). Curr. Opin. Chem. Biol.3, 207–219. [DOI] [PubMed] [Google Scholar]
- Schneider, T. R. & Sheldrick, G. M. (2002). Acta Cryst. D58, 1772–1779. [DOI] [PubMed] [Google Scholar]
- Simon, J. (2002). FEMS Microbiol. Rev.26, 285–309. [DOI] [PubMed] [Google Scholar]
- Sorokin, D. Y., Antipov, A. N. & Kuenen, J. G. (2003). Arch. Microbiol.180, 127–133. [DOI] [PubMed] [Google Scholar]
- Sorokin, D. Y., Touruva, T. P., Sjollema, K. A. & Kuenen, J. G. (2003). Int. J. Syst. Evol. Microbiol.53, 1779–1783. [DOI] [PubMed] [Google Scholar]
- Tikhonova, T. V., Slutsky, A., Antipov, A. N., Boyko, K. M., Polyakov, K. M., Sorokin, D. Y., Zvyagilskaya, R. A. & Popov, V. O. (2006). Submitted.
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst.30, 1022–1025. [Google Scholar]