The mutagenesis, crystallization and preliminary crystallographic analysis of M. arthritidis-derived mitogen is described.
Keywords: superantigens, MAM
Abstract
Mycoplasma arthritidis-derived mitogen (MAM) functions as a conventional superantigen (SAg). Although recombinant MAM has been crystallized by the hanging-drop vapour-diffusion method, the crystals diffracted poorly to only 5.0 Å resolution, with large unit-cell parameters a = 163.8, b = 93.0, c = 210.9 Å, β = 93.7° in the monoclinic space group P21. Unit-cell content analysis revealed that as many as 24 molecules could be present in the asymmetric unit. Systematic alanine mutagenesis was applied in order to search for mutants that give crystals of better quality. Two mutants, L50A and K201A, were crystallized under the same conditions as wild-type MAM (MAMwt). Crystals of the L50A mutant are isomorphous with those of MAMwt, while a new crystal form was obtained for the K201 mutant, belonging to the cubic space group P4132 with unit-cell parameters a = b = c = 181.9 Å. Diffraction data were collected to 3.6 and 2.8 Å resolution from crystals of the MAM L50A and K201A mutants, respectively. Molecular-replacement calculations suggest the presence of two molecules in the asymmetric unit for the MAM K201A mutant crystal, resulting in a V M of 5.0 Å Da−1 and a solvent content of 75%. An interpretable electron-density map for the MAM K201A mutant crystal was produced using the molecular-replacement method.
1. Introduction
Mycoplasma arthritidis-derived mitogen (MAM) is produced by M. arthritidis, a natural pathogen of rodents (Cole, 1991 ▶). MAM functions as a conventional superantigen (SAg). SAgs are functionally related immunoregulatory proteins that are generally produced by bacteria and viruses (Kappler et al., 1988 ▶; Kotzin et al., 1993 ▶; Li et al., 1999 ▶; White et al., 1989 ▶). Evidence has suggested that SAgs play important roles in a number of human diseases, including food poisoning, toxic shock syndrome and autoimmune diseases such as multiple sclerosis and rheumatoid arthritis (RA) (Abe et al., 1992 ▶; Conrad et al., 1994 ▶; Kotzin et al., 1993 ▶; McCormick et al., 2001 ▶; Renno & Acha-Orbea, 1996 ▶).
As an SAg, MAM can induce a spontaneous chronic arthritis, which resembles human RA, in genetically susceptible strains of rodents (Cole & Atkin, 1991 ▶; Cole & Griffiths, 1993 ▶). Although MAM, like other SAgs, interacts with T-cell receptors (TCRs) in a Vβ-restricted fashion, the binding of MAM to TCR is influenced by the CDR3 region of the TCR (Hodtsev et al., 1998 ▶). Thus, MAM could represent a new type of ligand for TCRs, distinct from both conventional peptide antigens and other known SAgs. In addition, we recently determined the crystal structure of MAM cocrystallized with its receptor, the HLA-DR1–HA complex (Zhao, Li, Drozd, Guo, Mourad et al., 2004 ▶). MAM displays a three-dimensional structure that is completely different from those of all other SAgs for which structures have been solved. All the pyrogenic toxin SAgs from Staphylococcus aureus and Streptococcus pyogenes have very similar three-dimensional structures composed of a β-grasp motif and a β-barrel (Li et al., 1999 ▶; Mitchell et al., 2000 ▶; Sundberg et al., 2002 ▶). Yersinia pseudotuberculosis-derived mitogen, an enteric SAg, is composed of a single domain with a nearly all β-sheet structure showing a jelly roll-like topology (Donadini et al., 2004 ▶). In contrast, MAM, when complexed with its receptor HLA-DR1–HA complex, adopts a novel fold composed of two completely α-helical domains (Zhao, Li, Drozd, Guo, Mourad et al., 2004 ▶). It is currently not known whether unliganded MAM has the same structure as it does in the complex. Thus, it is of great interest to determine the three-dimensional structure of apo MAM. This will allow us to address the question of whether conformational changes are associated with receptor binding. Here, we report the crystallization and preliminary crystallographic analysis of wild-type MAM (MAMwt) and MAM mutants.
2. Material and methods
2.1. Site-directed mutagenesis
Alanine-scanning mutagenesis was used to individually mutate selected MAM residues at the interface of the crystallographic MAM homodimer (Zhao, Li, Drozd, Guo, Mourad et al., 2004 ▶), as described in Li et al. (2006 ▶). These residues included Leu50, His58, Arg154, Arg192, Tyr193, Tyr194, Glu195, Asp197 and Lys201.
2.2. Expression and purification
Soluble MAM and MAM mutants were overexpressed in Escherichia coli using the pGEX-6P-1 GST fusion protein-expression system (GE Amersham Pharmacia) as described previously (Langlois et al., 2000 ▶; Zhao, Li, Drozd, Guo, Stack et al., 2004 ▶).
2.3. Crystallization
Purified wild-type and mutant MAM molecules were concentrated to about 10 mg ml−1 using a Millipore stirred cell. Initial crystallization conditions for MAMwt were established using Wizard Screens I and II (Jena Bioscience). Crystals appeared in two conditions, Nos. 14 and 18 of Wizard Screen II, in which PEG 1000 and PEG 3000 were used as the precipitants, respectively. These conditions were then further optimized. The best crystals were grown at room temperature in hanging drops by mixing 2 µl protein solution with an equal volume of reservoir solution containing 13–15% PEG 3350, 0.2 M NaCl, 5% ethanol glycol, 5% glycerol and 0.1 M potassium sodium phosphate pH 6.2.
Microseeding was used to produce large crystals of MAMwt as well as to produce crystals in a form similar to that of the MAM K201A mutant. After equilibration overnight, 0.5 µl of a stock solution of crystal seeds was introduced into the drops. Prior to microseeding, seed-stock solutions were produced by diluting crushed small crystals in the crystallization buffer to various degrees. Alternatively, a streak microseeding method was used. New crystals appeared within one week and continued to grow for 2–3 weeks.
2.4. Data collection
Prior to data collection, all crystals were transferred to a cryoprotectant solution (mother liquor supplemented with 25% glycerol) and were flash-cooled in a nitrogen-gas stream. Diffraction data for the MAM L50A and K201A mutant crystals were collected at 100 K at beamline X4A of the National Synchrotron Light Source (NSLS), Brookhaven National Laboratory (BNL). Data were processed, scaled and reduced using the HKL2000 (Otwinowski & Minor, 1997 ▶) and the CCP4 program suites (Collaborative Computational Project, Number 4, 1994 ▶).
2.5. Molecular replacement
The Phaser program (McCoy et al., 2005 ▶) was used for molecular-replacement (MR) calculations to determine the crystal structure of the MAMK201A mutant. CNS v.1.0 (Brünger et al., 1998 ▶) was used for structure refinement.
3. Results and discussion
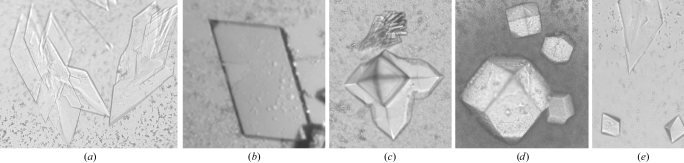
Thin plate-shaped crystals of MAMwt were first observed from condition Nos. 14 and 18 of Wizard Crystal Screen II, using PEG 1000 and PEG 3000 used as the precipitant, respectively. After optimization and microseeding, large crystals could be obtained with average dimensions of 0.5 × 0.4 × 0.1 mm (Fig. 1 ▶ a). However, these crystals diffracted poorly, even when a synchrotron X-ray source was used. These crystals belong to space group P21, with relatively large unit-cell parameters: a = 163.8, b = 93.0, c = 210.9 Å, β = 93.7°. Given the relatively small size of the protein (∼25.5 kDa), the large unit-cell parameters suggested that the MAM molecules can form oligomers within the asymmetric unit of the crystal. Indeed, unit-cell content analysis indicated that as many as 24 MAM molecules could be present in the asymmetric unit of the crystal lattice, with a reasonable Matthews coefficient (V M; Matthews, 1968 ▶) of 2.7 Å Da−1 and a solvent content of 54%. This may explain why the crystals diffracted poorly. With so many molecules in the asymmetric unit, small defects in molecular packing will become amplified, leading to poor crystal packing. Thus, good diffraction could not be obtained.
Figure 1.
Crystals of the wild-type MAM and two MAM alanine mutants (MAMK201A and MAML50A). (a) Plate-shaped MAMwt; (b) MAML50A; (c) MAMK201A; (d) MAMK201A; (e) MAMwt after seeding with the diamond-shaped MAMK201A crystals.
We previously observed a crystallographic dimer of MAM molecules when MAM was cocrystallized with its receptor, the HLA-DR1–HA complex (Zhao, Li, Drozd, Guo, Mourad et al., 2004 ▶). It is possible that a similar MAM dimer is formed in the apo MAMwt crystal and that multiple MAM dimers associate to form oligomers. Despite extensive attempts to optimize the crystallization conditions, these crystals still diffracted poorly.
In order to obtain crystals of high quality, we then chose to engineer MAM mutants that were less likely to oligomerize. Nine amino acids at the crystallographic dimer interface (Zhao, Li, Drozd, Guo, Mourad et al., 2004 ▶), namely Leu50, His58, Arg154, Arg192, Tyr193, Tyr194, Glu195, Asp197 and Lys201, were selected for alanine mutagenesis (Li et al., 2006 ▶). Of the nine mutants, five, L50A, R154A, R192A, E195A and K201A, gave yields compatible to that for MAMwt. Extensive crystallization conditions were searched for these five MAM mutants.
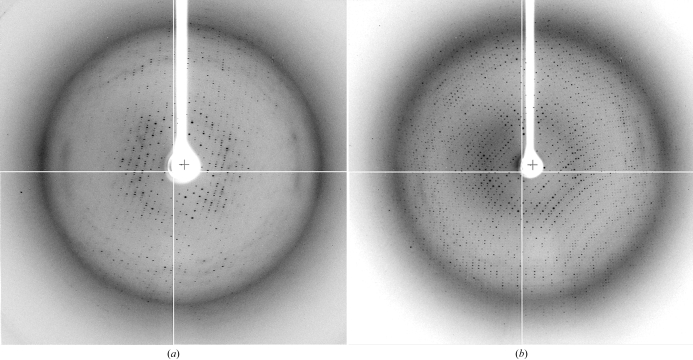
Under very similar conditions, the L50A mutant gave crystals with a form similar to that of MAMwt (Fig. 1 ▶ b). Diffraction analysis indicated that the L50A crystals are indeed isomorphous to those of MAMwt. These L50A crystals diffracted slightly better than the MAMwt crystals (Fig. 2 ▶ a). A set of diffraction data was collected at 3.6 Å resolution at the NSLS beamline X4A (Table 1 ▶). However, using the coordinates of MAMwt in the MAM–HLA-DR1–HA complex crystal as a search model, MR calculations using various programs failed to generate any clear solution.
Figure 2.
Diffraction images from crystals of (a) MAML50A and (b) MAMK201A.
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Crystal | MAM K201A | MAM L50A |
|---|---|---|
| Space group | P4132 | P21 |
| Unit-cell parameters (Å, °) | a = b = c = 181.9 | a = 164.4, b = 93.0, c = 211.5, β = 93.5 |
| Wavelength (Å) | 0.979 | 0.979 |
| Resolution (Å) | 2.8 (2.9–2.8) | 3.6 (3.7–3.6) |
| Redundancy | 6.9 (5.4) | 3.5 (3.2) |
| Completeness (%) | 95.9 (96.8) | 97 (91.5) |
| Average I/σ(I) | 9.6 (2.7) | 16 (4.7) |
| Rsym (%) | 11.7 (69.2) | 10.0 (35.8) |
In contrast to the L50A mutant, the MAM K201A mutant produced two distinct crystal forms in a single drop (Fig. 1 ▶ c). The clusters of plate-shaped crystals of MAMK201A are similar to those of MAMwt, while the diamond-shaped crystals apparently represent a new form for this molecule. Upon optimization, crystals of the latter form could be easily maximized to dimensions of 0.4 × 0.4 × 0.3 mm (Fig. 1 ▶ d). These crystals belong to the cubic space group P4132 or P4332, with unit-cell parameters a = b = c = 181.9 Å. Therefore, each asymmetric unit of the crystal lattice probably contains between one and five MAMK201A molecules. The V M values were accordingly calculated to be in the range 9.8–2.0 Å3 Da−1, corresponding to a solvent content in the range 87–37%.
Encouraged by the new crystal form, we tried to grow MAMwt crystals in the new form using the microseeding method. Small diamond-shaped MAMK201A crystals were crushed in the crystallization mother liquor. The crushed crystal solution was diluted 1000-fold to 100 000-fold with crystallization mother liquor. Prior to microseeding, the MAMwt crystallization trials were set up using 2 µl protein solution mixed with 2 µl reservoir solution. After overnight equilibration, 0.5 µl of the diluted microseed solution was introduced into the crystallization drops. Small diamond-shaped MAMwt crystals appeared along with the plate-shaped crystals in the same drop after a week of incubation (Fig. 1 ▶ e). Unfortunately, these diamond-shaped MAMwt crystals could not be grown as large as the MAMK201A crystals. Although these diamond-shaped MAMwt crystals were not of a suitable size for X-ray analysis, we expect that they are isomorphous with the diamond-shaped MAMK201A crystals.
The MAMK201A crystals diffracted much better than the plate-shaped crystals of MAMwt and MAML50A (Fig. 2 ▶ b). Diffraction data were measured to 2.8 Å resolution for the MAMK201A crystals at the NSLS X4A beamline (Table 1 ▶). With the crystal structure of MAM in the MAM–HLA-DR1–HA complex (Zhao, Li, Drozd, Guo, Mourad et al., 2004 ▶) as a search model, MR calculations for the MAM K201A with the program Phaser (McCoy et al., 2005 ▶) failed to generate a clear solution. The MAM model was therefore divided into two independent domains, with various truncations at the N- and C-termini and some flexible loop regions. These domains were individually subjected to MR calculations. Using this strategy, we obtained a clear solution in space group P4132 only when we used a model composed of the N-terminal 39–125 amino acids combined with a model including the C-terminal amino acids 128–168 and 178–196 in MR calculations. Cross-rotation and translation-function calculations confirmed that two MAM molecules are present in an asymmetric unit cell, which would indicate that the solvent content in the crystal is 75%, with a V M of 5.0 Å3 Da−1. With the best MR solution, one cycle of rigid-body refinement with CNS v.1.0 (Brünger et al., 1998 ▶) generated an R free of 42% and an R work of 39% for all K201A data between 100 Å and 2.8 Å resolution. Structure refinement is currently in progress.
Acknowledgments
This research was supported by the National Institutes of Health (NIH) grant AI50628 (to HL). We acknowledge the use of Wadsworth Center Core facilities: the Molecular Genetics Core for DNA sequencing and the Macromolecular Crystallography Core for crystal evaluation. We thank A. Verschoor for critical reading of the manuscript. We thank W. Mourad for the gift of MAM expression plasmid. Data for this study were measured in part at beamline X4A of NSLS, which is supported by the Department of Energy, by grants from the NIH and by the New York Structural Biology Center.
References
- Abe, J., Kotzin, B. L., Jujo, K., Melish, M. E., Glode, M. P., Kohsaka, T. & Leung, D. Y. (1992). Proc. Natl Acad. Sci. USA, 89, 4066–4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed] [Google Scholar]
- Cole, B. C. (1991). Curr. Top. Microbiol. Immunol.174, 107–119. [DOI] [PubMed] [Google Scholar]
- Cole, B. C & Atkin, C. L. (1991). Immunol. Today, 12, 271–276. [DOI] [PubMed] [Google Scholar]
- Cole, B. C. & Griffiths, M. M. (1993). Arthritis Rheum.36, 994–1002. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Conrad, B., Weidmann, E., Trucco, G., Rudert, W. A., Behboo, R., Ricordi, C., Rodriquez-Rilo, H., Finegold, D. & Trucco, M. (1994). Nature (London), 371, 351–355. [DOI] [PubMed] [Google Scholar]
- Donadini, R., Liew, C. W., Kwan, A. H., Mackay, J. P. & Fields, B. A. (2004). Structure, 12, 145–156. [DOI] [PubMed] [Google Scholar]
- Hodtsev, A. S., Choi, Y., Spanopoulou, E. & Posnett, D. N. (1998). J. Exp. Med.187, 319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappler, J. W., Staerz, U., White, J. & Marrack, P. C. (1988). Nature (London), 332, 35–40. [DOI] [PubMed] [Google Scholar]
- Kotzin, B. L., Leung, D. Y., Kappler, J. & Marrack, P. (1993). Adv. Immunol.54, 99–166. [DOI] [PubMed] [Google Scholar]
- Langlois, M. A., Etongue-Mayer, P., Ouellette, M. & Mourad, W. (2000). Eur. J. Immunol.30, 1748–1756. [DOI] [PubMed] [Google Scholar]
- Li, H. M., Llera, A., Malchiodi, E. L. & Mariuzza, R. A. (1999). Annu. Rev. Immunol.17, 435–466. [DOI] [PubMed] [Google Scholar]
- Li, H. M., Zhao, Y. W., Guo, Y., Van Vranken, S. J., Li, Z., Eisele, L. & Mourad, W. (2006). Submitted.
- McCormick, J. K., Yarwood, J. M. & Schlievert, P. M. (2001). Annu. Rev. Microbiol.55, 77–104. [DOI] [PubMed] [Google Scholar]
- McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C. & Read, R. J. (2005). Acta Cryst. D61, 458–464. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.245, 54–68. [DOI] [PubMed]
- Mitchell, D. T., Levitt, D. G., Schlievert, P. M. & Ohlendorf, D. H. (2000). J. Mol. Evol.51, 520–531. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Renno, T. & Acha-Orbea, H. (1996). Immunol. Rev.154, 175–191. [DOI] [PubMed] [Google Scholar]
- Sundberg, E. J., Li, Y. & Mariuzza, R. A. (2002). Curr. Opin. Immunol.14, 36–44. [DOI] [PubMed] [Google Scholar]
- White, J., Herman, A., Pullen, A. M., Kubo, R., Kappler, J. W. & Marrack, P. (1989). Cell, 56, 27–35. [DOI] [PubMed] [Google Scholar]
- Zhao, Y., Li, Z., Drozd, S., Guo, Y., Mourad, W. & Li, H. M. (2004). Structure, 12, 277–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y., Li, Z., Drozd, S., Guo, Y., Stack, R., Hauer, C. & Li, H. M. (2004). Acta Cryst. D60, 353–356. [DOI] [PMC free article] [PubMed] [Google Scholar]