Neutron diffraction data of hydrogenated recombinant urate oxidase enzyme (Rasburicase), complexed with a purine-type inhibitor 8-azaxanthin, was collected to 2.1 Å resolution from a crystal grown in D2O by careful control and optimization of crystallization conditions via knowledge of the phase diagram. Deuterium atoms were clearly seen in the neutron-scattering density map.
Keywords: urate oxidase, heavy water, phase diagram, neutron Laue diffraction
Abstract
Crystallization and preliminary neutron diffraction measurements of rasburicase, a recombinant urate oxidase enzyme expressed by a genetically modified Saccharomyces cerevisiae strain, complexed with a purine-type inhibitor (8-azaxanthin) are reported. Neutron Laue diffraction data were collected to 2.1 Å resolution using the LADI instrument from a crystal (grown in D2O) with volume 1.8 mm3. The aim of this neutron diffraction study is to determine the protonation states of the inhibitor and residues within the active site. This will lead to improved comprehension of the enzymatic mechanism of this important enzyme, which is used as a protein drug to reduce toxic uric acid accumulation during chemotherapy. This paper illustrates the high quality of the neutron diffraction data collected, which are suitable for high-resolution structural analysis. In comparison with other neutron protein crystallography studies to date in which a hydrogenated protein has been used, the volume of the crystal was relatively small and yet the data still extend to high resolution. Furthermore, urate oxidase has one of the largest primitive unit-cell volumes (space group I222, unit-cell parameters a = 80, b = 96, c = 106 Å) and molecular weights (135 kDa for the homotetramer) so far successfully studied with neutrons.
1. Introduction
Urate oxidase (Uox) is an enzyme which catalyses the oxidation of uric acid to allantoin, an inactive and soluble metabolite. It is inactivated in humans and higher primates. Urate oxidase from Aspergillus flavus (or Uricozyme) and rasburicase, a recombinant Uox (or Fasturtec), have been produced, purified and made commercially available by Sanofi–Aventis. Urate oxidase is used as a protein drug to reduce toxic uric acid accumulation and to resolve the hyperuricaemic disorders occurring during chemotherapy. The initial X-ray structure of the urate oxidase family was solved at 2.2 Å resolution in complex with a competitive inhibitor, 8-azaxanthin, and was further refined to 2.05 Å (Colloc’h et al., 1997 ▶). A putative mechanism for the oxidation of uric acid was then proposed; however, the precise ionization state of the substrate during the reaction was not definitively established. From recent X-ray structures of several urate oxidase–inhibitor complexes (Retailleau et al., 2004 ▶, 2005 ▶), including those with 8-azaxanthin, 9-methyl uric acid, oxonic acid, uracil, 5,6-diaminouracil, cymelarsan and guanine, as well as the ligand-free enzyme, a new position of the substrate (uric acid) was determined, i.e. a reversed position of the inhibitor in the active site with respect to the initial structure. This greatly improved our understanding of the enzymatic mechanism of urate oxidase. The mechanism, which converts uric acid into allantoin, is composed of many steps and has been extensively explored by Tipton and coworkers (Kahn et al., 1997 ▶; Kahn & Tipton, 1997 ▶; Imhoff et al., 2003 ▶). The first step is catalyzed by urate oxidase, which oxidizes uric acid to an intermediate compound. The enzyme-catalysed reaction occurs in the presence of an excess of molecular oxygen and releases hydrogen peroxide. The intermediate compound is then degraded to allantoin through a number of chemical steps. The recent X-ray structures of urate oxidase–inhibitor complexes have provided several new insights into this mechanism. Arg176, Gln228 and Phe159 hold the substrate in the active site via its six-membered ring, with Thr57 being linked to the five-membered ring of the inhibitor by the N7 atom. Moreover, two water molecules appear to play an important role in the enzymatic mechanism: a water molecule (W1) located above the C4—C5 bond, ideally positioned for hydroxylation, is stabilized by the Asn254 and Thr57 side chains, while another water molecule (W2) bonded to the N9 atom in 8-azaxanthin seems to be involved in charge transfer between the 3,7-dianion and the dioxygen molecule. Whilst the resolutions of the available X-ray urate oxidase structural complexes do not contain any information on the positions of the H atoms within the active site, neutron structure determination at similar resolutions should reveal the critical hydrogen-bonding network at the enzyme active site and therefore provide key insights into the proton shuttle involved in the urate oxidase-catalyzed oxidation of uric acid. Neutron diffraction studies have previously proved successful in addressing similar questions for endothiapepsin (Coates et al., 2001 ▶).
In order to directly observe the positions of the H atoms within the active site of the recombinant urate oxidase in complex with 8-azaxanthin, we have initiated neutron diffraction studies. Here, we report the details of the growth of large single crystals (volume >1 mm3) suitable for high-resolution neutron diffraction studies. The statistics of the neutron Laue diffraction data, as well as two examples of resulting neutron density maps, illustrate the quality of the data collected.
2. Crystallization
2.1. Purification and phase diagram of Uox
The recombinant Uox from A. flavus expressed in Saccharomyces cerevisiae (or rasburicase; trade name Fasturtec in the EU and Elitek in the USA) was supplied by Sanofi–Aventis. Complexed with a purine-type inhibitor (8-azaxanthin; MW ≃ 150 Da) purchased from Sigma–Aldrich, it was purified in 50 mM Tris buffer pH 8.5 by gel-filtration chromatography on Superdex S200PG with an ÄKTA basic system and was concentrated by ultrafiltration in an Amicon cell (Bonneté et al., 2001 ▶; Vivarès & Bonneté, 2002 ▶; Vivarès et al., 2006 ▶). A 4 M stock solution of sodium chloride (Sigma–Aldrich) and a solution of 40%(w/v) PEG 8000 (Hampton Research) were prepared in 50 mM Tris buffer pH 8.5. All experiments were performed with 100 mM sodium chloride at 293 K. The solubility, defined as the concentration of protein in solution in equilibrium with a crystal, was determined by seeding a supersaturated solution of urate oxidase (between 100 and 200 µl) with crushed microcrystals. The PEG concentrations were between 0.5 and 8%(w/v) and the buffer was 50 mM Tris pH 8.5 with 100 mM NaCl. The urate oxidase concentration was determined by measuring the UV absorbance at 280 nm using an experimental extinction coefficient of 2.2 ± 0.1 cm2 mg−1 and was followed over several weeks until equilibrium was reached. The same protocol was used to determine the solubility of urate oxidase in D2O buffer.
2.2. Crystallization in D2O
For the growth of Uox crystals in heavy water, the protein solution was exchanged with buffered D2O (50 mM Tris–DCl pD 8.5, 100 mM NaCl) containing 8-azaxanthin in a large excess (0.5–2 mg ml−1). The excess of 8-azaxanthin was then eliminated by gel-filtration chromatography using the same D2O buffer. The protein was then concentrated to 10 mg ml−1. The final Uox–8-AZA complex was crystallized by the sitting-drop vapour-diffusion method at room temperature (Bonneté et al., 2001 ▶; Vivarès & Bonneté, 2002 ▶) within 24–48 h in the presence of approximately 5% PEG 8000. The proper amounts of NaCl, PEG and Tris were dissolved in heavy water (Euriso-top, 99.92% D2O) to obtain solutions with the concentrations required for buffer exchange as well as for the crystallization process. The pD of the buffers were adjusted with NaOD (Euriso-top, 99% D atom) and DCl (Euriso-top, 99.8% D atom) according to the formula (pD = pHmeas + 0.3314n + 0.0766n 2, where n = %D2O; Lumry et al., 1951 ▶). All solutions were filtered through 0.22 µm Millipore filters. The final crystallization conditions were optimized via knowledge of the phase diagram and solubility measurements. Finally, crystals of about 50 µm in size were used as macroseeds and their size and quality were further improved using a temperature-control device developed at EMBL Grenoble (to be published).
2.3. Neutron data collection and reduction
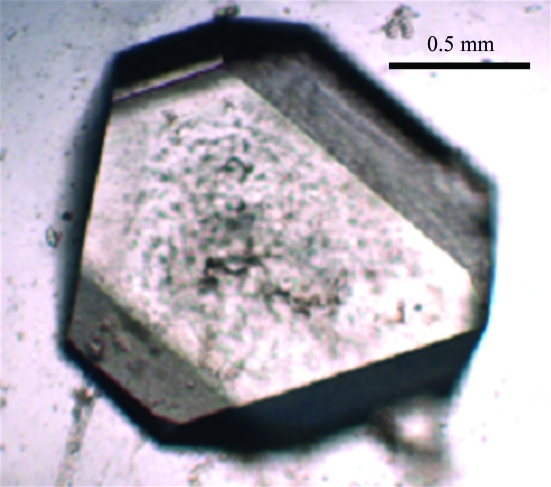
A crystal of volume 1.8 mm3 (1.5 × 1.5 × 0.8 mm; Fig. 1 ▶) was mounted in a quartz capillary and sealed with wax for data collection. Neutron Laue data were collected at 293 K on the LADI instrument which is installed on end-station T17 of the cold neutron guide H142 at ILL (Cipriani et al., 1994 ▶; Myles et al., 1998 ▶). The LADI instrument is equipped with a large neutron image-plate detector mounted on a cylindrical camera, which completely encircles the sample. An Ni–Ti multilayer wavelength filter was used to select a quasi-Laue bandpass (δλ/λ = 25%) centred at 3.45 Å. Laue data (stationary crystal, no oscillation during exposure) were collected in steps of 8° around the rotation axis (ϕ) of the instrument, with 14 frames collected from one crystal setting and nine frames from a second crystal setting in order to fill in the blind region. The exposure time for each frame was 36 h. Data extended to a nominal resolution of 2.0 Å, with subsequent analysis using data to 2.1 Å.
Figure 1.
Optical photograph of a Uox–8-AZA crystal, approximate dimensions 1.5 × 1.5 × 0.8 mm.
3. Results and discussion
3.1. Crystallization
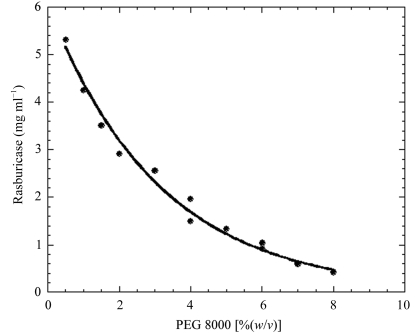
Original efforts to collect high-resolution neutron diffraction data were performed in 1998. The crystals provided by Sanofi–Synthélabo were prepared with hydrogenated protein and then exposed to vapour exchange in a capillary. Unfortunately, initial data-collection tests on LADI showed that these crystals were multiple and thus inadequate for structural analysis (Colloc’h, Mornon & Myles, unpublished results). Here, we have grown initial crystals of Uox in complex with 8-azaxanthin using the hydrogenated protein previously exchanged with buffered D2O (50 mM Tris–DCl pD 8.5, 100 mM NaCl) containing 8-azaxanthin (MW ≃ 150 Da). Crystallization conditions in D2O were found to be closely similar to those used for the hydrogenated complex. In agreement with previous studies with lysozyme and BPTI (Gripon et al., 1997 ▶; Budayova-Spano et al., 2000 ▶), we observed that urate oxidase solubility is lower in heavy water than in light water (work to be published). In particular, knowledge of the phase diagram (Fig. 2 ▶) has allowed us to control the nucleation and to optimize the growth conditions of Uox–8-AZA crystals in many crystallization conditions in both light and heavy water. By growing the crystals directly in D2O, the percentage of deuteration at exchangeable hydrogen positions in the protein is higher (approaching 80% of accessible protons) than by using the vapour-diffusion method. This improves the signal-to-noise ratio of the data and hence the resolution limit by significantly reducing the hydrogen incoherent scattering contribution to the background.
Figure 2.
Two-dimensional phase diagram representing the solubility of urate oxidase complex with 8-azaxanthin as a function of PEG 8000 percentage.
3.2. Neutron data processing
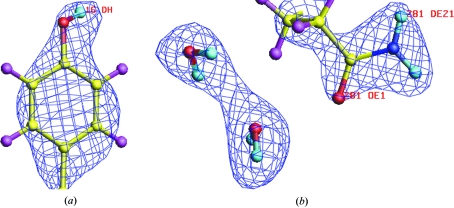
The observed Bragg reflections were indexed and integrated to a d min of 2.1 Å using LAUEGEN (Campbell et al., 1998 ▶). The program LSCALE (Arzt et al., 1999 ▶) was used to derive the wavelength-normalization curve using the intensities of symmetry-equivalent reflections measured at different wavelengths. The data were then scaled and merged using SCALA (Collaborative Computational Project, Number 4, 1994 ▶). Full statistics for processing of the neutron data set are provided in Table 1 ▶. The final data set for the urate oxidase–8-azaxanthin complex consisted of 45 937 reflections, which reduced to 15 192 unique reflections with a merging R factor of 15.1%. This data set is currently being used for refinement of the Uox–8-AZA complex. Two examples of typical neutron Fourier maps for these data are given in Fig. 3 ▶. Fig. 3 ▶(a) clearly shows that the H atom of the hydroxyl group of the tyrosine residue has exchanged with deuterium and thus is in positive nuclear density. Furthermore, the orientation of this OD group is now directly observed. Fig. 3 ▶(b) shows clearly that the orientation of the glutamine side chain is correct and also shows the positions and orientations of two water molecules as full D2O. The fully refined structure and the implications for the catalytic mechanism will be published shortly.
Table 1. Data-processing statistics for the neutron Laue data set collected from the urate oxidase–8-azaxanthin complex to 2.1 Å resolution.
| dmin (Å) | Rmerge | Rcum | I/σ(I) | Mean (I)/sd | Completeness (%) | Cumulative completeness (%) | Multiplicity |
|---|---|---|---|---|---|---|---|
| 6.64 | 0.095 | 0.095 | 6.3 | 8.0 | 89.5 | 89.5 | 3.7 |
| 4.70 | 0.143 | 0.124 | 4.5 | 9.0 | 93.5 | 92.1 | 4.9 |
| 3.83 | 0.166 | 0.141 | 3.9 | 8.6 | 92.0 | 92.0 | 4.7 |
| 3.32 | 0.161 | 0.145 | 4.1 | 7.3 | 84.3 | 89.3 | 3.7 |
| 2.97 | 0.160 | 0.147 | 4.3 | 6.1 | 75.0 | 85.3 | 3.0 |
| 2.71 | 0.167 | 0.148 | 3.9 | 5.1 | 61.2 | 79.5 | 2.4 |
| 2.51 | 0.164 | 0.149 | 4.4 | 4.6 | 53.9 | 74.2 | 2.2 |
| 2.35 | 0.166 | 0.149 | 4.3 | 4.2 | 51.9 | 70.2 | 2.1 |
| 2.21 | 0.174 | 0.150 | 4.1 | 3.9 | 49.5 | 66.9 | 2.1 |
| 2.10 | 0.179 | 0.151 | 4.0 | 3.6 | 48.3 | 64.1 | 2.0 |
| Total | 0.151 | 4.1 | 5.9 | 64.1 | 3.0 |
Figure 3.
Neutron scattering density map (2F o − F c at 1.5σ) superposed on the current model of Uox–8-AZA for (a) Tyr16 and (b) Glu281 with two D2O water molecules. Note the clear density for the D atoms as well as the orientations of the D2O water molecules.
4. Conclusion
This work illustrates the high quality of neutron diffraction data collected from crystals grown by careful control and optimization of crystallization conditions via knowledge of the phase diagram. An important point is that the crystal diffracted to high resolution even though it was obtained with hydrogenated protein (previously exchanged with buffered D2O) and possessed a relatively small crystal volume in comparison with other neutron structural projects (Coates et al., 2001 ▶; Maeda et al., 2004 ▶; Bau, 2004 ▶; Hanson et al., 2004 ▶). It is noteworthy that for a hydrogenated D2O-exchanged protein with such a large primitive unit-cell volume (407 040 Å3), this neutron diffraction study of urate oxidase represents one of the highest resolution neutron data sets thus far. In the cases of xylose isomerase (Hanson et al., 2004 ▶) and DHFR (Bennett et al., 2005 ▶), which both have slightly larger unit-cell volumes, the experimental neutron data extended to similar resolution to that of urate oxidase, but did not allow refinement of the protein structures. Refinement of the neutron structure is under way and clearly shows enhanced visibility for hydrogen isotope positions of the protein, solvent (as full D2O) and 8-azaxanthin. The direct determination of the protonation states of the residues and the orientation of the water molecules within the active site is crucial for a more complete understanding of the enzymatic mechanism of this protein drug. In addition, this study represents a significant advance in our work on urate oxidase. To date, the present neutron diffraction data obtained with the urate oxidase–8-azaxanthin complex are the only existing neutron structural data concerning this enzyme. Our future goal will focus on neutron diffraction studies of several other urate oxidase complexes. The supplementary information derived from an ensemble of neutron urate oxidase structures will further improve our understanding of the catalytic mechanism of this therapeutically important enzyme.
Acknowledgments
The authors thank the Institut Laue–Langevin for the generous provision of neutron beam time on the LADI diffractometer. Special thanks are given to Dr Stephen Cusack (EMBL Grenoble Outstation) and Dr Peter Timmins (ILL Grenoble), who have taken an active interest in this urate oxidase neutron crystallography project. This work has benefitted from the activities of the EU-funded DLAB project under contracts HPRI-CT-2001-50035 and RII3-CT-2003-505925.
References
- Arzt, S., Campbell, J. W., Harding, M. M., Hao, Q. & Helliwell, J. R. (1999). J. Appl. Cryst.32, 554–562. [Google Scholar]
- Bau, R. (2004). J. Synchrotron Rad.11, 76–79. [DOI] [PubMed] [Google Scholar]
- Bennett, B. C., Meilleur, F., Myles, D. A., Howell, E. E. & Dealwis, C. G. (2005). Acta Cryst. D61, 574–579. [DOI] [PubMed] [Google Scholar]
- Bonneté, F., Vivarès, D., Robert, C. & Colloc’h, N. (2001). J. Cryst. Growth, 232, 330–339.
- Budayova-Spano, M., Lafont, S., Astier, J. P., Ebel, C. & Veesler, S. (2000). J. Cryst. Growth, 217, 311–319.
- Campbell, J. W., Hao, Q., Harding, M. M., Nguti, N. D. & Wilkinson, C. (1998). J. Appl. Cryst.31, 496–502. [Google Scholar]
- Cipriani, F., Dauvergne, F., Gabriel, A., Wilkinson, C. & Lehmann, L. S. (1994). Biophys. Chem.53, 5–13. [DOI] [PubMed]
- Coates, L., Erskine P., T., Wood, S. P., Myles, D. A. & Cooper, J. B. (2001). Biochemistry, 40, 149–157. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Colloc’h, N., El Hajji, M., Bachet, B., L’Hermite, G., Schiltz, M., Prangé, T., Castro, B. & Mornon, J. P. (1997). Nature Struct. Biol.4, 947–952. [DOI] [PubMed] [Google Scholar]
- Gripon, C., Legrand, L., Rosenman, I., Vidal, O., Robert, M. C. & Boué, F. (1997). J. Cryst. Growth, 177, 238–247.
- Hanson, B. L., Langan, P., Katz, A. K., Li, X., Harp, J. M., Glusker, J. P., Schoenborn, B. P. & Bunick, G. J. (2004). Acta Cryst. D60, 241–249. [DOI] [PubMed] [Google Scholar]
- Imhoff, R. D., Power, N. P., Borrok, M. J. & Tipton, P. A. (2003). Biochemistry, 42, 4094–4100. [DOI] [PubMed] [Google Scholar]
- Kahn, K., Serfozo, P. & Tipton, P. A. (1997). J. Am. Chem. Soc.119, 5435–5442.
- Kahn, K. & Tipton, P. A. (1997). Biochemistry, 36, 4731–4738. [DOI] [PubMed] [Google Scholar]
- Lumry, R., Smith, E. L. & Glantz, R. R. (1951). J. Am. Chem. Soc.73, 4330–4340.
- Maeda, M., Chatake, T., Tanaka, I., Ostermann, A. & Niimura, N. (2004). J. Synchrotron Rad.11, 41–44. [DOI] [PubMed] [Google Scholar]
- Myles, D. A., Bon, C., Langan, P., Cipriani, F., Castagna, J. C., Lehmann, M. S. & Wilkinson, C. (1998). Physica B, 241–243, 1122–1130.
- Retailleau, P., Colloc’h, N., Vivarès, D., Bonneté, F., Castro, B., El Hajji, M., Mornon, J. P., Monard, G. & Prangé, T. (2004). Acta Cryst. D60, 453–462. [DOI] [PubMed] [Google Scholar]
- Retailleau, P., Colloc’h, N., Vivarès, D., Bonneté, F., Castro, B., El Hajji, M. & Prangé, T. (2005). Acta Cryst. D61, 218–229. [DOI] [PubMed] [Google Scholar]
- Vivarès, D. & Bonneté, F. (2002). Acta Cryst. D58, 472–479. [DOI] [PubMed] [Google Scholar]
- Vivarès, D., Veesler, S., Astier, J.-P. & Bonneté, F. (2006). Cryst. Growth Des.6, 287–292.