The class IV adenylyl cyclase from Y. pestis has been crystallized in an orthorhombic form suitable for structure determination.
Keywords: adenylyl cyclase, cyclic AMP, selenomethionine
Abstract
The class IV adenylyl cyclase from Yersinia pestis has been cloned and crystallized in both a triclinic and an orthorhombic form. An amino-terminal His-tagged construct, from which the tag was removed by thrombin, crystallized in a triclinic form diffracting to 1.9 Å, with one dimer per asymmetric unit and unit-cell parameters a = 33.5, b = 35.5, c = 71.8 Å, α = 88.7, β = 82.5, γ = 65.5°. Several mutants of this construct crystallized but diffracted poorly. A non-His-tagged native construct (179 amino acids, MW = 20.5 kDa) was purified by conventional chromatography and crystallized in space group P212121. These crystals have unit-cell parameters a = 56.8, b = 118.6, c = 144.5 Å, diffract to 3 Å and probably have two dimers per asymmetric unit and V M = 3.0 Å3 Da−1. Both crystal forms appear to require pH below 5, complicating attempts to incorporate nucleotide ligands into the structure. The native construct has been produced as a selenomethionine derivative and crystallized for phasing and structure determination.
1. Introduction
Adenylyl cyclase (AC) is the ubiquitous enzyme (EC 4.6.1.1) responsible for the synthesis from ATP of the signaling molecule cyclic adenosine 3′,5′-monophosphate (cAMP), which regulates key functions in virtually all cells. Six classes of ACs have been identified to date, representing different gene families and possessing widely different properties (Table 1 ▶). Class I ACs are soluble proteins in enteric bacteria such as Escherichia coli (Yang & Epstein, 1983 ▶) and the plague pathogen Yersinia pestis (Kim & Reddy, unpublished work). The E. coli class I AC is regulated by protein components of the sugar-transport system (Peterkofsky et al., 1993 ▶).
Table 1. Representatives of the six classes of adenylyl cyclase enzymes.
In most cases, the listed PDB crystal structures represent only the cyclase domain. Also, in most cases where crystal structures exist the listed PDB entry is only one of several closely related structures. TM, transmembrane. For references, see §1.
| Class | Source | Subunit size (kDa) | Soluble? | Oligomer | PDB code |
|---|---|---|---|---|---|
| I | E. coli | 97 | Soluble | Monomer | — |
| Y. pestis | 97 | Soluble | Monomer | — | |
| II | B. anthracis | 92 | † | Monomer | 1k8t |
| B. pertussis | 188 | † | Monomer | 1yru | |
| P. aeruginosa | 43 | Soluble | Monomer | — | |
| III | Mammalian (9 isoforms) | 120–150 | TM | Monomer‡ | 1ab8, 1azs |
| Mammalian (1 isoform) | 187 | Soluble | ? | — | |
| Trypanosome | 138 | TM | Monomer§ | 1fx2 | |
| M. tuberculosis (Rv1264) | 42 | Soluble | Homodimer | 1y10 | |
| M. tuberculosis (Rv1625c) | 47 | TM | Homodimer | — | |
| M. tuberculosis (Rv1900c) | 50 | ? | Homodimer | 1ybu | |
| S. platensis | ∼134 | Soluble | Homodimer | 1wc3 | |
| IV | A. hydrophila | 20 | Soluble | ? | — |
| Y. pestis | 20 | Soluble | Homodimer | — | |
| P. furiosus | 20 | ? | ? | — | |
| V | P. ruminicola | ∼67 | ? | ? | — |
| VI | R. etli | ∼38 | ? | ? | — |
These toxin ACs are initially soluble, then become anchored in the host-cell membranes.
Two nonidentical cytosolic domains interact to form a pseudodimeric catalytic site.
Thought to undergo regulation-dependent dimerization.
Class II ACs are also soluble proteins of prokaryotic origin. They are secreted by pathogens such as Bacillus anthracis (Baillie & Read, 2001 ▶), Bordetella pertussis (Ladant & Ullman, 1999 ▶) and Pseudomonas aeruginosa (Yahr et al., 1998 ▶). Upon translocation into the host cells, B. anthracis AC (EF, edema factor) and B. pertussis AC (cyaA) become integrated into the host-cell membrane (Guermonprez et al., 2001 ▶) and are activated by the host calmodulin. The P. aeruginosa AC (ExoY) is not activated by calmodulin, but is activated by a different factor from the host-cell cytosol (Yahr et al., 1998 ▶). The activation of class II ACs by host factors results in pathologically high concentrations of cAMP in the infected host cells.
Class III ACs are found in eukaryotes, including mammals (Sunahara et al., 1996 ▶; Litvin et al., 2003 ▶) and protozoa (Bieger & Essen, 2001 ▶), and also in some prokaryotes such as Mycobacterium tuberculosis (Guo et al., 2001 ▶; Reddy et al., 2001 ▶) and the cyanobacterium Spirulina platensis (Kasahara et al., 2001 ▶). Ten isoforms of class III ACs in mammals have been identified, of which nine are transmembrane proteins and one is soluble (Hanoune & Defer, 2001 ▶). This class of enzymes also includes both soluble and membrane-bound guanylyl nucleotide cyclases (Barbara & Garbers, 1998 ▶). The nine mammalian transmembrane ACs comprise two modules, each composed of a hydrophobic domain with six transmembrane helices and a cytosolic domain (Krupinski et al., 1989 ▶). The two heterologous cytosolic domains associate to form a single pseudodimeric catalytic site in the monomeric protein. The M. tuberculosis class III AC (Rv1625c), however, has only one module (composed of a hydrophobic domain with six transmembrane helices and a cytosolic domain) but functions as a homodimer with two catalytic sites (Guo et al., 2001 ▶). Among the class III ACs, four subclasses have been identified based on a diverse set of additional modules that regulate the adenylyl cyclase domains (Linder & Schultz, 2003 ▶).
Class IV ACs are found (along with class I enzymes) in the bacterial species Aeromonas hydrophila (Sismeiro et al., 1998 ▶) and Yersinia pestis (this work) and are also represented by homologous sequences (∼30% identical) in archaea. The class IV enzymes are soluble and encode about 180 residues, making them the smallest naturally occurring AC enzymes yet characterized. Class V AC was found in the anaerobic bacterium Prevotella ruminicola (Cotta et al., 1998 ▶) and class VI AC was found in the nitrogen-fixing bacterium Rhizobium etli (Téllez-Sosa et al., 2002 ▶).
Representatives of classes II and III have been analyzed by X-ray crystallography. The two structurally characterized AC classes II and III have distinct protein architectures. The three-dimensional structures of the class II AC from B. anthracis with and without calmodulin (PDB code 1k8t; Drum et al., 2002 ▶) and B. pertussis (PDB code 1yru; Guo et al., 2005 ▶) have been determined. These structures provide a classic example of divergence among homologous bacterial toxins in their manner of binding calmodulin. The catalytic core of the mammalian class III AC, which is formed by intramolecular association of the two cytosolic domains, has been described in two forms (PDB code 1ab8, Zhang et al., 1997 ▶; PDB code 11azs, Tesmer et al., 1997 ▶). Structures of two isozymes of trypanosomal class III ACs have been described (Bieger & Essen, 2001 ▶). Of the prokaryotic class III ACs, the structures of M. tuberculosis Rv1900c AC (Sinha et al., 2005 ▶) and a pH-sensing Rv1264 AC (Tews et al., 2005 ▶) provide examples of modulator diversity in M. tuberculosis class III ACs, while the structures of calcium- and bicarbonate-bound cyanobacterial class III AC reveal distinct catalytic states of the enzyme (Steegborn et al., 2005 ▶).
Class IV ACs are distinct from the other classes in size and in thermostability and appear to be unrelated by sequence to any proteins of known function. Bioinformatic analysis suggests an ancient kinship to thiamine triphosphatase and to a novel phosphate-binding superfamily (Iyer & Aravind, 2002 ▶). The Y. pestis genome sequence revealed two open reading frames for AC, one for a class I AC (YpAC1) and the other for a class IV AC (YpAC2) (Deng et al., 2002 ▶). We have expressed and characterized the class IV AC for structural investigation. The native protein has a length of 179 amino acids; its predicted molecular weight is 20 316 Da and its predicted pI is 4.9. We report here the purification and crystallization of the class IV AC from Y. pestis.
2. Materials and methods
2.1. Cloning, expression and purification of His-tagged YpAC2
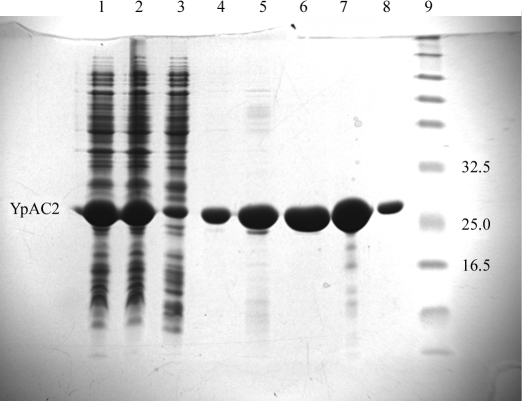
The gene encoding the class IV AC from Y. pestis (KIM 10+) genomic DNA was amplified by PCR using the forward primer 5′-G GAA TTC CAT ATG AGT GAG CAT TTT GTT G (NdeI restriction recognition sequence in bold) and the reverse primer 5′-CCG CTC GAG CTA AAA ACC CAA TAG TTG CCG (XhoI restriction recognition sequence in bold). 200 ng of Y. pestis chromosomal DNA (kindly provided by Dr Robert D. Perry, University of Kentucky) and 100 ng of primers were used in the amplification. Amplification conditions were 368 K for 60 s for initial melting of DNA, followed by 30 cycles of amplification, each cycle consisting of melting at 368 K for 45 s, annealing at 333 K for 60 s and polymerization at 345 K for 60 s. Polymerization was then continued for 10 min at 345 K. The PCR product was purified, digested with NdeI and XhoI restriction enzymes and cloned into a similarly digested pET15b vector. The recombinant plasmid was isolated from E. coli NovaBlue cells (Novagen, Madison, WI, USA) and introduced into E. coli BL21(DE3) cells for protein expression. Cells were grown at 310 K in LB medium containing ampicillin (50 µg ml−1) to A 600 ≃ 0.6. The culture was then cooled to 288 K and induced with 30 µM IPTG at 288 K for 16 h. The cells were collected by centrifugation and washed and 10 ml lysis buffer (50 mM Tris–HCl pH 8.0, 10 mM β-mercaptoethanol, 50 mM NaCl) was added per gram of wet cell pellet. The cell suspension was passed twice through a French press and the crude homogenate was centrifuged at 20 000g for 40 min. The supernatant was mixed with 1 ml Ni–NTA resin (Qiagen, Valencia, CA, USA) for 30 min at 277 K. The resin was poured into a column, washed successively with 20 ml of the lysis buffer, I10 buffer, I20 buffer and eluted twice with 15 ml I200 buffer (I10, I20 and I200 buffers are lysis buffer containing imidazole at 10, 20 or 200 mM, respectively). Most of the YpAC2 was eluted with I200 buffer and exceeded 98% purity. The I20 wash and I200 elution were pooled and imidazole was removed by dialysis overnight against 20 mM Tris–HCl buffer pH 8.0. The histidine tag was cleaved off from the protein by digestion with thrombin according to the manufacturer’s recommendation (Novagen, Madison, WI, USA) and the tag was removed by passing the protein through Ni–NTA. However, the resulting protein construct retains an extra Gly-Ser-His at the amino-terminus. Reaction components of thrombin digestion were removed by dialysis overnight against 20 mM Tris–HCl buffer pH 8.0, 1 mM DTT and the protein was concentrated in a Millipore centrifugal tube (Millipore Corporation, Bedford, MA, USA) to 30 mg ml−1. The purification profile of the His-tagged YpAC2 is presented in Fig. 1 ▶.
Figure 1.
15% SDS–PAGE analysis of the purification of YpAC2 on a Ni–agarose column. Cells from a 2 l culture (5 g wet weight) were suspended in 50 ml of the lysis buffer. Lane 1, 2 µl French press extract; lane 2, 2 µl 20 000g supernatant; lane 3, 2 µl flowthrough on Ni–NTA resin; lane 4, 8 µl wash with lysis buffer (no imidazole); lane 5, 8 µl wash with 10 mM imidazole; lane 6, 8 µl (16 µg protein) wash with 20 mM imidazole; lane 7, 2.5 µl (16 µg protein) eluate with 200 mM imidazole, fraction 1; lane 8, 2.5 µl (2.8 µg protein) eluate with 200 mM imidazole, fraction 2; lane 9, molecular-weight markers. Relevant molecular-weight markers (kDa) are labeled. YpAC2 is indicated.
2.2. Cloning, expression and purification of native YpAC2
The gene was amplified as described above. For native expression of the protein, the gene was cloned into the pET11a vector (Novagen, Madison, WI, USA). The pET11a vector was first digested with BamHI restriction endonuclease, filled in with the Klenow polymerase to make blunt ends and then digested with NdeI and treated with phosphatase. The PCR product with 5′ NdeI end and 3′ blunt end was inserted into the vector. Native protein was expressed as described above. Cells from 1 l culture (3 g) were suspended in 30 ml buffer A (50 mM Tris–HCl buffer pH 7.5 containing 1 mM EDTA) and passed twice through a French press. The homogenate was centrifuged at 20 000g for 30 min and the supernatant was loaded onto a 40 ml bed volume of DE52 anion-exchange resin (Whatman Laboratories). The column was washed with 50 ml buffer A and the protein was eluted with an NaCl gradient generated from 300 ml buffer A and 300 ml buffer A containing 0.4 M NaCl. Fractions containing YpAC2, as judged by SDS–PAGE, were pooled and concentrated to ∼10 ml. YpAC2 in the DE52 concentrate was further purified on a 500 ml Sephadex G-75 column equilibrated with buffer B (buffer A + 100 mM NaCl). Again, fractions were pooled for YpAC2 and concentrated. YpAC2 was ∼ 90% pure at this stage. Comparison with molecular-weight standards revealed that YpAC2 elutes at the position expected for a dimer. The protein was further purified on an Affigel Blue matrix. YpAC2 from the G-75 column was mixed with 5 ml Affigel Blue for 1 h and centrifuged to sediment the resin. The supernatant contained YpAC2 which was nearly homogeneous. The yield of YpAC2 from 1 l culture was 40 mg protein. The selenomethionine derivative of native YpCA2 was purified similarly, except that the protein was overproduced in an E. coli methionine auxotroph B834 (DE3). Cells were grown in LB medium to an A 600 of ∼0.6, harvested, washed with sterile water and suspended in minimal medium containing selenomethionine.
2.3. Crystallization and diffraction
The His-tagged form of the protein was prepared for crystallization by concentration to 30 mg ml−1 (0.7 mM dimer) in a buffer comprising 20 mM Tris–HCl pH 8.0, 1 mM DTT. Initial screening for crystal growth by vapor diffusion was attempted with various commercial screens at both room temperature (RT; ∼297 K) and 277 K using hanging drops. Several RT conditions involving pH between 4 and 5 gave clusters of thin triclinic plates (Fig. 2 ▶). The volume of the unit cell (78 300 Å3) is expected to accommodate one AC dimer with V M = 1.9 Å3 Da−1 (Matthews, 1968 ▶). No other crystal forms appeared despite extensive screening. The conditions were optimized to 150 mM ammonium sulfate, 100 mM sodium acetate pH 4.6, 12%(w/v) PEG 4K and microseeding was used to produce single crystals that could be grown to a thickness of 25 µm. One such crystal yielded a 1.9 Å data set using an R-AXIS IV rotating-anode diffractometer. Using a 0.5° scan width, 390 images were collected with a crystal-to-detector distance of 130 mm and exposure times of 6 min. These images were reduced by CrystalClear software to give the data set described in Table 2 ▶. Attempts to produce heavy-atom derivatives of these crystals for phasing were hampered by protein sensitivity, crystal fragility, nonisomorphism and poor binding, which are probably a consequence of the low pH.
Figure 2.
Photograph of two typical clusters of self-nucleated triclinic thin-plate crystals. Crystal thicknesses are about 4 µm. The crystal that gave the data set in Table 2 ▶ was similar but was cultivated to a thickness of 25 µm. Analysis of morphology and diffraction patterns indicates that the large slow-growing faces of these crystals correspond to the (001) plane.
Table 2. Statistics for AC class IV triclinic diffraction data set.
Data were processed using CrystalClear. Values in parentheses refer to the highest resolution shell.
| Space group | P1 |
| Unit-cell parameters (Å, °) | a = 33.49, b = 35.49, c = 71.79, α = 88.72, β = 82.49, γ = 65.50 |
| No. of observations | 42916 |
| No. of unique reflections | 20986 |
| Resolution (Å) | 20.0–1.90 (1.97–1.90) |
| Redundancy | 2.04 (2.04) |
| Coverage (%) | 89.3 (83.9) |
| Rmerge | 0.026 (0.170) |
| 〈I/σ(I)〉 | 15.7 (3.9) |
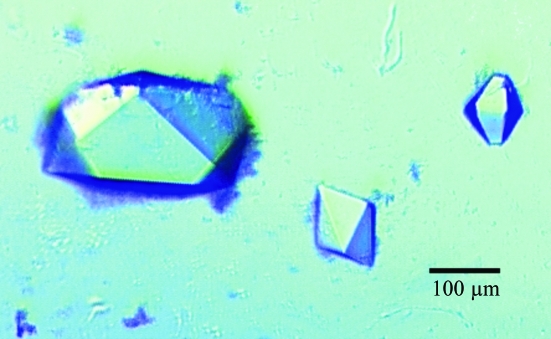
Native protein was prepared for crystallization by concentration to 30 mg ml−1 (0.7 mM dimer) in 20 mM Tris–HCl pH 7.5, 1 mM DTT. Screening revealed a single crystal form from low-pH and high-phosphate conditions that were optimized to 1.0 M sodium phosphate pH 4.2 at RT. These crystals usually grew singly by self-nucleation but behaved poorly in seeding. They are orthorhombic and grew to dimensions of 0.1–0.3 mm (Fig. 3 ▶). Using 2 M xylose as cryoprotectant (a 5 s dunk in an equal mix of 4 M xylose with reservoir solution), these crystals occasionally diffracted to 2.9 Å. They belong to space group P212121, with unit-cell parameters a = 56.8, b = 118.6, c = 144.5 Å. The resulting unit-cell volume leads to the prediction of four chains (two dimers) per asymmetric unit with V M = 3.0 Å3 Da−1 (Matthews, 1968 ▶).
Figure 3.
Photograph of orthorhombic crystals of the native enzyme. The faces of these crystals index are (110) and (011). The longest dimension of the largest crystal is 350 µm. The higher symmetry and easier cultivation of these crystals makes them preferable for structure determination despite their weaker diffraction.
3. Results and discussion
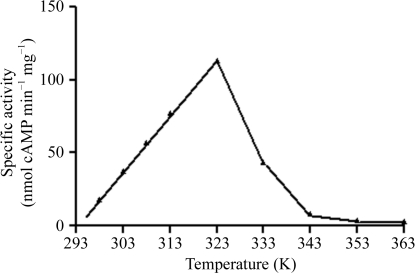
Adenylyl cyclases belonging to six identified classes serve key signaling and regulatory functions in all forms of life. Only two of the six classes (classes II and III) have been structurally characterized to date. We report here the crystallization of YpAC2 from Y. pestis, the first such report for a class IV AC. The enzyme as expressed in E. coli constitutes ∼20% of the total cellular protein and is soluble (Fig. 1 ▶). Purification of the native protein on a Sephadex G-75 (superfine) column showed the migration behavior of the protein as a dimer with a molecular weight of ∼40 kDa, equivalent to two subunits [the column was calibrated with bovine serum albumin (67 kDa), ovalbumin (43 kDa), carbonic anhydrase (30 kDa), trypsinogen (24 kDa) and cytochrome c (11 kDa); data not shown]. Unlike previously characterized ACs, which have maximum activity between 303 and 310 K, YpAC2 was found to have maximum activity near 323 K (Fig. 4 ▶), consistent with its close homology to sequences from thermophilic archaeal organisms (Sismeiro et al., 1998 ▶). YpAC2 was purified in two forms, His-tagged (Fig. 1 ▶) and native (data not shown), resulting in crystals of two forms with diffraction to 1.9 and 2.9 Å, respectively, and reasonable prospects for determination of the protein structure.
Figure 4.
Adenylyl cyclase activity analysis of the purified YpAC2 as a function of temperature. Assays using radiolabeled ATP substrate and monitoring formation of labeled cAMP were as described by Reddy et al. (2001 ▶).
The His-tagged YpAC2 was digested with thrombin to remove the His tag, which would leave the three residues Gly, Ser, His as a non-native N-terminal extension to the protein. Subsequent analysis by mass spectrometry revealed an unexpected thrombin-sensitive site after Arg171, resulting in a few percent content of a truncated protein with an apparent weight of 19.6 kDa along with the main peak at 20.6 kDa. The sequence at this site is EPR|SYR, differing from the conventional thrombin specificity. Mass-spectrometric analysis of crystals indicated that they contained only the full-length protein (20.6 kDa species). Addition of the thrombin inhibitor PPACK at 10 µM subsequent to the removal of the His tag protected the spurious site, but it was found that the PPACK-protected full-length protein produced inferior crystals unless a small amount of PPACK-untreated protein was added, suggesting that a low concentration of the truncated protein was necessary for optimal crystal growth. This development complicated efforts to cultivate reproducible crystals. Four variants of the His-tagged protein with mutations at conserved charged sites, produced for functional studies, were also screened for crystallization. Mutants E10A, E84A and E136A all gave crystals that were unsuitable owing to poor crystal growth or poor diffraction, while D143A did not crystallize.
The native construct was expressed using selenomethionine (there are six Met residues per protein subunit) and the orthorhombic crystals were reproduced. The SeMet crystals diffracted similarly to native crystals. Several such crystals were cryotreated, frozen by immersion in liquid nitrogen and shipped to Brookhaven National Laboratories for collaborative mail-in diffraction measurement to provide phasing to solve the structure in the orthorhombic form. If successful, the resulting structure will be transformed into the previously collected triclinic form in which diffraction data extend to 1.9 Å for refinement at this resolution.
ACs perform key functions in nearly all cells and the physiological significance of class II ACs as toxins from B. anthracis and B. pertussis is well established. In a number of other bacterial pathogens, cAMP has been shown to be involved in the virulence of the organisms. Y. pestis is the causative agent of the notorious historical bubonic and pneumonic plague epidemics. Antibiotics are effective against Y. pestis, but only if administered within 1–3 d of infection; thus, this organism is categorized as a class A pathogen. The existence of two classes of ACs, class I and class IV, in Y. pestis is intriguing. We have begun to characterize these ACs at the molecular level and to explore the role of the ACs in the virulence of Y. pestis.
Acknowledgments
The authors gratefully acknowledge the gift of Y. pestis chromosomal DNA from Dr Robert D. Perry, University of Kentucky, the collaboration and support of the Structural Biology Group at Brookhaven National Laboratories National Synchrotron Light Source and the logistical assistance of Sheila Williams of the Center for Advanced Research in Biotechnology. Disclaimer: identification of specific instruments and products in this paper is solely to describe the scientific procedures and does not imply recommendation or endorsement.
References
- Baillie, L. & Read, T. D. (2001). Curr. Opin. Microbiol.4, 78–81. [DOI] [PubMed] [Google Scholar]
- Barbara, J. W. & Garbers, D. L. (1998). Trends Endocrinol. Metab.9, 213–219. [DOI] [PubMed]
- Bieger, B. & Essen, L. (2001). EMBO J.20, 433–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotta, M. A., Whitehead, T. R. & Wheeler, M. B. (1998). FEMS Microbiol. Lett.164, 257–260. [DOI] [PubMed] [Google Scholar]
- Deng, W. et al. (2002). J. Bacteriol.184, 4601–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drum, C. L., Yan, S.-Z., Bard, J., Shen, Y.-Q., Lu, D., Soelaiman, S., Grabarek, Z., Bohm, A. & Tang, W.-J. (2002). Nature (London), 415, 396–402. [DOI] [PubMed] [Google Scholar]
- Guermonprez, P., Khelef, N., Blouin, E., Rieu, P., Ricciardi-Castagnoli, P. & Guiso, N. (2001). J. Exp. Med.193, 1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Y. L., Seebacher, T., Kurz, U., Linder, J. U. & Schultz, J. E. (2001). EMBO J.20, 3667–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Q., Shen, Y., Lee, Y. S., Gibbs, C. S., Mrksich, M. & Tang, W.-J. (2005). EMBO J.24, 3190–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanoune, J. & Defer, N. (2001). Annu. Rev. Pharmacol. Toxicol.41, 145–174. [DOI] [PubMed] [Google Scholar]
- Iyer, L. M. & Aravind, L. (2002). BMC Genomics, 3, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara, M., Unno, T., Yashiro, K. & Ohmori, M. (2001). J. Biol. Chem.276, 10564–10569. [DOI] [PubMed] [Google Scholar]
- Krupinski, J., Coussen, F., Bakalyar, H. A., Tang, W.-J., Feinstein, P. G., Orth, K., Slaughter, C., Reed, R. R. & Gilman, A. G. (1989). Science, 244, 1558–1564. [DOI] [PubMed] [Google Scholar]
- Ladant, D. & Ullman, A. (1999). Trends Microbiol.7, 172–176. [DOI] [PubMed] [Google Scholar]
- Linder, J. U. & Schultz, J. E. (2003). Cell. Signal.15, 1081–1089. [DOI] [PubMed] [Google Scholar]
- Litvin, T. N., Kamenetsky, M., Zarifyan, A., Buck, J. & Levin, L. R. (2003). J. Biol. Chem.278, 15922–15926. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Peterkofsky, A., Reizer, A., Reizer, J., Gollop, N., Zhu, P. P. & Amin, N. (1993). Prog. Nucleic Acids Res. Mol. Biol.44, 31–65. [DOI] [PubMed]
- Reddy, S. K., Kamireddi, M., Danireddi, K., Young, L., Davis, A. & Reddy, P. T. (2001). J. Biol. Chem.276, 35141–35149. [DOI] [PubMed] [Google Scholar]
- Sinha, S. C., Wetterer, M., Sprang, S. R., Schultz, J. E. & Linder, J. U. (2005). EMBO J.24, 663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sismeiro, O., Trotot, P., Biville, F., Vivares, C. & Danchin, A. (1998). J. Bacteriol.180, 3339–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steegborn, C., Litvin, T., Levin, L., Buck, J. & Wu, H. (2005). Nature Struct. Mol. Biol.12, 32–37. [DOI] [PMC free article] [PubMed]
- Sunahara, R. K., Dessauer, C. W. & Gilman, A. G. (1996). Annu. Rev. Pharmacol. Toxicol.36, 461–480. [DOI] [PubMed] [Google Scholar]
- Téllez-Sosa, J., Soberón, N., Vega-Segura, A., Torres-Márquez, M. E. & Cevallos, M. A. (2002). J. Bacteriol.184, 3560–3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesmer, J. J., Sunahara, R. K., Gilman, A. G. & Sprang, S. R. (1997). Science, 278, 1907–1916. [DOI] [PubMed] [Google Scholar]
- Tews, I., Findeisen, F., Sinning, I., Schultz A., Schultz, J. E. & Linder, J. U. (2005). Science, 308, 1020–1023. [DOI] [PubMed] [Google Scholar]
- Yahr, T. L., Vallis, A. J., Hancock, H. K., Barbieri, J. T. & Frank, D. W. (1998). Proc. Natl Acad. Sci. USA, 95, 13899–13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J. K. & Epstein, W. (1983). J. Biol. Chem.258, 3750–3758. [PubMed] [Google Scholar]
- Zhang, G., Liu, Y., Ruoho, A. E. & Hurley, J. H. (1997). Nature (London), 386, 247–253. [DOI] [PubMed] [Google Scholar]