A cytotoxin from Indian Russell’s viper (D. russelli russelli) venom having multifunctional activity has been crystallized in space group P41. Larger crystals diffracted to 1.5 Å but were found to be twinned; preliminary data were therefore collected (2.93 Å) from a smaller crystal.
Keywords: cytotoxins, Russell’s viper, antiproliferative activity
Abstract
A cytotoxin (MW 7.2 kDa) from Indian Russell’s viper (Daboia russelli russelli) venom possessing antiproliferative activity, cardiotoxicity, neurotoxicity and myotoxicity has been purified, characterized and crystallized. The crystals belong to the tetragonal space group P41, with unit-cell parameters a = b = 47.94, c = 50.2 Å. Larger crystals, which diffracted to 1.5 Å, were found to be twinned; diffraction data were therefore collected to 2.93 Å resolution using a smaller crystal. Molecular-replacement calculations identified two molecules of the protein in the asymmetric unit, which is in accordance with the calculated V M value.
1. Introduction
Snake-venom cytotoxins are typically small water-soluble proteins (60–74 amino-acid residues) that possess a diverse range of pharmacological activities including haemolysis, cardiotoxicity, neurotoxicity, depolarization of the membranes of excitable cells, membrane fusion and selective killing of certain type of tumour cells. Three-dimensional structure determinations of cytotoxins isolated from different venoms, mainly from those of cobras and kraits (Chen et al., 2005 ▶; Dubovskii et al., 2005 ▶; Lee et al., 2005 ▶; Paaventhan et al., 2003 ▶), have indicated that they all possess a common structure, popularly known as the three-fingered motif, which consists of β-sheets connected by four or five disulfide bridges with no helical content (Menez, 1998 ▶; Tsetlin, 1999 ▶). Despite the similarities in their backbone architecture, they show different biological activities even when isolated from the same venom source, which is believed to arise from subtle variations in their three-dimensional structures.
Although the functional profiles of cytotoxins from the venoms of Daboia russelli siamensis, Echis coloratus, Cerastes cerastes etc. from the Viperidae family have been studied, mostly on Yoshida sarcoma cells, KB cells, HeLa cells and EAC cell lines (Tu & Giltner, 1974 ▶; Maung-Maung et al., 1995 ▶; Mady, 2002 ▶; Abu-Sinna et al., 2003 ▶), no three-dimensional structure is presently available for any cytolytic and/or antitumour toxin isolated from a venom from the Viperidae family. Russell’s viper (D. russelli russelli) is one of the most dangerous snakes prevalent in eastern India, accounting for thousands of deaths each year. Envenomation by Russell’s viper causes oedema, haemolysis, haemorrhage, neurotoxicity, cardiotoxicity and kidney failure. Recently, we have purified and sequenced (20 N-terminal residues) a cytotoxin (hereafter referred to as drCT-I) from the venom of D. russelli russelli; its molecular weight was found to be 7.2 kDa. Together with cytotoxicity, drCT-I shows neurotoxicity, cardiotoxicity and myotoxicity. In addition, it exhibits antiproliferative activity in EAC and human cell lines. We have crystallized drCT-I and initiated its three-dimensional structure determination in order to address the diverse pharmacological functions of this protein. In this paper, we present the purification, crystallization and preliminary X-ray structural studies of drCT-I.
2. Purification
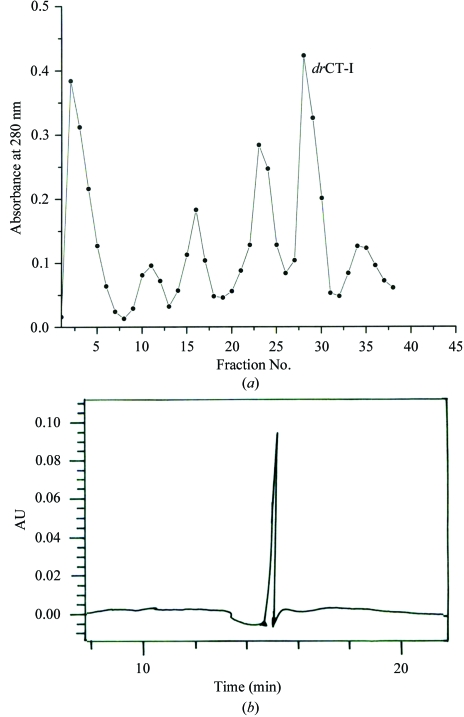
200 mg lyophilized venom was dissolved in 5 ml distilled water and after heat treatment at 353 K for 30 min to precipitate out the high-molecular-weight proteins present in the venom, the solution was centrifuged at 900g for 20 min and the supernatant was loaded onto a CM-cellulose (150 × 25 mm) column equilibrated with 0.02 M phosphate buffer pH 7.2. Fractions were eluted at a flow rate of 25 ml h−1 with a stepwise gradient of NaCl (0.02, 0.05, 0.1, 0.2 and 0.5 M in 0.02 M phosphate buffer pH 7.2). For each NaCl gradient, 30 ml of solution was used and the desired toxin (drCT-I) was obtained at an NaCl concentration of 0.2 M (Fig. 1 ▶ a). The purified drCT-I protein constitutes about 3.75% of the crude venom. The homogeneity of the protein was assessed by re-chromatography (Fig. 1 ▶ b) on a C-18 Nova Pak column (7.2 × 300 mm) equilibrated with 100 mM phosphate buffer pH 7.0 at a flow rate of 0.5 ml min−1 and also by SDS–PAGE (Fig. 2 ▶).
Figure 1.
(a) CM-cellulose chromatogram for the purification of drCT-I using a stepwise gradient of NaCl (0.02, 0.05, 0.1, 0.2 and 0.5 M). The peak containing drCT-I is indicated in the figure and is eluted at an NaCl concentration of 0.2 M. (b) HPLC profile of drCT-I in a C-18 Nova Pak column.
Figure 2.
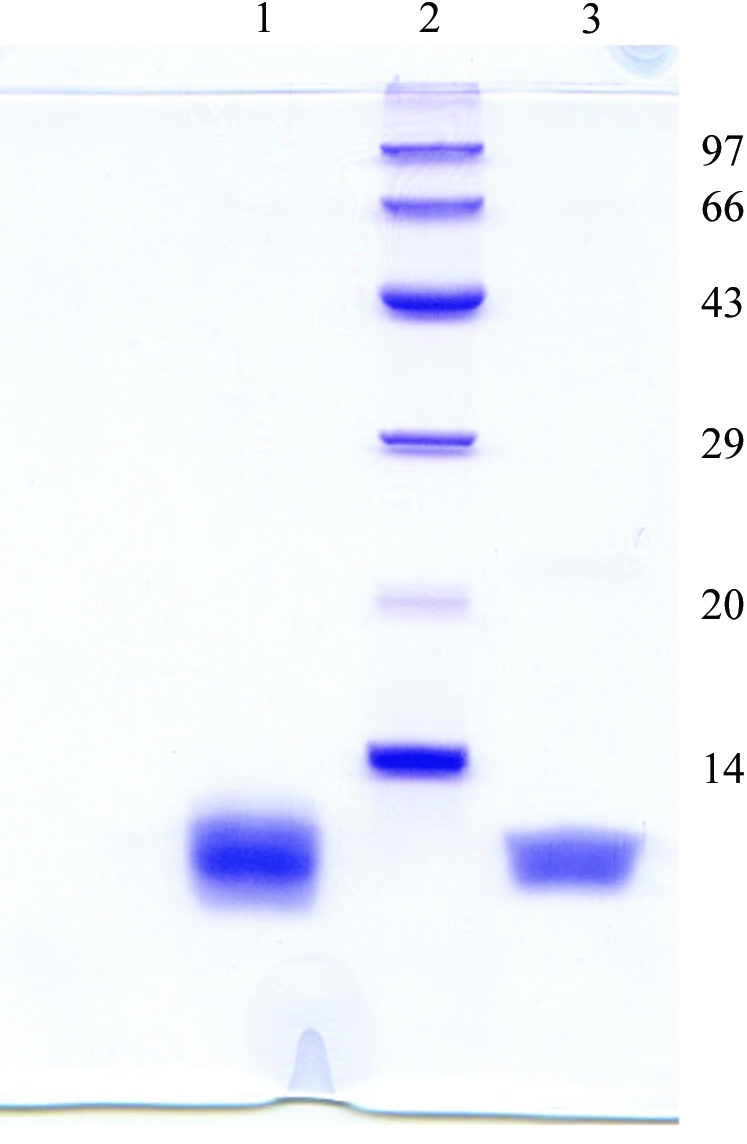
15% SDS–PAGE of drCT-I samples. Lane 1, purified from a CM-cellulose column; lane 3, crystals dissolved in SDS–PAGE sample buffer; lane 2, molecular-weight markers (kDa).
3. Crystallization
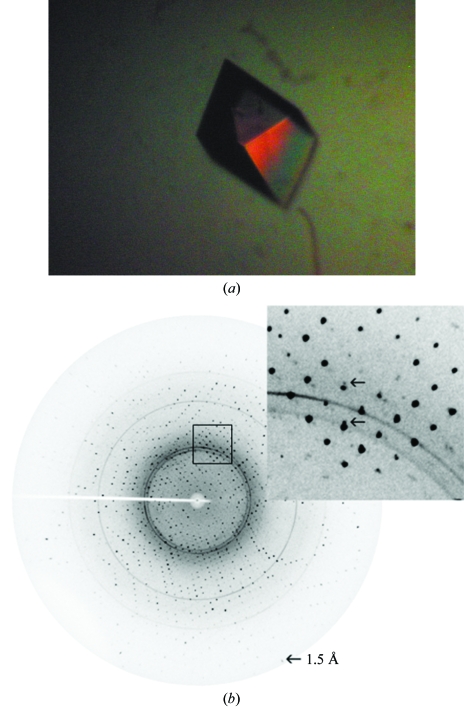
For crystallization, the protein was dialyzed against 10 mM Tris–HCl pH 7.2 containing 0.5 mM CaCl2 and concentrated with Amicon Centriprep centrifugal filtration units (3000 Da molecular-weight cutoff) to 6 mg ml−1. Crystallization was performed by the hanging-drop vapour-diffusion method using 24-well tissue-culture plates and initial trials for crystallizations were carried out using Crystal Screens I and II from Hampton Research. Typically, 2 µl protein solution at 6 mg ml−1 concentration was mixed with an equal volume of the screening solution and equilibrated over 700 µl of the latter as reservoir solution. The best crystals (Fig. 3 ▶ a) were obtained when 2 µl protein solution was mixed with an equal volume of reservoir solution consisting of 0.1 M sodium acetate pH 4.6, 30% PEG 4000, 0.2 M ammonium sulfate and 6% glycerol and equilibrated at 277 K for 10 d.
Figure 3.
(a) Crystal of drCT-I (0.5 × 0.4 × 0.35 mm) grown in PEG 4000 at 277 K. (b) Diffraction image of the crystal of drCT-1; the arrow indicates reflections at 1.5 Å. Inset: a portion of the image (marked in square) is enlarged to show that the reflections are split (shown by arrow).
4. Data collection and processing
Crystals of drCT-I grown in the presence of cryoprotectant were fished out from the crystallization drops and immediately flash-frozen in a stream of nitrogen (Oxford Cryosystem) at 100 K. X-ray diffraction data were collected to 2.93 Å resolution using a MAR Research 345 image-plate detector with Cu Kα radiation generated by a Bruker–Nonius FR591 rotating-anode generator equipped with Osmic MaxFlux confocal optics running at 50 kV and 90 mA. The larger crystals, which diffracted to 1.5 Å, showed a high degree of twinning (Fig. 3 ▶ b), with twin fractions of between 0.25 and 0.35 as calculated using the online test for determining twinning in protein crystals (http://nihserver.mbi.ucla.edu/Twinning; Yeates, 1997 ▶). The smaller crystals diffracted weakly but were found to be less twinned, with twin fractions of ∼0.1. The present data set was collected from such a smaller crystal with a low twin fraction. A total of 72 frames were collected with a crystal-to-detector distance of 290 mm. The exposure time for each image was 2 min and the oscillation range was maintained at 1°. Data were processed and scaled using the program suite AUTOMAR (http://www.marresearch.com/automar/run/htm). Data-collection and processing statistics are given in Table 1 ▶.
Table 1. Data-collection and data-processing parameters.
Valuse in parentheses are for the highest resolution shell.
| Space group | P41 |
| Unit-cell parameters (Å) | a = b = 47.94, c = 50.2 |
| Oscillation range (°) | 1.0 |
| Resolution (Å) | 2.93 (3.03–2.93) |
| No. of molecules per ASU | 2 |
| Matthews coefficient VM (Å3 Da−1) | 1.99 |
| Solvent content (%) | 37.0 |
| No. of observations | 8576 |
| No. of unique reflections | 2276 |
| Mosaicity | 0.38 |
| Completeness (%) | 95.2 (89.8) |
| Rmerge† (%) | 10.8 (24.2) |
| I/σ(I) for highest shell | 2.1 |
R
merge = 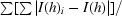
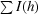 , where I(h)i is the observed intensity of the ith measurement of reflection h and I(h) is the mean intensity of reflection h calculated after scaling.
, where I(h)i is the observed intensity of the ith measurement of reflection h and I(h) is the mean intensity of reflection h calculated after scaling.
5. Results
The crystals belong to the tetragonal space group P41, with unit-cell parameters a = b = 47.94, c = 50.2 Å. The results of molecular-weight determination of drCT-I by MALDI–TOF and ESI–MS consistently produce a molecular weight of ∼7.2 kDa (see supplementary material1). Packing considerations, based on the molecular weight of 7.2 kDa, indicate the presence of two molecules in the asymmetric unit, corresponding to a Matthews coefficient (V M) of 1.99 Å3 Da−1 and a solvent content of 37%. The N-terminal sequence of the protein (20 amino acids only) has been determined and shows highest homology with cobra cardiotoxin A3 (PDB code 1xt3; Lee et al., 2005 ▶). Molecular-replacement calculations were performed by AMoRe (Collaborative Computational Project, Number 4, 1994 ▶) using the coordinates of cobra cardiotoxin A3 (PDB code 1xt3; Lee et al., 2005 ▶) as the search model. Two molecules in the asymmetric unit produced a correlation coefficient of 67.5% and an R factor of 44.9%, with data in the resolution range 10–3.8 Å. Subsequent rigid-body refinement followed by 20 cycles of positional refinement using CNS v.1.0 (Brünger et al., 1998 ▶) further reduced the R factor to 42.3% (R free = 44.3%) with a correlation coefficient of 69.8% using data in the resolution range 10–3.0 Å. Further refinement of this structure and model building using the program O (Jones et al., 1991 ▶) are under way.
Supplementary Material
Supplementary material file. DOI: 10.1107/S1744309106005963/pu5127sup1.pdf
Acknowledgments
SRC and US thank Dr J. Dasgupta and S. Khamrui for their help during crystallization and data collection.
Footnotes
Supplementary material has been deposited in the IUCr electronic archive (Reference: PU5127).
References
- Abu-Sinna, G., Esmat, A. Y., Al-Zahaby, A.-A. S., Soliman, N. A. & Ibrahim, T. M. (2003). Toxicon, 42, 207–215. [DOI] [PubMed] [Google Scholar]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed] [Google Scholar]
- Chen, T. S., Chung, F. Y., Tjong, S. C., Goh, K. S., Huang, W. N., Chien, K. Y., Wu, P. L., Lin, H. C., Chen, C. J. & Wu, W. G. (2005). Biochemistry, 44, 7414–7426. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Dubovskii, P. V., Lesovoy, D. M., Dubinnyi, M. A., Konshina, A. G., Utkin, Y. N., Efremov, R. G. & Arseniev, A. S. (2005). Biochem. J.387, 807–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991). Acta Cryst. A47, 110–119. [DOI] [PubMed] [Google Scholar]
- Lee, S. C., Guan, H. H., Wang, C. H., Huang, W. N., Tjong, S. C., Chen, C. J. & Wu, W. G. (2005). J. Biol. Chem.280, 9567–9577. [DOI] [PubMed] [Google Scholar]
- Mady, E. A. (2002). J. Venom. Anim. Toxins, 8, 283–296.
- Maung-Maung, T., Gopallakrishnakone, P., Yuen, R. & Tan, C. H. (1995). Toxicon, 33, 63–76. [DOI] [PubMed] [Google Scholar]
- Menez, A. (1998). Toxicon, 36, 1557–1572. [DOI] [PubMed] [Google Scholar]
- Paaventhan, P., Joseph, J. S., Nirthanan, S., Rajaseger, G., Gopalakrishnakone, P., Kini, M. R. & Kolatkar, P. R. (2003). Acta Cryst. D59, 584–586. [DOI] [PubMed] [Google Scholar]
- Tsetlin, V. I. (1999). Eur. J. Biochem.264, 281–286. [DOI] [PubMed] [Google Scholar]
- Tu, A. & Giltner, J. B. (1974). Res. Commun. Chem. Pathol. Pharmacol.9, 783–786. [PubMed] [Google Scholar]
- Yeates, T. O. (1997). Methods Enzymol.276, 344–358. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material file. DOI: 10.1107/S1744309106005963/pu5127sup1.pdf