Abstract
Identifying the genes that underlie phenotypic variation in natural populations is a central objective of evolutionary genetics. Here, we report the identification of the gene and causal mutation underlying coat colour variation in a free-living population of Soay sheep (Ovis aries). We targeted tyrosinase-related protein 1 (TYRP1), a positional candidate gene based on previous work that mapped the Coat colour locus to an approximately 15 cM window on chromosome 2. We identified a non-synonymous substitution in exon IV that was perfectly associated with coat colour. This polymorphism is predicted to cause the loss of a cysteine residue that is highly evolutionarily conserved and likely to be of functional significance. We eliminated the possibility that this association is due to the presence of strong linkage disequilibrium with an unknown regulatory mutation by demonstrating that there is no difference in relative TYRP1 expression between colour morphs. Analysis of this putative causal mutation in a complex pedigree of more than 500 sheep revealed almost perfect co-segregation with coat colour (χ2-test, p<0.0001, LOD=110.20), and very tight linkage between Coat colour and TYRP1 (LOD=29.50).
Keywords: coat colour, TYRP1, causal mutation, association mapping, linkage mapping
1. Introduction
Understanding the molecular basis of phenotypic variation in natural populations remains a fundamental, yet elusive, goal of evolutionary and developmental genetics (Erickson et al. 2004; Fitzpatrick et al. 2005; Slate 2005; Vasemagi & Primmer 2005). Fortunately, the increasing availability of genomics resources and cost-effective molecular profiling methods have enhanced the prospects of identifying genes that underlie phenotypic variation. In particular, colour variation in wild vertebrates has proved to be a tractable system for gene discovery studies (reviewed in Hoekstra 2006a), especially in birds (e.g. Theron et al. 2001; Mundy 2005), rodents (e.g. Hoekstra & Nachman 2003; Nachman et al. 2003; Hoekstra et al. 2006b) and reptiles (e.g. Rosenblum et al. 2004; Rosenblum 2006). One notable discovery from these studies is the repeated involvement of one gene, the melanocortin 1 receptor (MC1R), suggesting that parallel evolutionary changes can have a common underlying genetic cause (Mundy et al. 2004). The involvement of a single gene in colour polymorphism across species also indicates that a candidate gene approach may be suitable for gene discovery, particularly as the genetic pathways responsible for pigmentation are well established in the mouse model, and appear to be conserved across vertebrates (Bennett & Lamoreux 2003). The aim of this study was to identify the genetic basis of a stable colour polymorphism in an intensively studied vertebrate population for which data on individual fitness have been collected.
Coat colour in the free-living population of Soay sheep on St Kilda is either dark or light (figure 1), depending on inheritance of a single autosomal gene with two alleles where dark is completely dominant (Doney et al. 1974; Ryder et al. 1974; Coltman & Pemberton 2004). The ratio of dark to light sheep in the study population in Village Bay, Hirta is approximately 3 : 1. This polymorphism is interesting from an evolutionary perspective because it is temporally stable (Milner et al. 2004), in spite of periodic population crashes that occur as a consequence of resource limitation, parasitic infection and adverse winter weather (Coulson et al. 2001; Clutton-Brock & Pemberton 2004). Previous studies have suggested that there is differential over-winter survival between colour morphs; dark coats are generally favoured, but in some years mortality selection favours light females, or there is no difference in survival (Milner et al. 1999). Other studies have demonstrated that dark sheep are significantly heavier than light sheep (Clutton-Brock et al. 1997), and that body weight is genetically correlated with resistance to gastrointestinal nematodes (Coltman et al. 2001). This is likely to explain the generally superior survival of dark sheep, but it fails to account for the occasional reversals in favour of light females (Milner et al. 2004). It is also unclear why dark and light sheep should differ in weight. Identifying the gene and causal mutation underlying coat colour variation in Soay sheep would help to address these questions and determine how genetic variation for this trait is maintained.
Figure 1.
Dark (left) and light (right) coat colour morphs in Soay sheep.
We have previously established that the Coat colour locus in Soay sheep maps to an approximately 15 cM window on chromosome 2, where it co-segregates with the microsatellite BMS678 (Beraldi et al. 2006). No candidate pigmentation genes have been mapped to this chromosome in sheep, but tyrosinase-related protein 1 (TYRP1) is tightly linked to BMS678 on cattle chromosome 8 (data not shown), which is syntenic to sheep chromosome 2 (Slate et al. 2002). Independent of the genome scan described in Beraldi et al. (2006), we tested for association between coat colour in approximately 20 sheep and single nucleotide polymorphisms (SNPs) within intronic regions of five well-characterized candidate pigmentation genes (agouti signalling protein, ASIP; dopachrome tautomerase, DCT; melanocortin 1 receptor, MC1R; micropthalmia transcription factor, MITF; TYRP1; electronic supplementary material). This analysis revealed a significant association between coat colour and TYRP1 in Soay sheep. TYRP1 is therefore a strong positional candidate gene for colour variation in Soay sheep.
The role of TYRP1 in the vertebrate melanin synthesis pathway has been well characterized in mice (Silvers 1979; Bennett & Lamoreux 2003). TYRP1 is a type I membrane-bound protein that is expressed in both melanocytes and the retinal epithelium, where it is involved in the distal eumelanic pathway. Specifically, it acts in concert with DCT to convert dopaquinone, the common precursor of both eumelanin (dark pigment) and pheomelanin (light pigment), into eumelanin (Bennett & Lamoreux 2003). It also plays a role in stabilizing tyrosinase (TYR), which is the critical and rate-determining enzyme in melanogenesis (Kobayashi et al. 1998). Mutations in TYRP1 have been associated with albinism in humans (Boissy et al. 1996) and with colour variation in a number of domestic mammals, including mice (Zdarsky et al. 1990), cow (Berryere et al. 2003) and cat (Schmidt-Kuntzel et al. 2005), but not sheep; the only described mutation known to affect coat colour in domestic sheep is in the MC1R gene (Vage et al. 1999). Studies in mice have shown that mutations in TYRP1 can alter the density of eumelanin pigment deposited in melanosomes, leading to a dilution in overall coloration (e.g. Zdarsky et al. 1990). This is consistent with both studies in domestic sheep, where light coat colour is known to be due to a decrease in the ratio of eumelanin to pheomelanin, relative to black coat colour (Aliev et al. 1990), and the phenotype of hairs in Soay sheep. Indeed, as early as the 1970s, Ryder et al. (1974) suggested coat colour variation in Soays was attributable to the Brown locus, which in the mouse model was later shown to be TYRP1 (Jackson 1988). Coat colour variation in Soay sheep could be due to polymorphism in the coding sequence of TYRP1 or to the presence of a regulatory mutation. In this study, we characterized sequence diversity in the TYRP1 coding sequence and quantified relative expression of TYRP1 in the two coat colour morphs in order to determine whether TYRP1 is responsible for dark–light coat colour variation in Soay sheep.
2. Material and methods
(a) The study population
Soay sheep are the survivors of the earliest domestic sheep that spread across Europe in the Bronze Age (Clutton-Brock & Pemberton 2004). The study population in Village Bay, Hirta (Scotland, UK, 57°49′ N, 8°34′ W) was founded in 1932 with the introduction of 107 sheep from the neighbouring island of Soay and has existed unmanaged ever since. The Village Bay population has been studied on an individual basis since 1985. Each spring in excess of 95% of lambs born in the 230 ha study area have been caught, tagged, sampled for genetic analysis and phenotyped for coat colour (Clutton-Brock & Pemberton 2004).
(b) RNA isolation and cDNA synthesis
Isolation of RNA and synthesis of cDNA was undertaken to provide template for quantitative PCR and sequence analysis of the TYRP1 coding region (see §2c). We collected ear punches from dark and light Soay sheep into RNAlater solution (Ambion) during the summer of 2004. Total RNA from six dark and six light individuals was isolated using a standard Trizol (Invitrogen) protocol, following initial disruption of ear tissues using a TissueLyser (Qiagen) and then further homogenization using Qiagen QIAshredder columns. Total RNA was purified using Qiagen RNeasy columns and was quantified using a Agilent BioAnalyser. Complementary DNA (cDNA) was generated from 1 to 2 μg total RNA by reverse transcription using oligo(dT) primers (Ambion RETROscript). All procedures followed the manufacturers' instructions.
(c) Sequence diversity in the TYRP1 coding region
To determine whether coat colour variation in Soay sheep is due to polymorphism in the TYRP1 coding region, we amplified a 1611 bp fragment comprising exons II–VIII from cDNA using the polymerase chain reaction (PCR) in four overlapping fragments: (i) TYRP1-cDNA-F2b and TYRP1-cDNA-R3, (ii) TYRP1-cDNA-F2c and TYRP1-cDNA-R4a, (iii) TYRP1-cDNA-F3 and TYRP1-cDNA-R6, and (iv) TYRP1-cDNA-F5 and TYRP1-cDNA-R8 (table 1). Where possible primers were designed to traverse exon–exon boundaries, to ensure that they would not anneal to genomic DNA. PCR was performed in 25 μl reactions using approximately 20 ng cDNA, 0.5U Taq polymerase (Abgene) and a final concentration of 1× PCR buffer (AbGene), 2.0 mM MgCl2, 0.2 mM dNTPs and 0.5 μM of each primer. A two-stage touchdown PCR thermal cycling programme was used for all four fragments. In the first stage, the annealing temperature was decreased by 1°C per cycle from 60 to 50°C (a total of 11 cycles). In the second stage, the annealing temperature (Ta) was set to 50°C for an additional 24 cycles. In each case, one thermal cycle comprised 94°C for 30 s, Ta for 30 s and 72°C for 60 s. Initial denaturation of genomic DNA was carried out at 94°C for 3 min, and we included a final extension at 72°C for 5 min. DNA sequence extension products were generated directly from gel-purified PCR products (Promega Wizard gel extraction kit) using standard protocols (Applied Biosystems BigDye Terminator mix v. 1.1) and resolved by capillary electrophoresis on a 3730 instrument (Applied Biosystems). Chromatograms were visualized and edited using SeqScape v. 2.1 (Applied Biosystems) and aligned in Clustal X (Thompson et al. 1997).
Table 1.
Primers used for PCR and sequencing of cDNA, qPCR and SNP-SCALE genotyping.
| name | sequencea | positionb |
|---|---|---|
| qTYRP1-F1 | GGTAGATGTGAGGTGGCGATAG | 323–344 |
| qTYRP1-R1 | GTCCTGTTGAAGAAGCGTGTG | 429–409 |
| qGAPDH-F1 | ACCCAGAAGACTGTGGATGG | 98–117 |
| qGAPDH-R1 | CCAGTAGAAGCAGGGATGATG | 183–163 |
| TYRP1-cDNA-F2b | GAAATCTCCTACACTCCTCTCTC | 131–161 |
| TYRP1-cDNA-R3 | GAAGGTTTCTCCTGACTGTG | 536–517 |
| TYRP1-cDNA-F2c | CACGCTTCTTCAACAGGACA | 411–430 |
| TYRP1-cDNA-R4a | CATCAAGTCATCGGTGCAAA | 934–915 |
| TYRP1-cDNA-F3 | GAGGGACCAGCATTTCTCAC | 782–801 |
| TYRP1-cDNA-R6 | ATCAGCATTGTATCGCCTCA | 1401–1380 |
| TYRP1-cDNA-F5 | AGGTTACAGTGATCCCACAG | 1216–1235 |
| TYRP1-cDNA-R8 | CATAGACTGATTAGGGTTATGGA | 1744–1722 |
| TYRP1-ASO-G | GGGTTTTCCCAGTCACGACTTTCTCAATGGCGAGTAGTCTG | 983–1005 |
| TYRP1-ASO-T | CGGATAACAATTTCACACAGGATTTCTCAATGGCGAGTAGTCTT | 983–1005 |
| TYRP1-CRO | TGTTGCATAGGGTTCCCAGG | 1049–1030 |
| UFO-T1 | GGGTTTTCCCAGTCACGAC | n.a. |
| UFO-T2 | CGGATAACAATTTCACACAGGA | n.a. |
TYRP1-ASO-G and TYRP1-ASO-T have 5′ tails (bolded) corresponding to UFO-T1 and UFO-T2, respectively.
Numbered according to sequence NM174480 (TYRP1) and U94889 (GAPDH).
(d) Relative quantification of TYRP1 expression in dark and light Soay sheep
To examine the possibility that coat colour variation in Soay sheep is due to a regulatory mutation at TYRP1, we used relative quantitative PCR (qPCR) to compare gene expression in six dark and six light sheep. We quantified expression of TYRP1 relative to the GAPDH house-keeping gene. Our null hypothesis was for no difference in relative expression between coat colour morphs. Total RNA isolated from ear punches (described above) was treated with DNase I (Ambion TURBO DNA-free) to remove contaminating genomic DNA prior to cDNA synthesis. Complementary DNA was synthesized as previously described using approximately 1 μg DNase-treated total RNA and was then purified using the Promega Wizard SV Gel and PCR clean-up system. The purified cDNA was diluted 1 : 80 to provide template for qPCR. Three technical replicate amplifications were performed on each biological sample. Primer sequences for TYRP1 (qTYRP1-F1 and qTYRP1-R1) and GAPDH (qGAPDH-F1 and qGAPDH-R1) are provided in table 1. Quantitative PCR was performed in a total volume of 25 μl using 12.5 μl SensiMix (2×; Quantace), 0.5 μl SBYR Green (50×), 5 pmol of each primer and 5 μl cDNA. Reactions were performed on an Applied Biosystems 7700 real-time PCR instrument using a thermocycling programme that comprised an initial denaturation step at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Two DNase-treated RNA samples were included to confirm that all genomic DNA had been removed, and negative controls were included for both TYRP1 and GAPDH. Standard curves were generated for both target and control genes using Soay sheep genomic DNA of known concentration, with two technical replicates for each standard concentration.
(e) Testing for association between coat colour and TYRP1
Sequencing of the TYRP1 coding region revealed a non-synonymous single nucleotide polymorphism (SNP) that was an obvious candidate mutation for coat colour. We tested the legitimacy of this putative causal mutation by checking for co-segregation between coat colour and SNP genotype in a pedigree of 547 phenotyped sheep. The pedigree we used for this analysis, and to perform linkage mapping (see §2f), is described in detail in Beraldi et al. (2006). The ratio of dark to light animals in this pedigree (74% dark, 26% light, n=547) does not differ significantly from the Village Bay population as a whole (χ2(1df)=1.831, p=0.176). Genomic DNA was extracted from either ear punches or crude white blood fractions. For the former, we used Qiagen DNeasy kits or Qiagen Micro DNA kits, depending on the quantity and quality of starting material available. For the latter, we used Amersham's GFX blood kit. Genotyping was undertaken using the SNP-SCALE protocol described in Hinten et al. (in press). PCR was performed in 10 μl reactions using approximately 20 ng DNA, 0.25 U Taq polymerase (Abgene) and a final concentration of 1× PCR buffer (AbGene), 2.0 mM MgCl2 and 0.2 mM dNTPs. The final concentration of primers was 0.02 μM for the two-tailed allele-specific oligos (ASO; APS071-ASO-G, APS071-ASO-T), 0.2 μM for the common reverse oligo (CRO; APS071-CRO) and 0.1 μM for the two universal fluorescent oligos (UFO; UFO-T1, UFO-T2; table 1). The PCR thermal cycling programme comprised 40 cycles of 94°C for 30 s, annealing temperature for 30 s and 72°C for 30 s. The annealing temperature was set to 54°C for the first 10 cycles, 52°C for the next 15 cycles and 50°C for the last 15 cycles. Initial denaturation of genomic DNA was carried out at 94°C for 3 min and we included a final extension at 72°C for 5 min.
(f) Linkage mapping of TYRP1 and Coat colour
We added the putative causal SNP in the TYRP1 coding sequence to the Soay sheep genetic map described in Beraldi et al. (2006), using CRI-MAP v. 2.4 (Green et al. 1990). Prior to this mapping exercise, we used the programme Pedcheck (O'Connell & Weeks 1998) to screen for Mendelian inconsistencies and either resolved or removed the identified mismatches. We then used CRI-MAP v. 2.4 (Green et al. 1990) to conduct a linkage analysis for Coat colour on chromosome 2, assuming a single autosomal gene model with two alleles, where dark is dominant to light.
3. Results
(a) Sequence diversity in the TYRP1 coding region
We identified a total of six polymorphisms in the TYRP1 coding sequence, two of which (positions 869 and 1339) were non-synonymous (table 2). The first of these, a G→T transversion at nucleotide position 869 in exon IV (hereafter TYRP1 869G→T) was perfectly associated with coat colour; light sheep (n=6) were homozygous for T at this position, whereas dark sheep (n=6) were either homozygous for G or heterozygous (Fisher's exact test, p=0.002). This fits the most likely model of inheritance in which dark (G allele) is dominant to light (T allele). The TYRP1 869G→T mutation is predicted to cause the replacement of a cysteine (Cys) residue with phenylalanine at codon 290 (C290F). An amino acid alignment revealed that the Cys residue is conserved across all available vertebrates, suggesting that nucleotide position 869 may be under strong functional constraint and that dark coat colour is ancestral to light (figure 2). The other non-synonymous substitution at position 1339 was observed in one individual only and did not show an association with coat colour. Two other synonymous polymorphisms, at positions 1422 and 1470, also showed a significant association with coat colour. Light sheep were homozygous for a single haplotype across all six polymorphic sites (GenBank accession EF102109), whereas at least three additional haplotypes are segregating in dark sheep (data not shown; GenBank accessions EF102110–EF102112).
Table 2.
Sequence polymorphisms in the TYRP1 coding region in dark and light Soay sheep. (Non-synonymous mutations are highlighted in bold font. Dots indicate identity to the sequence in the top row. Codes for degenerate bases follow IUPAC conventions, in which K, M, R and Y represent G/T, A/C, A/G and C/T heterozygotes, respectively.)
| nucleotide position | |||||||
|---|---|---|---|---|---|---|---|
| ID no. | phenotype | 90 | 869 | 1107 | 1339 | 1422 | 1470 |
| 6048 | dark | C | G | C | A | T | A |
| 6034 | dark | Y | · | Y | R | . | M |
| 6047 | dark | T | K | · | · | Y | M |
| 6037 | dark | Y | · | · | · | · | · |
| 5551 | dark | Y | K | · | · | Y | M |
| 5591 | dark | Y | K | · | · | Y | M |
| 5919 | light | T | T | · | · | C | C |
| 5467 | light | T | T | · | · | C | C |
| 4308 | light | T | T | · | · | C | C |
| 5822 | light | T | T | · | · | C | C |
| 5632 | light | T | T | · | · | C | C |
| 5927 | light | T | T | · | · | C | C |
Figure 2.
Partial amino acid alignment for TYRP1 exon IV in dark and light Soay sheep and selected vertebrates (Capra hircus, AF136926; Bos Taurus, AF445638; Canis familiaris, AY052751; Homo sapiens, AF001295; Mus musculus, NM031202; Gallus gallus, NP990376; Xenopus laevis, BC043815; Takifugu rubripes, AF397401). The cysteine to phenylalanine substitution at position 290 (C290F) in Soay sheep is highlighted in bold font. Dots indicate shared identity with the amino acid sequence in dark Soay sheep. Numbers are amino acid residues in the TYRP1 coding sequence.
(b) Relative quantification of TYRP1 expression in dark and light Soay sheep
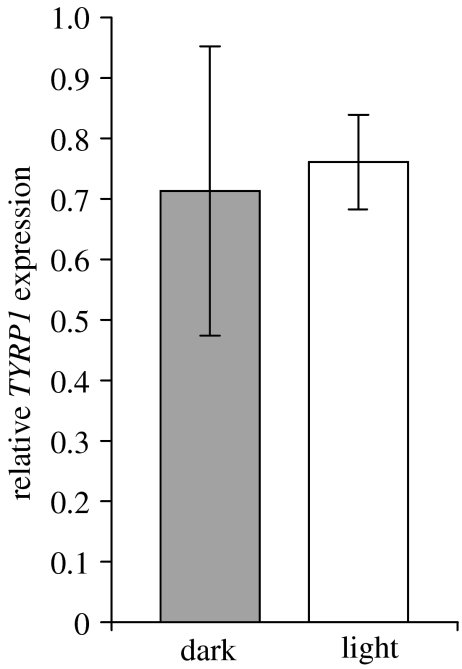
We quantified expression of TYRP1 in six dark and six light Soay sheep relative to the house-keeping gene GAPDH. We set an arbitrary fluorescence threshold in the exponential phase of amplification and recorded the threshold cycle (CT) for each sample cDNA (three replicates) and standard (two replicates). For each sample, and for each of TYRP1 and GAPDH, we took the mean CT and used the standard curve regression formulae for each gene to calculate the starting amount of cDNA. We then standardized TYRP1 expression relative to GAPDH expression and compared relative TYRP1 expression in dark and light sheep using a one-tailed t-test with unequal variance (the test was one-tailed because the alternative hypothesis was that TYRP1 was downregulated in light sheep). This analysis failed to reject the null hypothesis that there is no difference in TYRP1 expression between dark (0.71±0.24) and light (0.76±0.08) Soay sheep (p=0.33; figure 3).
Figure 3.
Expression of TYRP1 relative to GAPDH in dark and light Soay sheep.
(c) Testing for association between coat colour and TYRP1
We obtained TYRP1 869G→T SNP genotypes for 523 phenotyped sheep in the mapping pedigree. Analyses of these data revealed an almost perfect association with coat colour (99.4%, χ2-test, p<0.0001, LOD=110.20), with the exception of three individuals: one dark animal (ID no. 4428) with a genotype characteristic of light coats (T/T) and two light animals with genotypes normally associated with dark coats (ID nos. 2253 and 2257, both G/T). The frequencies of the TYRP1 869G→T alleles in the pedigree were almost equal (49% G, 51% T). There was no indication of a departure from Hardy–Weinberg equilibrium at TYRP1 869G→T (p=0.559).
(d) Linkage mapping of TYRP1 and Coat colour
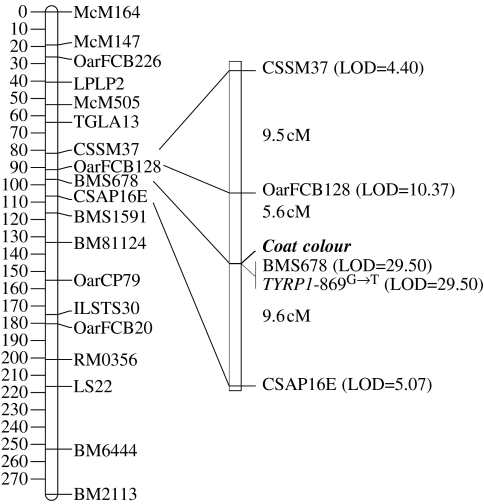
The TYRP1 869G→T SNP mapped to chromosome 2, in almost the same location as microsatellite marker BMS678 (two-point LOD for linkage to BMS678=65.84, recombination fraction=0.01). The linkage analysis on chromosome 2 revealed very tight linkage between Coat colour and TYRP1 (LOD=29.50, recombination fraction=0 cM; figure 4).
Figure 4.
Linkage map of Soay sheep chromosome 2, showing the location of TYRP1- 869G→T and Coat colour. LOD scores for linkage with the Coat colour locus are sex average computed. Marker intervals are reported in Kosambi centiMorgans.
4. Discussion
We have previously shown that the Coat colour locus in Soay sheep maps to a region of chromosome 2, which on the basis of conserved synteny between cattle and sheep is likely to contain the candidate gene TYRP1 (Beraldi et al. 2006). In this study, we confirmed that TYRP1 and Coat colour are tightly linked on chromosome 2 by mapping a putative causal mutation at nucleotide position 869 in exon IV of TYRP1. This mutation fitted the proposed single gene model of inheritance in which the dark allele is dominant to light (Doney et al. 1974; Coltman & Pemberton 2004), and it was almost perfectly associated with coat colour in a pedigree of more than 500 sheep (see below of the three observed mismatches for discussion). We demonstrated that no difference exists in relative TYRP1 expression between coat morphs and we can therefore eliminate the possibility that the TYRP1 869G→T-coat colour association is due to the presence of strong linkage disequilibrium (LD) with an unknown regulatory mutation in TYRP1.
The TYRP1 869G→T mutation is predicted to cause the replacement of a cysteine (Cys) residue with phenylalanine (C290F), a non-conservative amino acid substitution that involves a change in both polarity and hydrophobicity. The Cys residue involved shows a high degree of evolutionary conservation; it is conserved not only across vertebrates, but also in the two paralogues of TYRP1, DCT and TYR (Garcia-Borron & Solano 2002). Indeed, this is true for all 15 Cys residues that are common to the three members of the tyrosinase gene family (Garcia-Borron & Solano 2002). Cys residues in these genes are thought to be involved in the formation of internal disulphide bridges that are crucial for the successful transport, processing and maturation of synthesized proteins in the cytoplasm (Sarangarajan & Boissy 2001; Garcia-Borron & Solano 2002). Garcia-Borron & Solano (2002) note that almost all Cys residues in TYR appear to be essential for correct protein folding. Indeed, across all three tyrosinase family members, all known alterations involving either the loss or gain of a Cys residue, including mutations in mouse (Jackson & Bennett 1990; Yokoyama et al. 1990) and human TYR (Oetting & King 1999) and mouse TYRP1 (Zdarsky et al. 1990), result in an inactive protein. The TYRP1 869G→T polymorphism segregating in Soay sheep is novel, but these observations suggest that nucleotide position 869 is under strong functional constraint and is almost certainly a functional mutation. Taken together, these findings form a compelling argument that a single base mutation in TYRP1 is responsible for the coat-colour polymorphism in Soay sheep.
One concern is that there were three cases where TYRP1 869G→T did not co-segregate with coat colour in the Soay sheep pedigree: one instance of a dark animal with a genotype typically associated with light coats (T/T) and two cases of light animals with genotypes characteristic of dark coats (G/T). The existence of some mismatches should not be surprising because some errors in the pipeline from field to final genotype are unavoidable in studies of this type and magnitude (Bonin et al. 2004). Nonetheless, these discrepancies bear further investigation. We can envisage four possible explanations. The first is that they are due to genotyping errors or mislabelling of genetic samples. To address this possibility, we re-genotyped TYRP1 869G→T in the three mismatching sheep, initially using the original DNA extraction and then using freshly isolated DNA. In each case, the same unambiguous genotypes were obtained as in the initial genotyping effort. To assess the possibility that genetic samples had been mislabelled, we compared genotypes for the three mismatching sheep to the genotypes of their respective parents, at 247 microsatellites (data not shown); in all the three cases, parent–offspring genotypes mismatched at no more than one locus (i.e. less than 0.5% error). Clearly, the mismatches are not due to genotyping error or sample mislabelling.
A second possible explanation for the mismatches is that they are due to phenotyping errors. We addressed this possibility by re-examining field records for each of the three mismatching sheep. Interestingly, this revealed that the coat colour assigned to individuals 2253 and 2257 as lambs was consistent with their TYRP1 869G→T genotype (i.e. both were initially recorded as dark), but that in each case coat colour was amended to light during a follow-up census. Individual 4428 was always recorded as dark. Coat colour is usually unambiguous in yearlings and adults, but can be difficult to assign in newborn lambs (Doney et al. 1974). Consequently, the amended coat colour records, collected when the sheep were older, are more likely to be correct. Given that all three individuals were censused on multiple occasions as juveniles and adults (n=11 records for 2253; n=9 records for 2257 and n=46 records for 4428), it is unlikely that the mismatches are due to phenotyping error.
The third possible explanation for an imperfect association between TYRP1 869G→T and coat colour is that a mutation tightly linked to this site, rather than TYRP1 869G→T itself, is the true causal mutation. It is particularly important to consider this possibility given that long-range LD has been shown to confound the identification of functional mutations in livestock (de Koning 2006), and strong LD is predicted to extend several centiMorgans in this population (McRae et al. 2005). This scenario could involve a mutation in the TYRP1 coding sequence other than TYRP1 869G→T or a mutation in a tightly linked but unknown gene. A third possibility, the involvement of a linked regulatory mutation, can be ruled out because TYRP1 expression does not differ between dark and light Soay sheep. The involvement of polymorphisms in the TYRP1 coding sequence other than TYRP1 869G→T is unlikely, because the only other nucleotide positions that showed a perfect association with coat colour in the analysis of cDNA sequences (positions 1422 and 1470; n=12 sheep) are the site of synonymous mutations. Given that there is no difference in expression between colour morphs, it is difficult to envisage how these mutations could influence coat colour. Further evidence against their involvement is provided by alignments of the TYRP1 coding sequence in vertebrates, which show that contrary to expectations for a functional non-coding mutation (e.g. Smit et al. 2003; Sinha et al. 2006), these nucleotide positions are not evolutionarily conserved and do not appear to be under functional constraint; at position 1422, the site of a T to C transition in Soay sheep, bases C and T are both represented in the vertebrates across which TYRP1 869G→T is conserved and at position 1470, the site of an A to C transversion in our study population, bases A, C and T are represented in the same taxa (data not shown). The involvement of a tightly linked but unknown gene, although possible, is also implausible because TYRP1 is a known pigmentation gene and none of the more than 100 other recognized candidate genes for coat colour in mouse, nor their human homologues, map to the same chromosome as TYRP1 (Oetting & Bennett 2005).
The final explanation for the discrepancies that we can conceive of is that other low-frequency modifying mutations are segregating in the population in addition to the primary causal mutation, TYRP1 869G→T. This would require at least two modifying mutations operating in opposite directions: one that restores dark coat colour to sheep with a light TYRP1 genotype and another that causes sheep with a dark TYRP1 genotype to be light. We suggest that this is not implausible given that the melanin synthesis pathway in vertebrates involves a large number of genes, and studies in other vertebrates have revealed a multigenic basis for coat colour variation (e.g. Hoekstra et al. 2006b). However, it is difficult to assess this hypothesis because the mutations, if they exist, are clearly very rare and they may reside in one or more of a large number of candidate genes. Additionally, we do not know whether the mismatching individuals had unusual phenotypes that would be consistent with the action of other loci, because coat colour phenotyping in our study population is qualitative (i.e. dark or light), rather than quantitative. We acknowledge that the mismatches could be due to a source of error that we have failed to account for, but of the four possibilities we have considered, the involvement of rare modifying mutations is the most likely explanation for the observed discrepancies.
In systems such as ours where the construction of transgenic models to demonstrate the functionality of a candidate mutation may not be feasible or ethically desirable, causality can be inferred by combining multiple independent sources of evidence (e.g. Mackay 2001; de Koning 2006). We contend that the body of evidence in favour of TYRP1 869G→T, including highly significant association and linkage and the non-conservative and functionally important nature of the amino acid substitution identified is collectively very compelling. The novel TYRP1 869G→T mutation observed in Soay sheep causes the loss of a Cys residue that is required for correct protein function (Sarangarajan & Boissy 2001; Garcia-Borron & Solano 2002). Given that all other known mutations involving either the loss or gain of a Cys residue result in an inactive protein, we conclude that TYRP1 869G→T is the primary mutation underlying coat colour variation in Soay sheep, accounting for more than 99% of the trait variation in our study population. This discovery will facilitate future studies that aim to determine how genetic variation for coat colour is maintained in this population.
Acknowledgments
We thank JG Pilkington and many volunteers for collecting field data and genetic samples. We thank SA Johns for her helpful discussion and J Chittock, A Krupa and PJ Swarbrick for their technical support in the laboratory. We thank the National Trust of Scotland for granting permission to work on St Kilda and QinetiQ for logistical support. The long-term data collection on St Kilda has been supported by Natural Environment Research Council (NERC) and Wellcome Trust grants to T. H. Clutton-Brock, B. T. Grenfell, L. E. B. Kruuk, M. J. Crawley and J.M.P. This study was funded by NERC through its Environmental Genomics thematic programme (grant no. NER/T/S/2002/00189).
Supplementary Material
Initial candidate gene analysis
References
- Aliev G, Rachkovsky M, Ito S, Wakamatsu K, Ivanov A. Pigment types in selected color genotypes of Asiatic sheep. Pigment Cell Res. 1990;3:177–180. doi: 10.1111/j.1600-0749.1990.tb00286.x. doi:10.1111/j.1600-0749.1990.tb00286.x [DOI] [PubMed] [Google Scholar]
- Bennett D.C, Lamoreux M.L. The color loci of mice—a genetic century. Pigment Cell Res. 2003;16:333–344. doi: 10.1034/j.1600-0749.2003.00067.x. doi:10.1034/j.1600-0749.2003.00067.x [DOI] [PubMed] [Google Scholar]
- Beraldi D, McRae A.F, Gratten J, Slate J, Visscher P.M, Pemberton J.M. Development of a linkage map and mapping of phenotypic polymorphisms in a free-living population of Soay sheep (Ovis aries) Genetics. 2006;173:1521–1537. doi: 10.1534/genetics.106.057141. doi:10.1534/genetics.106.057141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berryere T.G, Schmutz S.M, Schimpf R.J, Cowan C.M, Potter J. TYRP1 is associated with dun coat colour in Dexter cattle or how now brown cow? Anim. Genet. 2003;34:169–175. doi: 10.1046/j.1365-2052.2003.00985.x. doi:10.1046/j.1365-2052.2003.00985.x [DOI] [PubMed] [Google Scholar]
- Boissy R.E, et al. Mutation in and lack of expression of tyrosinase-related protein-1 (TRP-1) in melanocytes from an individual with brown oculocutaneous albinism: a new subtype of albinism classified as ‘OCA3’. Am. J. Hum. Genet. 1996;58:1145–1156. [PMC free article] [PubMed] [Google Scholar]
- Bonin A, Bellemain E, Eidesen P.B, Pompanon F, Brochmann C, Taberlet P. How to track and assess genotyping errors in population genetics studies. Mol. Ecol. 2004;13:3261–3273. doi: 10.1111/j.1365-294X.2004.02346.x. doi:10.1111/j.1365-294X.2004.02346.x [DOI] [PubMed] [Google Scholar]
- Clutton-Brock T.H, Pemberton J.M. Cambridge University Press; Cambridge, UK: 2004. Soay sheep: dynamics and selection in an island population. [Google Scholar]
- Clutton-Brock T.H, Wilson K, Stevenson I.R. Density-dependent selection on horn phenotype in Soay sheep. Phil. Trans. R. Soc. B. 1997;352:839–850. doi: 10.1098/rstb.1997.0064. doi:10.1098/rstb.1997.0064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coltman D.W, Pemberton J.M. Inheritance of coat colour and horn type in Hirta Soay sheep. In: Clutton-Brock T.H, Pemberton J.M, editors. Soay sheep: dynamics and selection in an island population. Cambridge University Press; Cambridge, UK: 2004. pp. 321–327. [Google Scholar]
- Coltman D.W, Pilkington J, Kruuk L.E.B, Wilson K, Pemberton J.M. Positive genetic correlation between parasite resistance and body size in a free-living ungulate population. Evolution. 2001;55:2116–2125. doi: 10.1111/j.0014-3820.2001.tb01326.x. doi:10.1554/0014-3820(2001)055[2116:PGCBPR]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Coulson T, Catchpole E.A, Albon S.D, Morgan B.J.T, Pemberton J.M, Clutton-Brock T.H, Crawley M.J, Grenfell B.T. Age, sex, density, winter weather, and population crashes in Soay sheep. Science. 2001;292:1528–1531. doi: 10.1126/science.292.5521.1528. doi:10.1126/science.292.5521.1528 [DOI] [PubMed] [Google Scholar]
- de Koning D.-J. Conflicting candidates for cattle QTLs. Trends Genet. 2006;22:301–305. doi: 10.1016/j.tig.2006.04.006. doi:10.1016/j.tig.2006.04.006 [DOI] [PubMed] [Google Scholar]
- Doney J.M, Ryder M.L, Gunn R.G, Grubb P. Colour, conformation, affinities, fleece and patterns of inheritance of the Soay sheep. In: Jewell P.A, Milner C, Boyd J.M, editors. Island survivors: the ecology of the soay sheep of St Kilda. Athlone Press; London, UK: 1974. pp. 88–125. [Google Scholar]
- Erickson D.L, Fenster C.B, Stenoien H.K, Price D. Quantitative trait locus analyses and the study of evolutionary process. Mol. Ecol. 2004;13:2505–2522. doi: 10.1111/j.1365-294X.2004.02254.x. doi:10.1111/j.1365-294X.2004.02254.x [DOI] [PubMed] [Google Scholar]
- Fitzpatrick M.J, Ben-Shahar Y, Smid H.M, Vet L.E.M, Robinson G.E, Sokolowski M.B. Candidate genes for behavioural ecology. Trends Ecol. Evol. 2005;20:96–104. doi: 10.1016/j.tree.2004.11.017. doi:10.1016/j.tree.2004.11.017 [DOI] [PubMed] [Google Scholar]
- Garcia-Borron J.C, Solano F. Molecular anatomy of tyrosinase and its related proteins: beyond the histidine-bound metal catalytic center. Pigment Cell Res. 2002;15:162–173. doi: 10.1034/j.1600-0749.2002.02012.x. doi:10.1034/j.1600-0749.2002.02012.x [DOI] [PubMed] [Google Scholar]
- Green P, Falls K, Crooks S. Washington University; St Louis, MO: 1990. Documentation for Crimap. [Google Scholar]
- Hinten, G. N., Hale, M. C., Gratten, J., Mossman, J. A., Lowder, B. V., Mann, M. K. & Slate, J. In press. SNP SCALE: SNP scoring by colour and length exclusion. Mol. Ecol. Notes
- Hoekstra H.E. Genetics, development and evolution of adaptive pigmentation in vertebrates. Heredity. 2006;97:222–234. doi: 10.1038/sj.hdy.6800861. doi:10.1038/sj.hdy.6800861 [DOI] [PubMed] [Google Scholar]
- Hoekstra H.E, Nachman M.W. Different genes underlie adaptive melanism in different populations of rock pocket mice. Mol. Ecol. 2003;12:1185–1194. doi: 10.1046/j.1365-294x.2003.01788.x. doi:10.1046/j.1365-294X.2003.01788.x [DOI] [PubMed] [Google Scholar]
- Hoekstra H.E, Hirschmann R.J, Bundey R.A, Insel P.A, Crossland J.P. A single amino acid mutation contributes to adaptive Beach mouse color pattern. Science. 2006;313:101–104. doi: 10.1126/science.1126121. doi:10.1126/science.1126121 [DOI] [PubMed] [Google Scholar]
- Jackson I.J. A cDNA encoding tyrosinase-related protein maps to the Brown locus in mouse. Proc. Natl Acad. Sci. USA. 1988;85:4392–4396. doi: 10.1073/pnas.85.12.4392. doi:10.1073/pnas.85.12.4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson I.J, Bennett D.C. Identification of the albino mutation of mouse tyrosinase by analysis of an in vitro revertant. Proc. Natl Acad. Sci. USA. 1990;87:7010–7014. doi: 10.1073/pnas.87.18.7010. doi:10.1073/pnas.87.18.7010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Imokawa G, Bennett D.C, Hearing V.J. Tyrosinase stabilization by Tyrp1 (the brown locus protein) J. Biol. Chem. 1998;273:31 801–31 805. doi: 10.1074/jbc.273.48.31801. doi:10.1074/jbc.273.48.31801 [DOI] [PubMed] [Google Scholar]
- Mackay T.F. The genetic architecture of quantitative traits. Annu. Rev. Genet. 2001;35:303–339. doi: 10.1146/annurev.genet.35.102401.090633. doi:10.1146/annurev.genet.35.102401.090633 [DOI] [PubMed] [Google Scholar]
- McRae A.F, Pemberton J.M, Visscher P.M. Modelling linkage disequilibrium in natural populations: the example of the Soay sheep population of St. Kilda, Scotland. Genetics. 2005;171:251–258. doi: 10.1534/genetics.105.040972. doi:10.1534/genetics.105.040972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner J.M, Elston D.A, Albon S.D. Estimating the contributions of population density and climatic fluctuations to interannual variation in survival of Soay sheep. J. Anim. Ecol. 1999;68:1235–1247. doi:10.1046/j.1365-2656.1999.00366.x [Google Scholar]
- Milner J.M, Albon S.D, Kruuk L.E.B, Pemberton J.M. Selection on phenotype. In: Clutton-Brock T.H, Pemberton J.M, editors. Soay sheep: dynamics and selection in an island population. Cambridge University Press; Cambridge, UK: 2004. pp. 190–216. [Google Scholar]
- Mundy N.I. A window on the genetics of evolution: MC1R and plumage colouration in birds. Proc. R. Soc. B. 2005;272:1633–1640. doi: 10.1098/rspb.2005.3107. doi:10.1098/rspb.2005.3107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy N.I, Badcock N.S, Hart T, Scribner K, Janssen K, Nadeau N.J. Conserved genetic basis of a quantitative plumage trait involved in mate choice. Science. 2004;303:1870–1873. doi: 10.1126/science.1093834. doi:10.1126/science.1093834 [DOI] [PubMed] [Google Scholar]
- Nachman M.W, Hoekstra H.E, D'Agostino S.L. The genetic basis of adaptive melanism in pocket mice. Proc. Natl Acad. Sci. USA. 2003;100:5268–5273. doi: 10.1073/pnas.0431157100. doi:10.1073/pnas.0431157100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell J, Weeks D. PedCheck: a program for identifying genotype incompatibilities in linkage analysis. Am. J. Hum. Genet. 1998;63:259–266. doi: 10.1086/301904. doi:10.1086/301904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oetting, W. & Bennett, D. 2005 Mouse coat color genes. World Wide Web (URL: http://www.cbc.umn.edu/ifpcs/micemut.htm): International Federation of Pigment Cell Societies.
- Oetting W.S, King R.A. Molecular basis of albinism: mutations and polymorphisms of pigmentation genes associated with albinism. Hum. Mutat. 1999;13:99–115. doi: 10.1002/(SICI)1098-1004(1999)13:2<99::AID-HUMU2>3.0.CO;2-C. doi:10.1002/(SICI)1098-1004(1999)13:2<99::AID-HUMU2>3.0.CO;2-C [DOI] [PubMed] [Google Scholar]
- Rosenblum E.B. Convergent evolution and divergent selection: lizards in the White Sands ecotone. Am. Nat. 2006;167:1–15. doi: 10.1086/498397. doi:10.1086/498397 [DOI] [PubMed] [Google Scholar]
- Rosenblum E.B, Hoekstra H.E, Nachman M.W. Adaptive reptile color variation and the evolution of the Mc1r gene. Evolution. 2004;58:1794–1808. doi: 10.1111/j.0014-3820.2004.tb00462.x. doi:10.1554/03-741 [DOI] [PubMed] [Google Scholar]
- Ryder M.L, Land R.B, Ditchburn R. Coat colour inheritance in Soay, Orkney and Shetland sheep. J. Zool. 1974;173:477–485. [Google Scholar]
- Sarangarajan R, Boissy R.E. Tyrp1 and oculocutaneous albinism type 3. Pigment Cell Res. 2001;14:437–444. doi: 10.1034/j.1600-0749.2001.140603.x. doi:10.1034/j.1600-0749.2001.140603.x [DOI] [PubMed] [Google Scholar]
- Schmidt-Kuntzel A, Eizirik E, O'Brien S.J, Menotti-Raymond M. Tyrosinase and tyrosinase related protein 1 alleles specify domestic cat coat color phenotypes of the albino and brown loci. J. Hered. 2005;96:289–301. doi: 10.1093/jhered/esi066. doi:10.1093/jhered/esi066 [DOI] [PubMed] [Google Scholar]
- Silvers W.K. Springer; New York, NY: 1979. The coat colors of mice. [Google Scholar]
- Sinha H, Nicholson B.P, Steinmetz L.M, McCusker J.H. Complex genetic interactions in a quantitative trait locus. PLoS Genet. 2006;2:140–147. doi: 10.1371/journal.pgen.0020013. doi:10.1371/journal.pgen.0020013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slate J. Quantitative trait locus mapping in natural populations: progress, caveats and future directions. Mol. Ecol. 2005;14:363–379. doi: 10.1111/j.1365-294X.2004.02378.x. doi:10.1111/j.1365-294X.2004.02378.x [DOI] [PubMed] [Google Scholar]
- Slate J, et al. A deer (subfamily Cervinae) genetic linkage map and the evolution of ruminant genomes. Genetics. 2002;160:1587–1597. doi: 10.1093/genetics/160.4.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit M, et al. Mosaicism of solid gold supports the causality of a noncoding A-to-G transition in the determinism of the callipyge phenotype. Genetics. 2003;163:453–456. doi: 10.1093/genetics/163.1.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theron E, Hawkins K, Bermingham E, Ricklefs R.E, Mundy N.I. The molecular basis of an avian plumage polymorphism in the wild: a melanocortin-1-receptor point mutation is perfectly associated with the melanic plumage morph of the bananaquit, Coereba flaveola. Curr. Biol. 2001;11:550–557. doi: 10.1016/s0960-9822(01)00158-0. doi:10.1016/S0960-9822(01)00158-0 [DOI] [PubMed] [Google Scholar]
- Thompson J.D, Gibson T.J, Plewniak F, Jeanmougin F, Higgins D.G. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. doi:10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vage D.I, Klungland H, Lu D, Cone R.D. Molecular and pharmacological characterization of dominant black coat color in sheep. Mamm. Genome. 1999;10:39–43. doi: 10.1007/s003359900939. doi:10.1007/s003359900939 [DOI] [PubMed] [Google Scholar]
- Vasemagi A, Primmer C.R. Challenges for identifying functionally important genetic variation: the promise of combining complementary research strategies. Mol. Ecol. 2005;14:3623–3642. doi: 10.1111/j.1365-294X.2005.02690.x. doi:10.1111/j.1365-294X.2005.02690.x [DOI] [PubMed] [Google Scholar]
- Yokoyama T, Silversides D.W, Waymire K.G, Kwon B.S, Takeuchi T, Overbeek P.A. Conserved cysteine to serine mutation in tyrosinase is responsible for the classical albino mutation in laboratory mice. Nucleic Acids Res. 1990;18:7293–7298. doi: 10.1093/nar/18.24.7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdarsky E, Favor J, Jackson I.J. The molecular basis of Brown, an old mouse mutation, and of an induced revertant to wild-type. Genetics. 1990;126:443–449. doi: 10.1093/genetics/126.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Initial candidate gene analysis