Abstract
Lon protease and members of the Clp family of molecular chaperones and protease regulatory subunits contain homologous regions with properties expected for substrate-binding domains. Fragments corresponding to these sequences are stably and independently folded for Lon, ClpA, and ClpY. The corresponding regions from ClpB and ClpX are unstable. All five fragments exhibit distinct patterns of binding to three proteins that are protease substrates in vivo: the heat shock transcription factor σ32, the SOS mutagenesis protein UmuD, and Arc repressor bearing the SsrA degradation tag. Recognition of UmuD is mediated through peptide sequences within a 24-residue N-terminal region whereas recognition of both σ32 and SsrA-tagged Arc requires sequences at the C terminus. These results indicate that the Lon and Clp proteases use the same mechanism of substrate discrimination and suggest that these related ATP-dependent bacterial proteases scrutinize accessible or disordered regions of potential substrates for the presence of specific targeting sequences.
Keywords: protein degradation, disassembly chaperone, molecular recognition, sensor- and substrate-discrimination domains, energy-dependent proteases
Recognition of the correct partners or substrates is critical for almost all biological transactions. For irreversible processes like protein degradation, choosing the appropriate molecular targets is especially important because cleavage of the wrong polypeptides could be damaging or even fatal. Moreover, degradation of the proper substrates is required to eliminate misfolded or defective proteins and to control the cellular activities of critical regulatory proteins (1–3). A bacterium such as Escherichia coli contains ≈4,300 different proteins, and its intracellular proteases must differentiate among these diverse molecules, selecting only a small subset for destruction.
In E. coli and other bacteria, many of the intracellular proteases must hydrolyze ATP to degrade protein substrates; this class includes single-chain enzymes, such as Lon, and two-chain proteases, such as ClpAP, ClpXP, and ClpYQ (HslUV) (1–3). For the latter enzymes, ClpP or ClpQ is the peptidase subunit, and ClpA, ClpX, or ClpY is the ATPase and substrate-binding subunit. The peptidase subunits form hexameric or heptameric rings, which combine in a two-tier stack (4–6); the regulatory ATPase subunits form six-member rings that can bind at both ends of the protease stack (6–8). The resulting assembly sequesters the protease active sites in a central cavity, preventing inadvertent cleavage of the wrong proteins. This architecture requires that the Clp ATPase subunits act as gatekeepers, recognizing the proper substrates and mediating their delivery to the proteolytic cavern (9). By themselves, ClpA and ClpX can function as disassembly chaperones to catalyze the dissociation of certain multimeric proteins (10–13). Other Clp family ATPases, including E. coli ClpB and yeast Hsp104, are not known to interact with protease subunits and may function solely as chaperones (2, 3, 14).
An understanding of the molecular determinants of substrate recognition by intracellular proteases requires two types of information: (i) which features of the protein substrates are recognized; and (ii) how the proteases mediate this recognition. Some unstable proteins in E. coli are degraded by a particular protease whereas others are cleaved by several different proteases. For example, UmuD, a protein involved in regulation of the SOS response to damaged DNA, is degraded by Lon protease in a reaction that depends on internal sequences in a region of 24 N-terminal residues (15). λ repressor’s N-terminal domain tagged with the SsrA degradation peptide is subject to C-terminal specific degradation by ClpXP and ClpAP, as well as by FtsH, a membrane-bound ATP-dependent protease of the AAA family (16, 17). The heat-shock transcription factor, σ32, is proteolyzed by both ClpYQ (18) and FtsH (19, 20), again in reactions that seem to involve C-terminal sequence determinants (ref. 21; C. Herman and C. Gross, personal communication). Thus, some proteolytic substrates in E. coli appear to be recognized via signals at their C terminus whereas others are recognized by internal sequences.
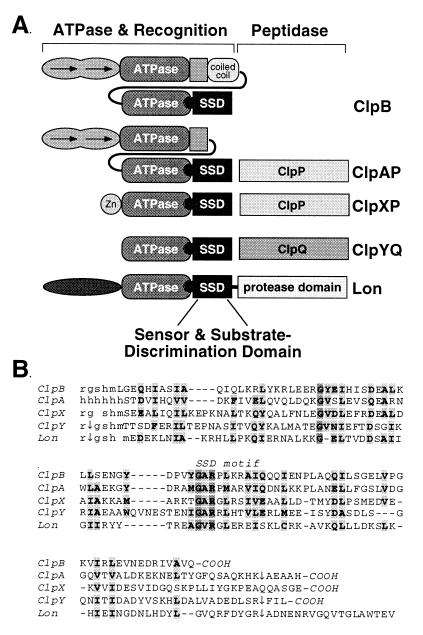
The domain structures of Lon protease and the Clp proteins are shown in Fig. 1A. In ClpX, the C-terminal domain binds proteins in a fashion that parallels the substrate specificity of the intact enzyme (22, 23). Homologous sequences are present in the other Clp ATPases and also in Lon protease (Fig. 1B). The most conserved motif in this region, G-Φ-R-X-Φ (Φ = hydrophobic), is part of signature sequence V of Clp family ATPases (24) and is present in all Lon homologs. Neuwald et al. (25) have aligned this Clp/Lon motif with the sensor-2 motif G/P-X-Φ-R-X-Φ in a superfamily of ATPases. Partial three-dimensional structures are known for two of these superfamily members (26–28), revealing that side chains in the sensor-2 motif are part of an α-helical domain and are positioned to interact with bound nucleotide in a nearby ATPase domain.
Figure 1.
(A) Domain structures of Lon and Clp proteins. Approximate domain boundaries are from this work for SSD domains, Neuwald et al. (25) for ATPase domains, and homology with the purified, stably folded N-domain of ClpA (J. Lo, personal communication) and coiled-coil domain of ClpB (C.K.S., unpublished work). (B) Alignment of the E. coli Lon and Clp SSD sequences. Lowercase letters represent sequences from His tags added during cloning. Arrows indicate domain boundaries determined by tryptic digestion.
Here, we examine the structural properties of the putative substrate-binding domains from ClpA, ClpB, ClpX, ClpY, and Lon and assay their binding to three pairs of proteins that are degraded differentially by proteases in E. coli. These domains display distinct but overlapping binding preferences, which are affected by C-terminal peptide sequences for two substrates and by peptide sequences near the N terminus for another substrate. We propose that these homologous “sensor- and substrate-discrimination” or SSD domains play critical roles in the mechanism by which energy-dependent bacterial proteases recognize the correct substrates.
MATERIALS AND METHODS
Plasmids.
A plasmid expressing the ClpA SSD domain was constructed by PCR subcloning of DNA encoding E. coli ClpA residues 654–758 between the NdeI and BamHI sites of pET800 (29). The resulting plasmid contains the sequence NH2-MH6 fused to the ClpA SSD domain. DNA encoding the SSD domains of E. coli ClpB (residues 766–857), ClpX (residues 318–424), ClpY (residues 336–443), or Lon (residues 495–607) was PCR amplified and cloned between the NdeI and XhoI sites of pET15b (Novagen), generating plasmids with NH2-MGSSH6SSGLVPRGSHM fused to the SSD domains. Plasmid constructions were confirmed by restriction mapping and DNA sequencing.
Proteins.
The His-tagged SSD domains were purified from E. coli strain BL21 (DE3) (Novagen) containing the appropriate plasmid. Cells were grown at 37°C to an A600 of 0.6 in Luria–Bertani broth, were induced by addition of 1 mM isopropyl β-d-thiogalactoside, were grown for 3 h more, were harvested by centrifugation, and were lysed with 200 μg/ml lysozyme at 4°C for 1 h in 50 mM Tris (pH 8) and 10% sucrose. The cell paste was sonicated to reduce viscosity and then was centrifuged at 20,000 × g for 30 min. The supernatant was purified by chromatography on Ni–nitrilotriacetic acid–agarose (Qiagen, Chatsworth, CA) under native conditions, according to the manufacturer’s recommendations. Proteins were concentrated in a centriprep 3 concentrator (Amicon) and were further purified by gel filtration on Superdex 75 HR 10/30 (Amersham Pharmacia). Purified proteins were >95% pure as determined by Coomassie blue staining after SDS/PAGE and had masses predicted from the sequence after post-translational removal of the N-terminal methionines of the Lon, ClpB, ClpX, and ClpY fragments.
Arc-ssrA contains the C-terminal extension H6KNQHEAANDENYALAA fused to wild-type Arc, where the italicized sequence is the SsrA-degradation tag (30). Arc-ssrA-DD has the C-terminal sequence H6KNQHEAANDENYALDD. These proteins were expressed at 32°C in E. coli strain SG1146a (clpP−, lon−, DE3) (16) transformed with pET3a-Arc-ssrA or pET3a-Arc-ssrA-DD (22). Both proteins were purified by Ni–nitrilotriacetic acid chromatography under denaturing conditions (31). UmuD, UmuD′, and UmuD antisera were gifts from Mark Sutton and Graham Walker (Massachusetts Institute of Technology). σ32, σ32-DD, and σ32 antisera were gifts from Christophe Herman and Carol Gross (University of California at San Francisco).
Spectroscopy.
Fluorescence spectra were collected on a Hitachi (Tokyo) F4500 fluorescence spectrophotometer by using 2 μM protein in 50 mM potassium phosphate (pH 7.6) and 100 mM NaCl at 20°C. Circular dichroism (CD) experiments were performed by using an Aviv Associates (Lakewood, NJ) 60DS spectrapolarimeter in 25 mM potassium phosphate (pH 7.5), 50 mM NaCl, and 10% glycerol (with 0.5 mM β-mercaptoethanol for Lon SSD). Far ultraviolet wavelength scans were performed at 20°C, and ellipticity measurements were averaged for 15 s at each wavelength. The α-helical content was calculated by assuming a mean residue ellipticity of 34,000 for 100% helix. Thermal denaturation was monitored by changes in CD ellipticity at 222 nm by using 2–3 μM protein. Samples were equilibrated for 1.5 min between temperature changes, and ellipticity readings were averaged for 30 s at each temperature. Fraction native was determined as (ɛ-ɛD)/(ɛN-ɛD), where ɛ is the observed ellipticity, and ɛD and ɛN are the ellipticities of the denatured and native baselines, respectively. Near ultraviolet wavelength scans were the average of five measurements taken at 20°C in 25 mM Tris⋅HCl (pH 8.0) and 50 mM NaCl. Ellipticity measurements were averaged for 15 s at each wavelength.
Limited Proteolysis.
Trypsin or chymotrypsin were used to digest a 500-fold molar excess of the SSD domains (60–100 μM) at room temperature in 25 mM Tris (pH 8.0), 50 mM NaCl, and 1 mM CaCl2. Reactions were stopped by adding phenylmethylsulfonyl fluoride and freezing in liquid nitrogen. Samples were analyzed by electrophoresis on 15% Tris tricine SDS gels and by matrix-assisted laser desorption ionization–time-of-flight mass spectroscopy using a PerSeptive Biosystems (Framingham, MA) Voyager DE-STR instrument. The program paws 8.1.1 (ProteoMetrics, New York) was used to correlate masses with sequences.
Oligomeric State.
Analytical ultracentrifugation at 20°C was performed by using a Beckman Coulter XL-A ultracentrifuge and rotor speeds of 16,000 and 24,000 rpm. For the ClpA, ClpX, ClpY, and Lon domains, the buffer was 25 mM potassium phosphate (pH 7.6), 100 mM NaCl, and 0.1 mM EDTA (with 2 mM β-mercaptoethanol for Lon). For the ClpB domain, the buffer was 50 mM sodium acetate (pH 6.3), 100 mM NaCl, and 0.1 mM EDTA. Proteins were passed through 0.2-μm filters before centrifugation. After reaching equilibrium (15–22 h), five scans at wavelengths of 276 or 288 nm were collected and averaged. Data were fitted by nonlinear least squares analysis (Gnuplot) by using partial specific volumes calculated by using the program sednterp (32). In most cases, a function including monomers, dimers, and trimers was required to fit the centrifugation data with random distributions of residuals. For these multispecies fits, the masses were fixed at values calculated from the monomer molecular weight.
Binding Assays.
For indirect ELISAs, the procedure of Levchenko et al. (22) was used with minor variations: 9 μg of each SSD domain was immobilized; wells were blocked by using 0.3% BSA in PBS buffer; horseradish peroxidase-conjugated donkey (Fab′)2 anti-rabbit antibody (Amersham Pharmacia) was used; and immunocomplexes were detected by using TMB Micro Well (Kirkegaard & Perry Laboratories). Nonspecific binding, determined by omitting the SSD domains from the ELISA, was subtracted from the readings obtained with the SSD domains present. There was no significant cross-reactivity of antibodies with the SSD domains, and each antisera reacted equally well with the wild-type and control protein variants.
RESULTS
Previous studies of a ClpX fragment containing residues 327–424 showed that this region had the same binding specificity as full length ClpX for two known substrates (22). To characterize the homologous regions of other Clp ATPases and Lon, we cloned fragments containing the ClpA, ClpB, ClpY, and Lon sequences shown in Fig. 1B. Because the original 327–424 ClpX fragment did not fold cooperatively, we also cloned a longer ClpX fragment in the hope of improving stability. We refer to these Clp and Lon fragments as SSD domains. These SSD fragments contain 9 or 10 additional N-terminal residues relative to the original 327–424 fragment of ClpX and extend to the natural C termini of the Clp proteins. The Lon fragment contained 13 C-terminal residues past the region of Clp homology because of uncertainty in the domain boundary. Each SSD fragment was cloned with an N-terminal His-tag for affinity purification.
Stable Folding of the ClpA, ClpY, and Lon SSD Domains.
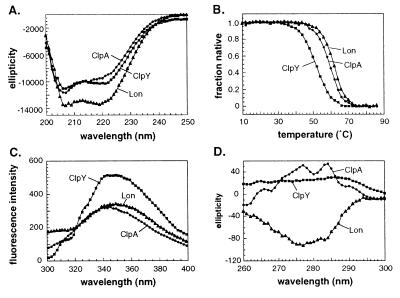
The purified SSD fragments from ClpA, ClpY, and Lon behaved as stable, independently folded, native domains. Far ultraviolet CD revealed α-helical secondary structure ranging from 26–38% for individual domains (Fig. 2A), which was lost cooperatively during thermal denaturation (Fig. 2B). Fluorescence spectra showed blue shifts of 1.5–6.3 nm expected for partial burial of tryptophan (Fig. 2C), and strong and distinct near ultraviolet CD spectra were observed, providing evidence for tight tertiary packing of aromatic side chains (Fig. 2D).
Figure 2.
(A) Far UV CD spectra at 20°C. (B) Thermal denaturation monitored by the change in CD ellipticity at 222 nm. (C) Fluorescence emission spectra at 20°C (excitation 280 nm). (D) Near UV CD spectra at 20°C. In A and D, the units of ellipticity are deg⋅cm2/dmol.
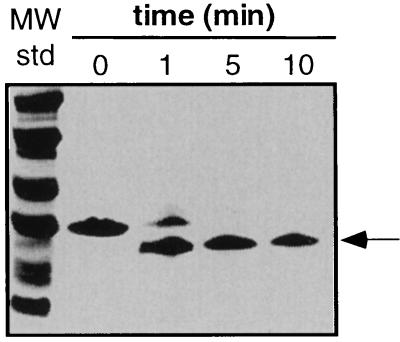
The SSD fragments of ClpA, ClpY, and Lon were largely resistant to protease digestion, again suggesting a stable tertiary fold. A time course of tryptic digestion of the ClpY fragment, monitored by SDS/PAGE, revealed cleavage to produce a slightly smaller species (Fig. 3). Analysis of the digestion products by mass spectrometry showed that trypsin removed a portion of the N-terminal His-tag and three C-terminal residues. As depicted by the arrows in Fig. 1B, trypsin also cut the Lon SSD fragment within the His-tag region and cleaved both the Lon and ClpA domains at positions analogous to the C-terminal ClpY cleavage site. Digestion of the Lon fragment trimmed this domain to approximately the same size as the ClpA and ClpY SSD domains.
Figure 3.
Limited tryptic digestion of the ClpY SSD domain assayed by SDS/PAGE.
Non-Native Properties of the ClpB and ClpX Fragments.
The SSD fragments of ClpB and ClpX were not stably folded and had properties consistent with significant non-native character. For example, both fragments were digested to short peptides by trypsin and chymotrypsin, showed less CD ellipticity than the other SSD domains (18 and 12% helix, respectively), had poorly cooperative (ClpB) or noncooperative (ClpX) thermal melts, and exhibited no significant near ultraviolet CD spectrum (data not shown).
Solution Molecular Weight.
The intact Clp ATPases and Lon protease function as oligomers (1–3, 14). When assayed by analytical ultracentrifugation, the SSD domains did not form stable oligomers but sedimented as expected for mixtures composed predominantly of monomers (Table 1) with some dimeric and trimeric species.
Table 1.
Oligomeric form probed by analytical ultracentrifugation
| SSD domain | Monomer Mr | Weight-average Mr |
|---|---|---|
| ClpA (104 μM) | 12,615 | 16,789 |
| ClpB (60 μM) | 12,635 | 12,649 |
| ClpX (50 μM) | 13,943 | 13,245 |
| ClpY (20 μM) | 14,227 | 15,226 |
| Lon (102 μM) | 15,062 | 18,484 |
SSD Domains Discriminate Between Protein Substrates.
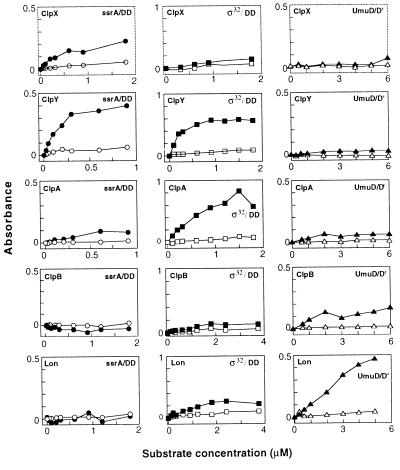
In binding assays (Fig. 4), the SSD domains from ClpA, ClpB, ClpX, ClpY, and Lon showed distinct patterns of interaction with three potential substrates: σ32, UmuD, and Arc repressor with the SsrA-degradation tag (Arc-ssrA). Each of these proteins is degraded rapidly in E. coli (19, 30, 33, 34). No binding was detected to the σ32-DD, UmuD′, or Arc-ssrA-DD control variants, which are comparatively stable to intracellular degradation. The DD variants have different C-terminal dipeptides than their counterparts, and UmuD′ lacks the N-terminal 24 residues of UmuD. Of the 15 combinations of SSD domains and protein pairs, five cases of strong, specific binding were detected. The SSD domains of ClpA and ClpY bound well to σ32 but not to σ32-DD; those of ClpY and ClpX bound to Arc-ssrA but not Arc-ssrA-DD; and the SSD domain of Lon bound to UmuD but not to UmuD′. Specific but lower-level binding also was observed for several other combinations of SSD domains and test proteins (e.g., Lon/σ32; ClpA/Arc-ssrA; ClpA/UmuD; and ClpB/UmuD). As discussed below, many of these binding interactions mimic degradation preferences observed in vivo. We note that the ClpX and ClpB SSD domains bind some potential substrates even though these fragments do not fold stably by themselves. This observation may mean that stable folding is not a prerequisite for protein binding or that binding of the ClpX and ClpB fragments to substrates or other components in the assay stabilizes their native structures.
Figure 4.
ELISA assays of the binding of SSD domains to Arc-ssrA (left column), σ32 (center column), and UmuD (right column) are shown in closed symbols. Binding to the control proteins Arc-ssrA-DD, UmuD′, and σ32-DD are shown in open symbols. Data points are the average of a minimum of four duplicate samples.
DISCUSSION
We have shown that Lon protease and members of the Clp family of ATPases and protease regulatory subunits contain homologous sequences of ≈100 amino acids that mediate binding to and discrimination among different protein substrates. These sequences also contain a signature sequence related to the sensor-2 motif found in clamp-loading subunit of DNA polymerase III and the AAA+ superfamily of ATPases (25, 26). We refer to these Lon/Clp sequences as sensor- and substrate-discrimination or SSD domains. Protein fragments corresponding to the SSD regions of the ClpA, ClpY, and Lon proteins of E. coli fold independently as autonomous structural domains by the criteria of cooperative thermal denaturation, evidence for well packed hydrophobic cores, and resistance to digestion by trypsin and chymotrypsin. The SSD sequences of the ClpX and ClpB proteins of E. coli were poorly folded as isolated fragments. However, this region of ClpX is protease resistant in the intact protein (I. Levchenko and T.A.B., unpublished work), suggesting that interactions with other ClpX domains are required for stability.
The purified SSD domains of Lon and the Clp ATPases showed distinct patterns of binding to three proteins—UmuD, σ32, and SsrA-tagged Arc—that are rapidly degraded in E. coli. In each case, a closely related protein that is comparatively resistant to degradation served as a negative control. For example, UmuD but not but UmuD′ bound strongly to the SSD domain of Lon. UmuD is a substrate for Lon degradation in vivo and in vitro; UmuD′, which lacks 24 N-terminal residues of UmuD, is not degraded by Lon (15). Agreement between degradation specificity and SSD-binding specificity also is observed for the heat-shock transcription factor σ32. ClpYQ has been implicated in σ32 degradation in the cell (18), and the wild-type σ32 protein but not the σ32-DD mutant was recognized by the ClpY SSD domain. As previously shown and confirmed here for Arc-ssrA, there are also strong correlations between the binding specificity of the ClpX SSD domain and the known protease and chaperone specificities of the intact ClpX and ClpXP enzymes (16, 22, 23). As a whole, these results support the conclusion that the SSD domains of Lon and the Clp ATPases function in the recognition of the specific protein substrates of these enzymes.
The observed binding interactions of the SSD domains do not always parallel established proteolytic specificities. For example, the SSD domain of ClpA binds σ32, but ClpAP is not known to degrade σ32. Similarly, the SSD domain of ClpY binds Arc-ssrA, but ClpYQ has not been implicated in degradation of other proteins bearing the SsrA tag (16). Several explanations for these discrepancies are possible. First, binding of the Lon or Clp enzymes to a particular protein substrate may be necessary but not sufficient to trigger degradation because one or more additional signals also are required for substrate unfolding or translocation to the protease active sites. Second, substrates that are bound in vitro may not be degraded in vivo because the enzyme and substrate are localized in different regions of the cell or are expressed at different times or under different growth conditions. Finally, further experiments may show that some of these proteins are indeed substrates for the proteases whose SSD domains bind to them.
What features of substrates are recognized by the SSD domains? C-terminal binding determinants are clearly important for σ32 and Arc-ssrA because substituting their C-terminal amino acids reduces binding by the SSD domains of ClpA, ClpX, or ClpY. By contrast, determinants near the N terminus are indicated for UmuD, as removal of 24 residues to generate UmuD′ prevents binding by the Lon SSD domain. Alanine-substitution experiments suggest that Lon recognizes an internal site in this region of UmuD that includes the sequence F15-P16-L17-F18 (15). Thus, SSD domains, as a class, appear capable of recognizing both internal and C-terminal peptide sequences. Because ClpAP has been implicated in degradation of N-end rule substrates in E. coli (35), SSD-recognition sequences also may include the extreme N-terminal amino acids of some substrates. The number of substrate residues that comprise a recognition site is unknown. However, adding tags of 7–11 residues is sufficient to target several proteins for degradation (23, 36, 30).
Are the SSD domains of specific enzymes specialized for C-terminal, internal, or N-terminal peptide recognition? The available data are too sparse to answer this question definitively, but we suspect that the answer may be no. We note, for example, that the Lon SSD domain discriminates between UmuD and UmuD′, which differ in sequences near the N terminus, and also between σ32 and σ32-DD, which differ at the C terminus. Moreover, ClpAP-dependent degradation in vivo involves the C termini of some substrates and sequences near the N termini of others (16, 35, 37). Obviously, the exact nature of the substrate peptide sequences must play critical roles in determining specificity, but the recognition rules for any given SSD domain are still obscure. The SsrA tag is presented as an unstructured peptide (16, 22), and the UmuD recognition site also is thought to be highly accessible to proteases (15). Hence, it seems likely that the SSD domains of the Clp ATPases and Lon search disordered or accessible regions for the presence of specific sequences that can adopt a complementary structure in the complex.
It was proposed that the substrate-binding region of ClpX contained tandem “PDZ-like” sequences, which are peptide-binding domains in eukaryotic signaling proteins and periplasmic bacterial proteases (22). Our studies here, however, have provided a better definition of the SSD domain boundaries and are inconsistent with this model. Specifically, the regions of cooperatively folded structure in the SSD domains of ClpY, ClpA, and Lon contain all of one and part of another sequence that had been modeled as separate PDZ-like domains. Neuwald et al. (25) have predicted that the SSD regions of the Clp and Lon ATPases resemble helical domains in the structures of the δ′ subunit of E. coli DNA polymerase III (26) and the D2 subfragment of NSF, the N-ethylmaleimide-sensitive factor involved in vesicle fusion (27, 28). Both of these proteins, like the Clp ATPases, are thought to change the conformations of other proteins as part of their mechanisms. NSF is also a hexameric ATPase in which a lysine in a sensor-2 motif interacts with bound nucleotide in an ATPase domain. An interaction of this type between the arginine in the G-Φ-R-X-Φ motif of the Clp/Lon proteins and the ATPase domains of these enzymes might provide a way to couple substrate binding to the SSD domain with nucleotide binding or hydrolysis. This interaction, in turn, could initiate substrate unfolding or translocation reactions. The stable SSD domains characterized here are partly α-helical, as judged by their CD spectra, but the extent of possible structural homology with the sensor-2 domains remains to be determined.
Our finding that Lon and the ClpAP, ClpXP, and ClpYQ proteases use related mechanisms of substrate recognition extends the functional parallels between these energy-dependent, oligomeric proteases (1–3, 14, 38) and suggests that Lon actually can be considered to be part of an extended Clp family. Consistent with this model, the Lon homolog from yeast, like the Clp ATPases, is hexameric and seems to function as a chaperone as well as a protease (39, 40).
Acknowledgments
We thank C. Gross, C. Herman, G. Walker, M. Sutton, and J. Flanagan for their generous gifts of proteins, antibodies, and clones. We thank I. Levchenko for invaluable advice. This work was supported by National Institutes of Health Grant AI-16892 and the Howard Hughes Medical Institute. T.A.B. is an employee of the Howard Hughes Medical Institute. C.K.S. is a Pfizer Life Sciences Research Foundation Post-doctoral Fellow.
ABBREVIATION
- SSD domain
sensor- and substrate-discrimination domain
References
- 1.Gottesman S. Annu Rev Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- 2.Sherman M Y, Goldberg A L. EXS. 1996;77:57–78. doi: 10.1007/978-3-0348-9088-5_5. [DOI] [PubMed] [Google Scholar]
- 3.Wawrzynow A, Banecki B, Zylicz M. Mol Microbiol. 1996;21:895–899. doi: 10.1046/j.1365-2958.1996.421404.x. [DOI] [PubMed] [Google Scholar]
- 4.Wang J, Hartling J A, Flanagan J M. Cell. 1997;91:447–456. doi: 10.1016/s0092-8674(00)80431-6. [DOI] [PubMed] [Google Scholar]
- 5.Bochtler M, Ditzel L, Groll M, Huber R. Proc Natl Acad Sci, USA. 1997;94:6070–6074. doi: 10.1073/pnas.94.12.6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rohrwild M, Pfeifer G, Santarius U, Muller S A, Huang H C, Engel A, Baumeister W, Goldberg A L. Nat Struct Biol. 1997;4:133–139. doi: 10.1038/nsb0297-133. [DOI] [PubMed] [Google Scholar]
- 7.Kessel M, Maurizi M R, Kim B, Kocsis E, Trus B L, Singh S K, Steven A C. J Mol Biol. 1995;250:587–594. doi: 10.1006/jmbi.1995.0400. [DOI] [PubMed] [Google Scholar]
- 8.Beuron F, Maurizi M, Belnap D M, Kocsis E, Booy F P, Kessel M, Steven A C. J Struct Biol. 1998;123:248–259. doi: 10.1006/jsbi.1998.4039. [DOI] [PubMed] [Google Scholar]
- 9.Hoskins J R, Pak M, Maurizi M, Wickner S. Proc Natl Acad Sci USA. 1998;95:12135–12140. doi: 10.1073/pnas.95.21.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wickner S, Gottesman S, Skowyra D, Hoskins J, McKenney K, Maurizi M R. Proc Natl Acad Sci USA. 1994;91:12218–12222. doi: 10.1073/pnas.91.25.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levchenko I, Luo L, Baker T A. Genes Dev. 1995;9:2399–2408. doi: 10.1101/gad.9.19.2399. [DOI] [PubMed] [Google Scholar]
- 12.Wawrzynow A, Wojtkowiak D, Marszalek J, Banecki B, Jonsen M, Graves B, Georgopoulos C, Zylicz M. EMBO J. 1995;14:1867–1877. doi: 10.1002/j.1460-2075.1995.tb07179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Konieczny I, Helinski D R. Proc Natl Acad Sci USA. 1997;94:14378–14382. doi: 10.1073/pnas.94.26.14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottesman S, Wickner S, Maurizi M R. Genes Dev. 1997;11:815–823. doi: 10.1101/gad.11.7.815. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez M, Frank E, Levine A S, Woodgate R. Genes Dev. 1998;12:3889–3899. doi: 10.1101/gad.12.24.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottesman S, Roche E, Zhou Y, Sauer R T. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herman C, Thévenet D, Bouloc P, Walker G C, D’Ari R. Genes Dev. 1998;12:1348–1355. doi: 10.1101/gad.12.9.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanemori M, Nishihara K, Yanagi H, Yura T. J Bacteriol. 1997;179:7219–7225. doi: 10.1128/jb.179.23.7219-7225.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herman C, Thévenet D, Bouloc P, D’Ari R, Bouloc P. Proc Natl Acad Sci USA. 1995;92:3516–3520. doi: 10.1073/pnas.92.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomoyasu T, Gamer J, Bukau B, Kanemori M, Mori H, Rutman A J, Oppenheim A B, Yura T, Yamanaka K, Niki H, et al. EMBO J. 1995;14:2551–2560. doi: 10.1002/j.1460-2075.1995.tb07253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biaszczak A, Georgopoulos C, Liberek K. Mol Microbiol. 1999;31:157–166. doi: 10.1046/j.1365-2958.1999.01155.x. [DOI] [PubMed] [Google Scholar]
- 22.Levchenko I, Smith C K, Walsh N P, Sauer R T, Baker T A. Cell. 1997;91:939–947. doi: 10.1016/s0092-8674(00)80485-7. [DOI] [PubMed] [Google Scholar]
- 23.Levchenko I, Yamauchi M, Baker T A. Genes Dev. 1997;11:1561–1572. doi: 10.1101/gad.11.12.1561. [DOI] [PubMed] [Google Scholar]
- 24.Schirmer E C, Glover J R, Singer M A, Lindquist S. Trends Biochem Sci. 1996;21:289–296. [PubMed] [Google Scholar]
- 25.Neuwald A F, Iyer A, Spouge J L, Koonin E V. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 26.Guenther B, Onrust R, Sali A, O’Donnell M, Kuriyan J. Cell. 1997;91:335–345. doi: 10.1016/s0092-8674(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 27.Lenzen C U, Steinmann D, Whiteheart S W, Weis W I. Cell. 1998;94:525–536. doi: 10.1016/s0092-8674(00)81593-7. [DOI] [PubMed] [Google Scholar]
- 28.Yu R C, Hanson P I, Jahn R, Brünger A T. Nat Struct Biol. 1998;5:803–811. doi: 10.1038/1843. [DOI] [PubMed] [Google Scholar]
- 29.Milla M E, Brown B M, Sauer R T. Nat Struct Biol. 1994;1:518–523. doi: 10.1038/nsb0894-518. [DOI] [PubMed] [Google Scholar]
- 30.Keiler K C, Waller P R H, Sauer R T. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 31.Milla M E, Brown B M, Sauer R T. Protein Sci. 1993;2:2198–2205. doi: 10.1002/pro.5560021219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laue T M, Shah B D, Ridgeway T M, Pelletier S L. In: Analytical Ultracentrifugation in Biochemistry and Polymer Science. Harding S E, Rowe A J, Hortons J C, editors. Cambridge, U.K.: R. Soc. Chem.; 1992. pp. 90–125. [Google Scholar]
- 33.Straus D, Walter W, Gross C A. Nature (London) 1987;329:348–351. doi: 10.1038/329348a0. [DOI] [PubMed] [Google Scholar]
- 34.Frank E G, Ennis D G, Gonzalez M, Levine A S, Woodgate R. Proc Natl Acad Sci USA. 1996;93:10291–10296. doi: 10.1073/pnas.93.19.10291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tobias J W, Shrader T E, Rocap G, Varshavsky A. Science. 1991;254:1374–1377. doi: 10.1126/science.1962196. [DOI] [PubMed] [Google Scholar]
- 36.Laachouch J E, Desmet L, Geuskens V, Grimaud R, Toussaint A. EMBO J. 1996;15:437–444. [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L, Elliott M, Elliott T. J Bacteriol. 1999;181:1211–1219. doi: 10.1128/jb.181.4.1211-1219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gottesman S, Wickner S, Jubete Y, Singh S K, Kessel M, Maurizi M. Cold Spring Harbor Symp Quant Biol. 1995;60:533–548. doi: 10.1101/sqb.1995.060.01.057. [DOI] [PubMed] [Google Scholar]
- 39.Kutejova E, Durcova G, Surovkova E, Kuzela S. FEBS Lett. 1993;329:47–50. doi: 10.1016/0014-5793(93)80190-6. [DOI] [PubMed] [Google Scholar]
- 40.Rep M, van Dijl J M, Suda K, Schatz G, Grivell L A, Suzuki C K. Science. 1996;274:103–106. doi: 10.1126/science.274.5284.103. [DOI] [PubMed] [Google Scholar]