Abstract
ZEB is an active transcriptional repressor that regulates lymphocyte and muscle differentiation in vertebrates. Its homologue in Drosophila (zfh-1) is also essential for differentiation of somatic and cardiac muscle. Here, we demonstrate that ZEB and zfh-1 interact with the corepressor CtBP to repress transcription. ZEB and zfh-1, both contain the sequence PLDLS in the same region of the repressor domain, and we demonstrate that this sequence binds CtBP-1 and -2. In vertebrate species, ZEB contains two additional CtBP-like binding sites (variations of the PLDLS sequence) that also bind CtBP proteins and are required for full repressor activity. The three sites have an additive effect, and mutation of all three sites is necessary to abolish both binding to CtBP and repressor activity. Finally, we demonstrate that the interaction of CtBP with ZEB at the promoter is necessary for repressor activity.
Transcriptional repression is essential to establish patterns of gene expression during development and differentiation along the phylogenetic scale from Drosophila to mammals (1, 2). A number of transcriptional activators regulate transcription through interaction with a coactivator protein, the best studied of which is CBP/p300 (3). And in an analogous fashion, several repressors have been shown to recruit corepressors. Examples of such corepressors are the sin3/NCoR/histone deacetylase complex, retinoblastoma protein/histone deacetylase complex, and Groucho and Nab proteins (4–7). Another corepressor is CtBP, which was discovered by the Chinnadurai group as a protein that interacts with the PLDLS sequence located in the C-terminal region of the E1A protein from adenovirus types 2 and 5 (8, 9). Deletion of this sequence increased the efficiency of E1A to collaborate with ras in transformation of cells (8). It also increases the tumorigenic and metastatic capacity of the transformed cells, suggesting a regulatory role for CtBP in cell proliferation (8).
CtBP does not bind directly to DNA but when tethered directly to the promoter through a Gal4 DNA binding domain CtBP efficiently represses transcription, demonstrating that the protein is a corepressor (10). CtBP recently has been shown to bind to another mammalian protein, CtIP (11). And, CtIP has been shown to interact with the breast-associated tumor suppressor, BRCA-1 (12, 13). The significance of this interaction remains unclear; however, interaction between BRCA-1 and CtIP is abolished by tumor-associated mutations on BRCA-1, suggesting that the BRCA-1/CtIP interaction may be required for tumor suppression by BRCA-1. Recently, a CtBP-related gene, termed CtBP-2, has been cloned (14, 15). Less is known about CtBP-2 although it has been shown to interact with the basic Krüppel–like factor (BKLF) in in vitro assays and in yeast two-hybrid analysis (15).
We and others previously have characterized the vertebrate zinc finger/homeodomain ZEB as an active transcriptional repressor that regulates both muscle and lymphoid differentiation (16–20). Its homologue in Drosophila, zfh-1, regulates somatic and cardiac myogenesis during Drosophila embryogenesis (A.A.P., E. Ward, J. B. Skeath, and D.C.D., unpublished work). ZEB and zfh-1 share a number of similarities with the repressor snail (A.A.P., E. Ward, J. B. Skeath, and D.C.D., unpublished work). Both ZEB/zfh-1 and snail are also zinc finger repressors that bind to a subset of E box (and E box-like) sequences with the highest affinity for the sequence CACCTG (refs. 16, 17, and 22; A.A.P., E. Ward, J. B. Skeath, and D.C.D., unpublished work). snail regulates mesodermal formation and differentiation and establishes the boundary between mesoderm and neurogenic ectoderm in the Drosophila embryo by preventing ectopic expression of genes in the mesoderm (23, 24). snail is required for zfh-1 expression, but zfh-1 persists after snail is down-regulated (23–25), and our results suggests that zfh-1 functions downstream of snail in a snail-like fashion to regulate differentiation of mesodermal derivatives (21). Recently, it was shown that transcriptional repression by snail depends on its binding to CtBP (10), and that CtBP is essential for mesoderm formation and patterning in early Drosophila embryos (26). CtBP also functions as a corepressor with two other Drosophila repressors that are critical during Drosophila embryogenesis, hairy and knirps (10, 26). Here, we demonstrate that like snail, ZEB and zfh-1 interact with CtBP-1 and -2 to repress transcription.
METHODS
Cell Culture, Transient Transfections, and Chloramphenicol Acetyltransferase (CAT) Assays.
The C33a cervical carcinoma cell line (American Type Culture Collection) was maintained in DMEM (Life Technologies, Grand Island, NY) containing 5% FCS and 5% calf serum (Life Technologies).
C33a cells were transfected by the calcium phosphate method as described (18, 19). After 48 hr, lysates were collected, and CAT assays were performed as described (18, 19).
Plasmid Construction.
Flag-tagged DB-ZEB, DB-zfh-1, ZEB, and zfh-1 were made as described (18, 19). snail was amplified by PCR from the pPac-snail vector (obtained from T. Yp, University of Massachusetts, Worcester) and cloned into the EcoRI/XbaI sites of pCI-neo (Promega). A Flag tag sequence (18) was included in the forward primer. The vectors refered to as “soluble” simply lack the DNA binding domain of yeast Gal4 protein.
To construct G-ZEB-1–1120, G-ZEB-302–903, and G-ZEB-700–776 (referred in this paper as G-ZEB), the region between the corresponding amino acids of ZEB was amplified by PCR and cloned into the BamHI/XbaI sites of the Gal4 DNA binding domain PM1 vector as described (18). G-ZEBmut (containing the sequence between amino acids 700 and 776 with a mutation of the CtBP site at amino acid 734 to the sequence ASASA) and G-ZEB-3 mut (containing the sequence of ZEB between amino acids 700 and 776 with the CtBP sites at amino acids 705, 734, and 767 mutated to the sequence ASASA) were obtained as follows. First, the region of ZEB cDNA between amino acids 546 and 740 was amplified by using a reverse oligonucleotide where the CtBP site (amino acid 734) was mutated from PLDLS to ASASA. The corresponding fragment then was cloned back into G-ZEB. The mutated region of ZEB (amino acids 700–776) then was amplified by PCR using oligonucleotides corresponding to the wild-type (to construct G-ZEBmut) or mutated (to construct G-ZEB-3 mut) sequences for the sites at amino acids 700 and 776. Forward and reverse oligonucleotides contained BamHI and XbaI sites, respectively, which were used to clone the PCR fragments into the corresponding sites of PM1.
To construct G-zfh-1, the region of zfh-1 between amino acids 765 and 821 was amplified by PCR and cloned into the EcoRI/XbaI site of PM1. To create G-zfh-1-mut, we amplified that region by using a forward primer with an EcoRI site and a reverse primer containing a mutation of the PLDLS sequence to ASASA sequence and including the SacI site located at 2,420 bp and a flanking SalI site. This fragment was cloned into the EcoRI/SalI of PM1. Then with a second PCR we amplified the rest of the region by using a forward primer with a SacI site and a reverse primer containing a XbaI site. The resulting fragment then was cloned into the SacI/XbaI site of the previous construct.
CtBP-1 (G. Chinnadurai, St. Louis University, St. Louis, MO) was cloned in-frame in the EcoRI/XbaI site of the myc-tagged CS2-MT expression vector (R. Kopan, Washington University School of Medicine, St. Louis, MO). CtBP-2 (M. Crossley, University of Sydney, Australia) was cloned in the NcoI/XbaI site of CS2-MT.
Coimmunoprecipitation and Western Blot Analysis.
Forty-eight hours after transfection, C33a cells were lysed in ELB (150 mM NaCl/50 mM Hepes, pH 7.0/5 mM EDTA/0.1% NP-40), sonicated briefly, and centrifuged. The lysates then were immunoprecipitated with anti-M2 anti-Flag mAb (IBI-Kodak, New Haven, CT) for 2 hr at room temperature, washed three times with ELB, twice with PBS, and boiled in sample buffer-5% β-mercapto-ethanol (βME). The samples then were loaded onto a 4–15% polyacrylamide gradient gel (Bio-Rad) and transferred overnight to a poly(vinylidene difluoride) membrane (Millipore) in 10 mM 3-cyclohexylamino-l-propane sulfonic acid (CAPS) (pH 11.0) buffer. The membrane was serially incubated with anti-9E10 anti-myc tag mAb (Santa Cruz Biotechnology) and anti-mouse Ig G-HRP secondary antibody (Jackson ImmunoResearch). After several washes, the membrane then was developed by the chemiluminiscence method (NEN Life Science) according to manufacturer’s instructions. After that, the antibodies were stripped from the membrane by incubation at 55°C for 10 min in stripping solution (62 mM of Tris⋅HCl, pH 6.8/2% SDS/100 mM of βME). After stripping, membrane was incubated with anti-Flag polyclonal antibody (Santa Cruz Biotechnology), secondary anti-rabbit Ig G-horseradish peroxidase (Jackson ImmunoResearch), and developed again by the chemiluminiscence method.
RESULTS
ZEB and zfh-1 Bind to CtBP-1 and CtBP-2.
We have found that zfh-1 and ZEB are functionally related to snail, a repressor that plays a key role in early stages of Drosophila embryogenesis and that represses transcription by interacting with CtBP (refs. 10, 23, and 24; A.A.P., E. Ward, J. B. Skeath, and D.C.D., unpublished work). We wondered whether ZEB and zfh-1 also might use CtBP as a corepressor.
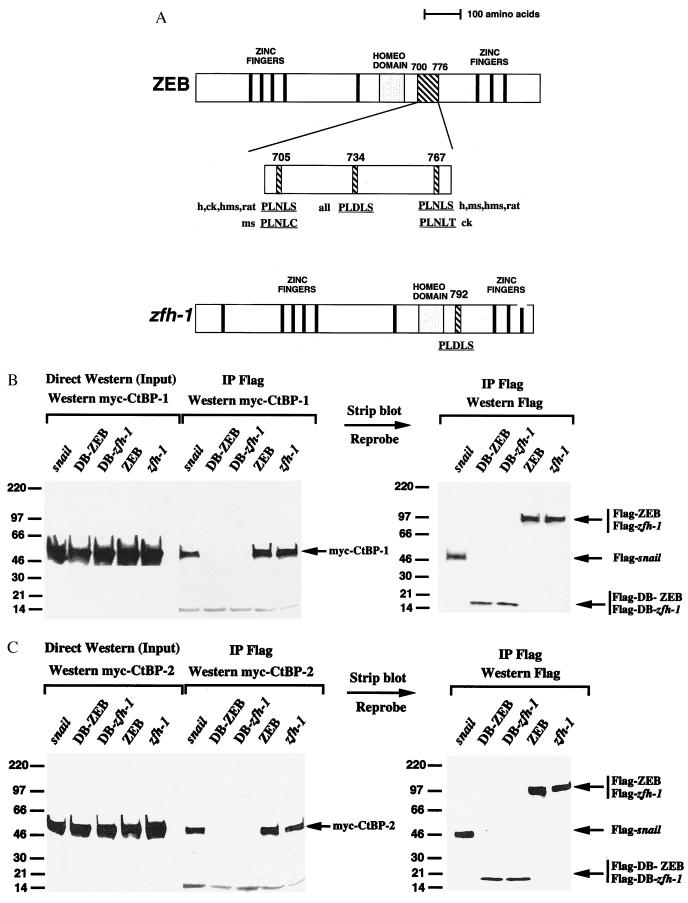
We have localized the repressor domain of ZEB and zfh-1 to the central region of the molecule (amino acids 302–903), between the zinc fingers (DNA binding domain) at either end of the protein (18). The homeodomain is located in the region but is not required for the repressor activity (18). ZEB and zfh-1 have a high level of sequence similarity in the sequence of their zinc finger and homeodomain regions, but there is little evidence of such extended sequence similarity in the repressor domain (16, 27). Nevertheless, ZEB and zfh-1 repress the same set of transcription factors and their activities in vivo overlap significantly, suggesting that their repressor domains and mechanisms of action must be similar (A.A.P., E. Ward, J. B. Skeath, and D.C.D., unpublished work). Despite the lack of similarity between ZEB and zfh-1 in the repressor domain, we found that both proteins contain a potential CtBP binding site (PLDLS sequence) in the same region of the repressor domain (amino acid 734 in ZEB and 792 in zfh-1) (Fig. 1A), suggesting conservation of this site during evolution. This site also is conserved in all of the vertebrate species where ZEB has been cloned, including hamster, rat, mouse, chicken, and human (16, 28–31).
Figure 1.
ZEB and zfh-1 bind to CtBP-1 and CtBP-2. (A) ZEB and zfh-1 contain CtBP binding sites. Drosophila zfh-1 and ZEB from various vertebrate species (h: human; ck: chicken; hms: hamster; ms: mouse and rat) contain a PLDLS CtBP binding sequence in their repressor domain. In addition, vertebrates contain two additional PLDLS-like sequences, which vary slightly from species to species. (B) Flag-tagged constructs for snail, the repressor domains of ZEB and zfh-1 (the region between both zinc finger domains) as well as the DNA binding domains (C-terminal zinc fingers) of ZEB (DB-ZEB) and zfh-1 (DB-zfh-1) were cotransfected in C33a cells with myc-tagged CtBP-1. After 48 hr, cells were lysed and after immunoprecipitation with Flag antibody, binding to CtBP-1 was detected by Western blot using 9E10 anti-myc mAb as described in Materials and Methods. Ten percent of the lysate was run without immunoprecipitation as input control. Blots then were stripped and incubated with anti-Flag antibody to detect levels of snail, ZEB and zfh-1. (C) As in B but using myc-tagged CtBP-2.
We used a coimmunoprecipitation assay to test for interaction between ZEB/zfh-1 and CtBP-1. As shown in Fig. 1B, we found that the repressor domain of both ZEB and zfh-1 interacts with CtBP-1 as efficiently (or even more efficiently) than snail. As a control, the DNA binding domain of both proteins (DB-ZEB and DB-zfh-1) did not bind to CtBP-1 (Fig. 1B). In view of the sequence similarity between CtBP-1 and CtBP-2 we also tested the ability of ZEB and zfh-1 to interact with CtBP-2. As shown in Fig. 1C, both ZEB and zfh-1 also interact very efficiently with CtBP-2.
The CtBP-Binding Region of ZEB Mediates Transcriptional Repression.
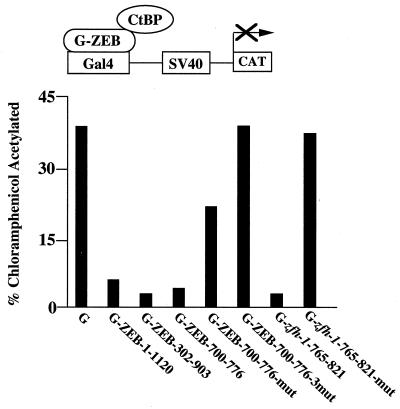
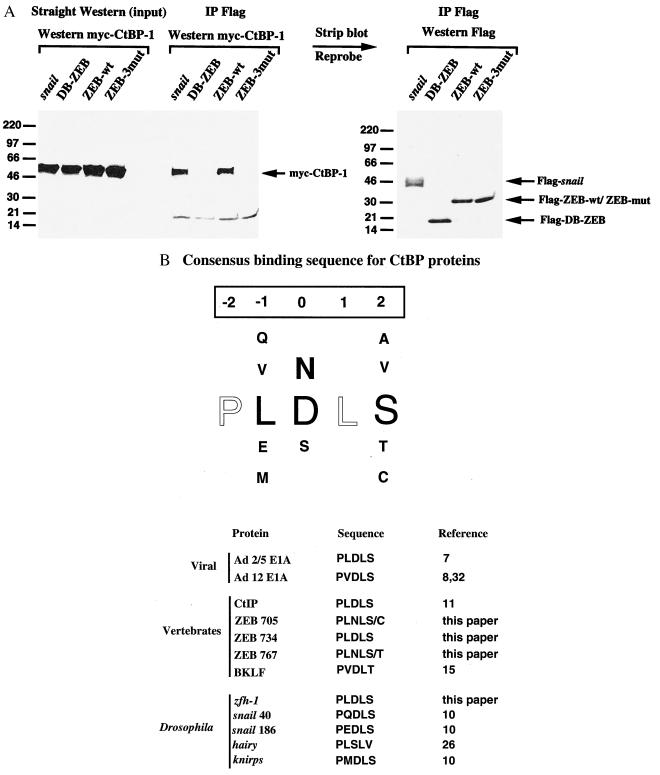
We previously identified that the region between both zinc finger domains of ZEB (amino acids 302–903) contains the repressor activity (18). We next asked whether the region of ZEB containing the PLDLS sequence was sufficient for repressor activity. This region (amino acids 700–776) was fused to the DNA binding domain of the yeast protein Gal4 and tested for its ability to repress the activity of the simian virus 40 (SV40) enhancer/promoter reporter in transfection assays. As shown in Fig. 2, this region of ZEB (G-ZEB-770–776) repressed transcription as efficiently as full-length ZEB (G-ZEB-1–1120) or the area between both zinc fingers (G-ZEB-302–903). In the adenoviral 2/5 E1A protein, mutations of the PLDLS sequence to ASASA completely abolish binding to CtBP-1 (8, 9). The same mutation in the PLDLS sequence of ZEB (G-ZEB-mut) decreased the repressor activity, but it did not eliminate it (Fig. 2). In addition, when we checked this mutant for its ability to bind CtBP-1, we found that there was still binding activity (data not shown). Examination of the ZEB cDNA sequence revealed two additional CtBP-like sites (Fig. 1A). Therefore, we mutated all three sites and fused this mutated sequence to the Gal4 binding domain (G-ZEB-3 mut). G-ZEB-3 mut had no repressor activity (Fig. 2), and mutation of all three CtBP sites also completely abolished binding to CtBP-1 (Fig. 3A). We conclude that all three sites participate in repression of ZEB through binding to the CtBP proteins. Mutation of the unique PLDLS site in zfh-1 also completely blocked the ability of G-zfh-1 (amino acids 765–821) to repress transcription (Fig. 2).
Figure 2.
Binding of ZEB to CtBP mediates transcriptional repression. The region of ZEB corresponding to amino acids 1–1120, 302–903, and 700–776 were fused to the DNA binding domain of the yeast Gal4 protein and tested for its ability to repress transcription of the SV40 promoter/enhancer. Two micrograms of the Gal4 PM1 empty vector, 2 μg of wild-type G-ZEB-700–776 (amino acids 700–776), 2 μg of G-ZEB-700–776-mut (amino acids 700–776 with mutation of the CtBP site at 734), 2 μg of G-ZEB3 mut (amino acids 700–776 with mutation of CtBP sites at 705, 734, and 767), 3 μg of G-ZEB-302–903 (amino acids 302–903), 4 μg of G-ZEB-1–1120 (full-length ZEB), 2 μg of wild-type G-zfh-1–765-821 (amino acids 765–821) and 2 μg of G-zfh-1–765-821-mut (amino acids 765–821 with a mutation of the CtBP site at 792) were cotransfected with 0.8 μg of a reporter containing the SV40 enhancer/promoter (18). Transfection and assessment of CAT activity was performed as described in Materials and Methods.
Figure 3.
(A) Mutation of all three CtBP binding sites in ZEB abolishes binding to CtBP. Flag-tagged expression vectors for snail, the DNA binding domain of ZEB, and the region of ZEB between amino acids 700 and 776 (either wild type or mutated in all three CtBP binding sites, as described in Fig. 2) were cotransfected along with myc-tagged CtBP-1. Cells were collected, and binding to CtBP-1 was detected by Western blot using 9E10 anti-myc mAb as described in Materials and Methods. The blot then was stripped and reprobed with anti-Flag antibody to check expression levels of the Flag proteins. (B) Consensus binding sequence for CtBP. Amino acids at positions −2 and +1 are constant in all proteins and species (outlined). The residue at position 0 is also highly conserved with Asp and Asn in most cases. The residues at positions −1 and +2 show great variability. The size of the residue indicates the frequency for the presence of that residue. The consensus sequence was established by accounting all proteins so far known to interact with CtBP proteins: E1A regions of adenovirus types 2, 5, and 12, CtIP, ZEB in all species where it has been cloned (see Fig. 1A), BKLF, zfh-1, hairy, snail, and knirps.
Our results also expand the previously known consensus binding sequence of CtBP. As shown in Fig. 3B, CtBP recognizes a 5-aa core (8, 32) in which amino acids in positions −2 and +1 are conserved in all CtBP-binding proteins (8–11, 15, 26, 32). The amino acid in position 0 in all proteins is Asp or Asn except in hairy where it is a Ser residue (26). However, amino acid at position −1 shows wide variability from protein to protein. Finally, the residue at position +2 is usually a Ser except for hairy (Val), BKLF (Thr), and ZEB [mouse amino acid 705 (Cys) and chicken amino acid 767 (Thr)] (Fig. 1A) (15). By using synthetic peptides for the CtBP binding site in E1A from adenovirus 5 and 12, it has been shown that although the major binding region corresponds to these five amino acids the sequence around this core also contributes to the binding (32). Although there is not a consensus for the sequence around the five core amino acids, C terminal of it there are usually Glu or Gln residues, whereas in the N-terminal region Lys residues are common.
CtBP Interacts with ZEB at the Promoter to Repress Transcription.
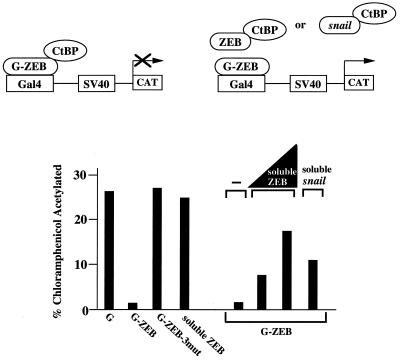
ZEB must bind to the promoter to repress transcription (when a DNA binding site is not present, ZEB did not repress transcription) (Fig. 4). We reasoned that if repression by G-ZEB (Fig. 2) depends on the interaction of G-ZEB with CtBP at the promoter, overexpression of a soluble form of the repressor domain of ZEB (lacking the Gal4 DNA binding domain) should bind and titrate out endogenous CtBP proteins, preventing their interaction with G-ZEB bound to the promoter. Indeed, as shown in Fig. 4, overexpression of increasing amounts of soluble ZEB efficiently blocked the repression by G-ZEB and worked as a dominant negative. Likewise, overexpression of soluble snail (without a Gal4 DNA binding domain) inhibited significantly repressor activity by ZEB (Fig. 4). And conversely, repression by snail was inhibited by overexpression of soluble ZEB (data not shown). These results indicate that the repressor domain of ZEB and snail interacts with CtBP at the promoter to repress transcription. Also, they suggest that the binding sites on snail and ZEB are similar such that they compete for binding to CtBP. Beyond simply demonstrating that proteins containing PLDLS-like sites can compete for binding to CtBP, this assay may reflect a competitive mechanism occurring in vivo. For example, it has been demonstrated with the p300/CBP coactivator that its interaction with STAT-1α can titrate p300 limiting and regulating its availability for other p300-binding factors such as AP-1 (21).
Figure 4.
CtBP and ZEB must interact at the promoter to repress transcription. Soluble ZEB (amino acids 700–776, without the Gal4 DNA binding domain) is not able to repress transcription of the reporter containing Gal4 binding sites upstream of the SV40 promoter/enhancer. Two micrograms of either G-ZEB (amino acids 700–776 of ZEB fused to Gal4 binding domain), G-ZEB-3 mut (amino acids 700–776 of ZEB fused to Gal4 binding domain but with all three CtBP sites at positions 705, 734, and 767 mutated) or soluble ZEB were cotransfected with 0.8 μg of the SV40 promoter/enhancer reporter. However, overexpression of soluble ZEB and soluble snail inhibit repression by G-ZEB (amino acids 700–776) bound to the promoter. Increasing amounts of the soluble ZEB expression vector (2.3 and 4.7 μg) or soluble snail expression vector (6.2 μg) were cotransfected with 0.8 μg of a SV40 enhancer/promoter reporter and 2 μg of the G-ZEB (amino acids 700–776) expression vector.
DISCUSSION
In this paper, we have shown that interaction of ZEB with CtBP is important in transcriptional repression by ZEB, and that ZEB interacts with CtBP-1 and-2 in vivo. Several different transcription factors have been shown to interact with CtBP. Most of them are known transcriptional repressors in Drosophila (snail, hairy, knirps) (10, 26). CtBP is critical in early stages of Drosophila embryogenesis where these repressors function (26). Other CtBP-binding proteins like the human CtIP protein are still of unknown function (11). The interaction of CtBP with E1A is critical in regulating the ability of E1A to mediate cell transformation (8). Taken together, these results demonstrate that CtBP is a important corepressor whose number of repressor partners is likely to increase significantly as further studies are completed. It is of note that all repressors that have been shown to date to interact with CtBP are zinc finger proteins (10, 15, 26). It is then interesting to speculate that there is a more general evolutionary linkage between these repressors that extends beyond the PLDLS (or PLDLS-like) sequence and their recruitment of CtBP. It is also interesting that this sequence is also present in viral proteins such as E1A (8), suggesting that binding to CtBP is an important regulatory mechanism that also has been used by viruses. The PLDLS sequence itself and its position within the repressor domain of ZEB and zfh-1 is conserved from Drosophila to human (16, 27–31). This finding together with the fact that both proteins repress an identical set of factors and that their functions appear to be conserved, suggests that CtBP is a critical protein that serves a common function of ZEB and zfh-1 that has been maintained evolutionarily.
It is interesting that ZEB, in addition to the PLDLS sequence, contains two PLDLS-like sequences that interact with CtBP. All three sites are required for full repressor activity and all three sites have to be mutated to block binding. Contrary to the PLDLS sequence, which is conserved in ZEB/zfh-1 from Drosophila to humans, these two additional CtBP-like sites are present only in vertebrates and show some variations in the third and fifth amino acid of the core sequence (16, 27–31) (Fig. 1A). The significance and relevance of these three binding sites in ZEB remains to be determined. It is possible that the three sites simply act to recruit multiple CtBP proteins and therefore increase repressor activity. Alternatively, multiple sites may have additional biological significance. Conceivably, these three different sites on ZEB may be able to discriminate among different CtBP members or related proteins in vivo. So far only one protein has been described in Drosophila (10, 26) whereas in mammals there are at least two members (8, 9, 14, 15).
It would be of interest to study whether the lack of CtBP would interfere with the function of zfh-1 and ZEB in vivo. zfh-1 is down-regulated during Drosophila embryogenesis, allowing muscle differentiation to proceed (A.A.P., E. Ward, J. B. Skeath, and D.C.D., unpublished work; ref. 25). In the same way, we have provided evidence that ZEB regulates the onset of mammalian myogenesis (18). Maintenance of zfh-1 expression later in Drosophila development (by using a heat shock construct) results in severe block of somatic and cardiac myogenesis (A.A.P., E. Ward, J. B. Skeath, and D.C.D., unpublished work). The contribution of CtBP to the regulation of myogenesis by zfh-1 is difficult to address at this point because embryos lacking CtBP show severe defects in mesodermal differentiation before zfh-1 is expressed (25, 26).
CtBP-1 and -2 are quite similar in their sequence (9, 14, 15). Our results demonstrate that both proteins can recognize the same repressor in vivo. It is unclear whether CtBP-1 and -2 may be able to discriminate among binding partners, or whether their targets are completely overlapping. It is also unclear yet whether CtBP-1 and -2 will repress the same set of transcription factors and thus the same set of genes. The pattern of expression for CtBP-1 and -2 in vivo is also unknown, so it is not known whether they may be involved in distinct biological processes. Further, study of this family of corepressors and their repressor binding partners should provide further insight into the mechanism surrounding transcriptional repression.
Acknowledgments
We are grateful to Drs. G. Chinnadurai, M. Crossley, N. Katsanis, R. Kopan, and T. Yp for providing us with plasmids and other reagents. A.A.P. is supported by a fellowship from the Leukemia Society of America. This work was funded by grants from the National Institutes of Health to D.C.D.
ABBREVIATIONS
- SV40
simian virus 40
- CAT
chloramphenicol acetyltransferase
References
- 1.Ogbourne S, Antalis T M. Biochem J. 1998;331:1–14. doi: 10.1042/bj3310001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gray S, Levine M. Curr Opin Cell Biol. 1998;8:358–364. doi: 10.1016/s0955-0674(96)80010-x. [DOI] [PubMed] [Google Scholar]
- 3.Eckner R. Biol Chem. 1996;377:685–688. [PubMed] [Google Scholar]
- 4.Alland L, Muhle R, Hou H, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Nature (London) 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 5.Luo R X, Postigo A A, Dean D D. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 6.Fisher A L, Caudy M. Genes Dev. 1998;12:1931–1940. doi: 10.1101/gad.12.13.1931. [DOI] [PubMed] [Google Scholar]
- 7.Swirnoff A H, Apel E D, Svaren J, Sevetson B R, Zimonjic D B, Popescu N C, Milbrandt J. Mol Cell Biol. 1998;18:512–524. doi: 10.1128/mcb.18.1.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyd J M, Subramanian T, Schaeper U, La Regina M, Bayley S, Chinnadurai G. EMBO J. 1993;12:469–478. doi: 10.1002/j.1460-2075.1993.tb05679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schaeper U, Boyd J M, Verman S, Uhlmann E, Subramanian T, Chinnadurai G. Proc Natl Acad Sci USA. 1995;92:10467–10471. doi: 10.1073/pnas.92.23.10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nibu Y, Zhang H, Levine M. Science. 1998;280:101–104. doi: 10.1126/science.280.5360.101. [DOI] [PubMed] [Google Scholar]
- 11.Schaeper U, Subramanian T, Lim L, Boyd J M, Chinnadurai G. J Biol Chem. 1998;273:8549–8552. doi: 10.1074/jbc.273.15.8549. [DOI] [PubMed] [Google Scholar]
- 12.Yu X, Wu L C, Bowcock A M, Aronheim A, Baer R. J Biol Chem. 1998;273:25388–25392. doi: 10.1074/jbc.273.39.25388. [DOI] [PubMed] [Google Scholar]
- 13.Wong A K, Ormonde P A, Pero R, Chen Y, Lian L, Salada G, Berry S, Lawrence Q, Dayananth P, Ha P, et al. Oncogene. 1998;17:2279–2285. doi: 10.1038/sj.onc.1202150. [DOI] [PubMed] [Google Scholar]
- 14.Katsanis N, Fisher E M. Genomics. 1998;47:294–299. doi: 10.1006/geno.1997.5115. [DOI] [PubMed] [Google Scholar]
- 15.Turner J, Crossley M. EMBO J. 1998;17:5129–5140. doi: 10.1093/emboj/17.17.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Genetta T, Ruezinsky D, Kadesch T. Mol Cell Biol. 1994;14:6153–6163. doi: 10.1128/mcb.14.9.6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sekido R, Murai K, Funahashi J, Kamachi Y, Fujisawa-Sehara A, Nabeshima K, Kondoh H. Mol Cell Biol. 1994;14:5692–5700. doi: 10.1128/mcb.14.9.5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Postigo A A, Dean D C. EMBO J. 1997;16:3935–3943. doi: 10.1093/emboj/16.13.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Postigo A A, Sheppard A M, Mucenski M L, Dean D C. EMBO J. 1997;16:3924–3934. doi: 10.1093/emboj/16.13.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Higashi Y, Moribe H, Takagi T, Sekido R, Kawakami K, Kikutani H, Kondoh H. J Exp Med. 1997;185:1467–1479. doi: 10.1084/jem.185.8.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horvai A E, Xu L, Korzus E, Brard G, Kalafus D, Mullen T M, Rose D W, Rosenfeld M G, Glass C K. Proc Natl Acad Sci USA. 1997;94:1074–1079. doi: 10.1073/pnas.94.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuse N, Hirose S, Hayashi S. Development (Cambridge, UK) 1996;122:1059–1067. doi: 10.1242/dev.122.4.1059. [DOI] [PubMed] [Google Scholar]
- 23.Leptin M. Genes Dev. 1991;5:1568–1576. doi: 10.1101/gad.5.9.1568. [DOI] [PubMed] [Google Scholar]
- 24.Gray S, Levine M. Genes Dev. 1996;10:700–710. doi: 10.1101/gad.10.6.700. [DOI] [PubMed] [Google Scholar]
- 25.Lai Z, Fortini M E, Rubin G M. Mech Dev. 1991;34:123–134. doi: 10.1016/0925-4773(91)90049-c. [DOI] [PubMed] [Google Scholar]
- 26.Poortinga G, Watanabe M, Parkhurst S M. EMBO J. 1998;17:2067–2078. doi: 10.1093/emboj/17.7.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fortini M E, Lai Z C, Rubin G M. Mech Dev. 1991;34:113–122. doi: 10.1016/0925-4773(91)90048-b. [DOI] [PubMed] [Google Scholar]
- 28.Funahashi J, Seikido R, Murai K, Kamachi Y, Kondoh H. Development (Cambridge, UK) 1993;119:433–446. doi: 10.1242/dev.119.2.433. [DOI] [PubMed] [Google Scholar]
- 29.Franklin A J, Jetton T L, Shelton K D, Magnuson M A. Mol Cell Biol. 1994;14:6773–6788. doi: 10.1128/mcb.14.10.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Genetta T, Kadesch T. Gene. 1996;169:289–290. doi: 10.1016/0378-1119(95)00824-1. [DOI] [PubMed] [Google Scholar]
- 31.Cabanillas A M, Darling D S. DNA Cell Biol. 1996;15:643–651. doi: 10.1089/dna.1996.15.643. [DOI] [PubMed] [Google Scholar]
- 32.Molloy D P, Milner A E, Yakub I K, Chinnadurai G, Gallimore P H, Grand R. J Biol Chem. 1998;273:20867–20876. doi: 10.1074/jbc.273.33.20867. [DOI] [PubMed] [Google Scholar]