Abstract
Pleiotrophin (PTN) is an 18-kDa heparin-binding secretory growth/differentiation factor for different cell types. Its gene is differentially expressed in both mesenchyme and central nervous system during development and highly expressed in a number of different human tumors. Recently, a PTN mutant was found to act as a dominant-negative effector of PTN signaling. We have now used homologous recombination to introduce the dominant-negative PTN mutant into embryonic stem cells to generate chimeric mice. All highly chimeric male mice with germinal epithelium exclusively derived from embryonic stem cells with the heterologous PTN mutation were sterile. Their testes were uniformly atrophic, and the spermatocytes were strikingly apoptotic at all stages of development. The results support a central role of PTN signaling in normal spermatogenesis and suggest that interruption of PTN signaling may lead to sterility in males.
Pleiotrophin (PTN) (1, 2), also known as HB-GAM (3), is a heparin-binding protein of 168 aa with a 32-aa signal sequence (2). It was purified from bovine uterus (1) as a mitogen for murine fibroblasts and from neonatal rat brain (1, 3) as a neurite outgrowth promoter. Transcripts of the Ptn gene are differentially expressed in both mesenchyme and cells of the central nervous system during mouse development (2, 4, 5), whereas stable but significantly lower levels of PTN gene expression limited to fewer cell types are seen in adults (6). An exception in adults is found in regenerating tissues in which PTN transcripts are sharply increased in specific cell types, for example, in endothelial cells, macrophages, and activated astrocytes after ischemic injury in brain and in cells stimulated by platelet-derived growth factor (7, 8).
PTN is also a mitogen for brain capillary endothelial cells (8–10) and SW-13 (adrenal carcinomas) cells (10) and is angiogenic in the rabbit corneal pocket assay (8, 9). PTN induces differentiation responses and neoantigen expression in oligodendrocyte progenitors (H.-J.Y. and T.F.D., unpublished data), is expressed in bone (11, 12), and stimulates proliferation and induction of differentiation markers in osteoblasts in culture (13).
The human, bovine, rat, and mouse Ptn cDNAs predict extraordinarily conserved mature proteins of 136 aa with clusters of N- and C- terminal lysine residues that may account for the ability of PTN to bind tightly to heparin (2, 4) and extracellular matrix (2). PTN has nearly 50% amino acid sequence identity and perfect conservation of 10 cysteine residues with midkine (MK) (2), a recently isolated retinoic acid-induced heparin-binding differentiation factor (14, 15), and together, PTN and MK establish a unique family of heparin-binding growth/differentiation factors (2) that is believed to have a broad role in both growth and differentiation.
PTN transforms NIH 3T3 cells (16), and its transcripts have been identified in many tumor specimens and tumor cell lines (10, 17). Recently, a mutant PTN that contains only the first N-terminal 40 aa (PTN 1–40) was shown to be a dominant-negative effector of PTN signaling, because its expression effectively blocked transformation of NIH 3T3 cells by wild-type (wt) PTN and formed disulfide linked heterodimers with wt PTN when expressed in human breast cancer (MDA-MB-231) cells that express high levels of endogenous PTN. Furthermore, PTN 1–40 effectively reversed the malignant phenotype of these cells, indicating that it functionally blocks endogenous PTN signaling and that endogenous PTN signaling is required to maintain the malignant phenotype of MDA-MB-231 cells (18).
MATERIALS AND METHODS
Generation of Highly Chimeric Mice Carrying a Ptn Dominant-Negative Mutation Allele.
The fragments of the mouse Ptn gene were isolated by screening a λ Dash (Stratagene) mouse genomic library derived from the 129/SV mouse strain, by using a 350-bp SmaI coding region fragment (base pair 1–325) of mouse Ptn cDNA as a probe, and a single positive clone was subcloned into pBluescript II SK(+) plasmid vector (Stratagene). Restriction mapping, DNA sequencing of coding portions and exon–intron junctions, and hybridization with exon-specific oligonucleotide probes were used to construct the restriction map shown in Fig. 1A. To introduce the Ptn dominant-negative construct into the endogenous mouse Ptn gene, a Ptn gene targeting vector was prepared as follows. A 4.5-kb KpnI fragment of the mouse Ptn gene (including intron 2, exon 3, intron 3, exon 4, and intron 4) was subcloned into the KpnI site of pBluescript II SK(+), yielding a 4.5-kb pBluescript II plasmid. This plasmid was cut with SmaI and blunt-ligated with a HindIII–XhoI fragment containing PGK/neomycin gene from pPGK-Neo-bpA digested by HindIII and XhoI. After restriction mapping, the resultant plasmid with the PGK promoter 5′ to the exon 2 of mouse Ptn gene was designated as pPtn4.5/Neo. The pPtn4.5/Neo was cut with HindIII–BamHI, filled in, and blunt-ligated to itself to generate pPtn4.5/Neo-blunt. The plasmid pPtn4.5/Neo-blunt was digested by KpnI and blunt-ligated with an XhoI–HindIII fragment containing pHSV-TK derived from the plasmid pMCI-TK cut by XhoI and HindIII. This new plasmid was named pmPtn-1. The 10.5-kb EcoRI fragment from the mouse Ptn λ genomic clone was assembled into pBluescript II SK(+) with the 5′ end ligated to the T3 promoter. This resultant plasmid is called pPtn-10.5. The 10.5-kb XhoI–NotI fragment derived from pPtn-10.5 cut by XhoI and NotI replaced the 3-kb XhoI–NotI fragment of pmPtn-1, yielding a Ptn gene targeting vector.
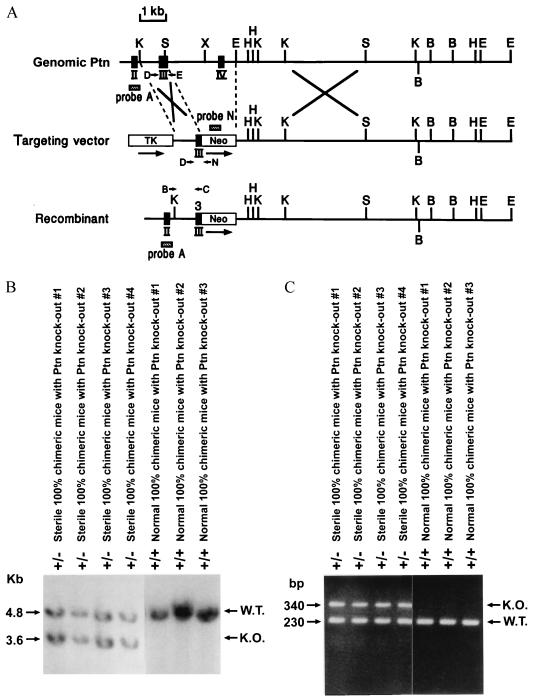
Figure 1.
Targeted dominant-negative mutation of mouse Ptn gene. (A) Restriction map of a part of the mouse Ptn gene, the targeting vector, and the structure at the mutated locus following the homologous recombination. The coding exons are depicted by black boxes and are numbered. Genomic fragments from the Ptn gene and neomycin gene used as probes for Southern blotting are depicted as hatched boxes (probe A and probe N); arrows denote PCR primers (B, C, D, E, and N) used for genotyping the ES cells and the chimeric animals. Neo, the neomycin transferase gene; TK, thymidine kinase gene; arrows depict the transcriptional orientation of these genes. Only relevant restriction sites are shown: K, KpnI; S, SmaI; X, XhoI; E, EcoRI; H, HindIII; B, BamHI. (B) Southern blot analysis of genomic DNA extracted from the tails of the sterile, highly chimeric mice with a Ptn dominant-negative mutation and normal highly chimeric mice without Ptn gene disruption. The DNA was digested with EcoRI and hybridized with probe A. The sizes of wt (W.T.) and disrupted (K.O.) alleles are shown. (C) PCR amplification products of the DNA extracted from the mouse tails. The sizes of amplification products of wt (W.T.) and disrupted (K.O.) alleles are shown.
The NotI-linearized targeting vector was electroporated into E14 (kindly provided by D. Y.Loh, Washington University, St. Louis]) and RW4 cells [the RW-4 embryonic stem (ES) cell line was established from strain SV129 and kindly provided by R. L. Wesselschmidt and T. J. Ley, Washington University, St. Louis]. G418 and gancyclovir double-resistant ES clones were selected, expanded, and frozen. The homologous recombination event is predicted to replace a 2.8-kb SmaI–EcoRI fragment of the Ptn gene with a neomycin-resistance gene. DNA was isolated from 280 double-resistant E14 ES cell clones and 120 RW4 ES cell clones, digested with EcoRI, blotted onto Hybond-N membranes, and probed with the 250-bp NotI–KpnI fragment of the Ptn gene. The probe hybridizes to a 4.8-kb EcoRI fragment derived from the wt allele, whereas the targeted locus is predicted to yield a 3.6-kb fragment. Of the 28 E14 clones and 14 RW4 clones heterozygous at the Ptn locus, 6 and 8, respectively, were expanded, and their genetic status was validated. The presence of additional integration sites was ruled out by probing EcoRI digests of their genomic DNA with a PstI–PstI neomycin-resistance gene probe. Genomic DNA from the positive clones was also analyzed by using PCR. E14 wt ES cells, RW4 wt ES cells, both G418 and gancyclovir double-resistant E14 clones and RW4 clones without Ptn gene disruption, three Ptn heterozygous E14 clones, and eight Ptn heterozygous RW4 clones were injected into blastocyst-stage C57BL/6 mouse embryos (≈15 cells per blastocyst) and implanted into pseudopregnant ICR females. The resulting chimeras were bred with C57BL/6 mice. For the genotyping, genomic DNA was isolated from the tails of chimeric animals, digested with EcoRI, and analyzed by Southern blotting using probe A or by PCR. The PTN-specific primers were: D, 5′-AAA AAA AGG TGA AAA AGT CTG ACT GTG GAG-3′; E, 5′-CTC CAA ACT GCT TCT TCC AG-3′); and a neomycin-resistance gene-specific primer N, 5′-CAC GAG ACT AGT GAG ACG TG-3′).
Northern Blot Assay.
Total RNAs were isolated from testis tissue by the guanidinium-thiocyanate method as described (19). Twenty micrograms of total RNA was separated on 1.2% formaldehyde-agarose gels, blotted onto nylon membrane (Micron Separations, Westboro, MA), prehybridized, and hybridized with an [α-32P]dCTP-labeled SmaI fragment of mouse Ptn cDNA. After overnight hybridization, blots were washed twice with 0.5× SSC and 0.1% SDS for 20 minutes at room temperature, washed once for 20 minutes at 68°C, and autoradiographed with intensifying screens at −70°C.
Fertility of the Chimeric Male Mice.
C57BL/6 female mice were mated with chimeric males generated from different Ptn heterozygous ES clones, wt ES cells, Ptn targeting vector randomly integrated ES clones. In the morning, plugs from mated C57BL/6 female mice were harvested and placed in Eppendorf tubes with 100 μl of PBS, suspended, and smeared onto glass plate. Spermatozoal quantity and quality were examined by using a microscope. Meanwhile, each highly chimeric male was bred with four C57BL/6 female mice for 4 months to test fertility.
Anatomy Pathological and Histological Examination of Mice.
Tissues were collected from euthanized mice for gross and histological examination, fixed in formalin, processed, and embedded in paraffin. Sections (6 μm) were cut and stained with hematoxylin and eosin by using standard procedures.
Immunocytochemistry.
Sections of testes of sterile and fertile chimeric mice were stained with the Apop Tag Plus-Peroxidase (Oncor, catalog no. S7101) to detect apoptotic cells in situ as described (20) according to the manufacturer’s instructions. The stained sections were mounted and observed by using a light microscope after counterstaining with 1% methyl green.
In Situ hybridization.
In situ hybridization was performed on paraffin sections of testes from the sterile and fertile males as described (21). Standard protocols for collecting, embedding, and sectioning the tissues were used. pBluescript II mouse Ptn cDNA and pPGK-Neo-bpA were linearized with EcoRI and NcoI, respectively (22), and antisense transcripts were generated by using T7 polymerase. Specimens were hybridized with [35S]thio-UTP-labeled riboprobes. Both pBluescript II mouse Ptn cDNA and pPGK-Neo-bpA were also cut by XhoI and XbaI, respectively, and sense probes were produced by T3 polymerase and used as controls.
RESULTS
Generation of Highly Chimeric Mice Carrying a Ptn Dominant-Negative Mutant.
To determine the roles of PTN signaling in vivo, a Ptn dominant-negative mutant allele that encodes PTN amino acid residues −32 to 40 (referred to as PTN 1–40, after removal of the signal peptide) was generated in mice by introducing a Ptn mutant targeting vector into ES cells through homologous recombination. The Ptn targeting vector (Fig. 1A) was introduced into both 280 E14 and 120 RW4 ES cell lines, and the transfected cells were selected. Southern blot and PCR analysis of DNA established that 28 E14 clones and 14 RW4 clones were heterozygous at the Ptn locus. Three E14 Ptn heterozygous ES clones, eight RW4 Ptn heterozygous ES clones, one positive-negative selected E14 clone without mutation of the Ptn gene (as control), one positive-negative selected RW4 clone without targeting of the Ptn gene (as control), and E14 wt cells and RW4 wt cells (as controls) were expanded and injected into mouse blastocysts. All highly chimeric founder mice were analyzed. Genotypes were identified by Southern blot and PCR amplification of tail DNA (Fig. 1 B and C).
Fertility and gross pathological examination of the chimeric male mice. Male mice with >90% chimerism were bred but failed to produce offspring. For this reason, mice were tested for fertility over a 6-month period, including 30 males with the dominant-negative Ptn mutation produced from 11 ES clones (including both E14 and RW 4) heterozygous at the Ptn locus, 4 males generated from double-selected E14 and RW4 cells without disruption of the Ptn gene, and 4 males derived from E14 and RW4 wild- type ES cells. Of the 30 male mice with the Ptn dominant-negative mutation, 22 failed to sire offspring. However, each produced many vaginal plugs in multiple matings with B6 female mice. The 8 remaining highly chimeric male mice (derived from 3 Ptn heterozygous RW4 clones and 1 Ptn heterozygous E14 clone) were fertile, but none of these were established as germ-line transmission as indicated by inheritance of agouti color and genotyping (data not shown). When the control highly chimeric male mice (generated from wt E14 or RW4 cells and from the double-selected E14 or RW4 clones that lacked the homologously recombined Ptn gene) were bred with B6 females, all sired agouti offspring, indicating germ-line transmission.
Although the sterile male chimeric mice with the Ptn dominant-negative mutation produced vaginal plugs, sperm were never detected. The testes of these sterile chimeric mice were normally descended in the scrotum and the genital ducts (i.e., epididymis, vas deferens) and accessory glands (i.e., seminal vesicle, prostate and bulbourethral glands) were macroscopically and histologically normal. The remainder of the testis was markedly atrophic, and the testes of the sterile male chimeric mice weighed only ≈10% that of the normal control animals and those of the fertile chimeric males (i.e., the mice with the PTN dominant-negative mutation that failed to establish germ-line transmission) (Table 1).
Table 1.
Characteristics of chimeric PTN mutant mice
| Reproductive status | Body weight, g | Testis, mg | Epididymis, mg | Seminal vesicle, mg | Epididymis sperm count, ×106 |
|---|---|---|---|---|---|
| Fertile | 33 ± 5 | 138 ± 18 | 38 ± 5 | 98 ± 16 | 19 ± 3 |
| Sterile | 33.5 ± 6.8 | 22 ± 9 | 20 ± 3 | 95 ± 11 | 0 |
Values given are mean ± SD of six fertile or six sterile chimeric mice.
Detection of Ptn and Its Mutant Gene Expression.
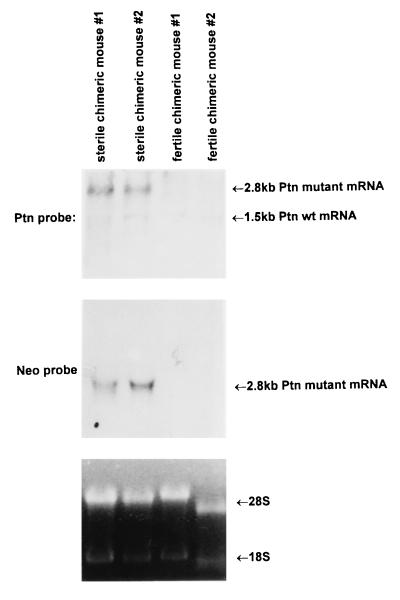
Expression of the dominant-negative mutant gene in chimeric mice was confirmed by Northern blot analysis of total RNA extracted from testes of sterile males (see below) in which both the 1.5-kb endogenous Ptn mRNA species and a larger Ptn transcript of 2.8 kb generated from the disrupted Ptn gene were found (Fig. 2). The larger transcript uses the polyadenylation site of the neomycin-resistance gene that was fused with the Ptn gene at the SmaI site in the middle of exon 3. The fidelity of the Ptn transcript encoded by the mutant Ptn gene was validated by using reverse transcription–PCR and DNA sequence analysis (data not shown). It encodes 118 aa to include amino acid residues −32 to 40 of PTN and the additional 46 amino acids encoded by the PGK promoter region terminated by a stop codon in the pPGK–neomycin cassette. Western blots with a chicken polyclonal anti-PTN antibody detected the mutant Ptn gene product only in high chimeric mice carrying the Ptn dominant-negative mutation and not in the control, highly chimeric mice from normal control ES cells that were without disruption of the Ptn gene (data not shown).
Figure 2.
Northern blot analysis of total RNA prepared from the testes of the sterile and the fertile chimeric mice with a Ptn dominant-negative mutant allele. The blot was probed with a SmaI fragment of mouse cDNA. Bands corresponding to Ptn transcripts are indicated.
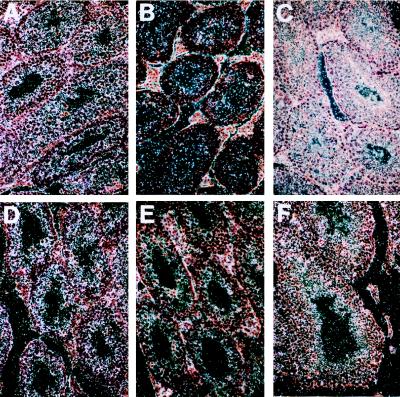
We also examined the pattern of expression of the neomycin gene in the testes of the sterile and the fertile chimeric mice with the Ptn dominant-negative mutation by using in situ hybridization. The transcripts of the neomycin gene were found only in the sterile chimeric mice, indicating that the spermatogonia of the sterile chimeric males with the PTN dominant-negative mutation generated from ES cells contained the homologously recombined Ptn gene. The results also indicated that the spermatogonia of the fertile chimeric mice were not derived from ES cells containing the PTN dominant negative mutation (Fig. 3 A–C).
Figure 3.
Expression of neomycin and Ptn mRNA in the testes of the sterile and fertile chimeric males with a targeted Ptn dominant-negative mutant allele. (A) Neomycin gene transcript was detected in all kinds of germ cells during spermatogenesis in the sterile chimeric mice using neomycin gene antisense as a riboprobe. (B) The signal was absent in sections of testes from the sterile animals when neomycin sense was used as a riboprobe. (C) The neomycin gene signal was absent in sections of testes from the fertile chimeric by using a neomycin gene antisense riboprobe for detection. (D) Intensive expression of Ptn mRNA in seminiferous epithelium was found in sections of testes from the sterile chimeric males by using mouse Ptn antisense as a riboprobe. (E) The signal was absent in the sterile animals by using a Ptn sense riboprobe. (F) Ptn signal was detected in the testes of the fertile chimeric mice with a mouse Ptn antisense riboprobe.
To test whether spermatocytes express the endogenous Ptn gene during spermatogenesis, we used a mouse Ptn antisense riboprobe to examine Ptn gene expression in the seminiferous epithelium from sterile and fertile chimeric mice with the Ptn dominant-negative mutation by using in situ hybridization. The results indicate that production of Ptn mRNA is more intense in the seminiferous epithelium of the sterile mice than the normal mice. It is possible that the more intense hybridization signal seen in the seminiferous epithelium of the sterile mice (Fig. 2) may reflect greater mRNA stability in these animals. The cDNA containing the Ptn dominant-negative mutant allele was fused to the neomycin gene at the 3′ end and does not contain three ATTTA repeats implicated in mRNA instability that are found in wt PTN 3′ untranslated mRNA. Ptn gene expression was present in spermatocytes during all stages of spermatogenesis, although the intensity of staining was lower in the normal fertile chimeric mice than in the sterile animals, suggesting that PTN signaling may be important in spermatogenesis (Fig. 3 D–F).
Histological Examination and Analysis of Apoptosis.
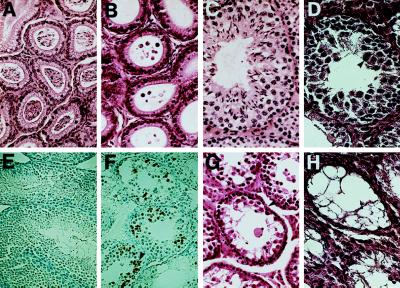
Histological examination revealed that the epididymis of the adult wt mice and the fertile chimeric mice were packed with sperm (Fig. 4A); in striking contrast, differentiated spermatozoa were not seen in the epididymides of adult sterile mice (Fig. 4B). A thorough histological analysis of the testis was then done; spermatogenesis and spermiation in 10 of the highly chimeric 2- to 4-month-old fertile male mice (including mice derived from wt E14 cells or RW4 cells, from double-selected E14 or RW4 clones without disruption of the Ptn gene, and from the Ptn mutant heterozygotic E14 and RW4 cell clones) occur normally in the seminiferous tubules (Fig. 4C).
Figure 4.
Histology and immunohistochemical staining of the epididymis of adult sterile and fertile chimeric mice with a Ptn dominant negative mutation. (A and B) Hematoxylin and eosin-stained sections of epididymis of the fertile and sterile animals. The epididymes of the fertile male mice are packed with spermatozoa (A), whereas those of the sterile male mice lacked detectable spermatozoa (B). (C and D) Hematoxylin and eosin-stained section of the fertile and sterile chimeric mouse testes. Normal spermatogenesis was seen in the seminiferous tubules of the 2- to 4-month-old fertile chimeric mice (C), and degeneration of seminiferous epithelium was observed in 2- to 4-month-old sterile chimeric animals generated from Ptn dominant-negative mutation heterozygous E14 or RW4 clones (D). The multinucleated giant cells, as noted by arrowheads, and an expanded layer of cells, indicated by a double-headed arrow, have a nuclear morphology characteristic of premeiotic cells. (E and F) Testis sections of sterile and fertile chimeric mice were stained with the Apop Tag Plus Peroxidase Kit to detect apoptotic cells in situ. The assay shows clusters of apoptotic germ cells and a marked increase in apoptosis in the testes of the sterile mice (F) as compared with the extent of cell death known to occur in normal spermatogenesis (E). The labeled cells are denoted by an arrow. (G and H) In older mutant males, progressive degeneration of the germinal epithelium occurred, ending with the formation of striking vacuolization of tubules that were observed in testis from the sterile males at 6 months (G) and 8 months (H). Sertoli cells and Leydig cells appear normal and are denoted by arrows.
However, degeneration of the seminiferous epithelium was observed in all 10 highly chimeric 2- to 4-month-old sterile male mice (generated from E14 or RW4 clones heterozygous for the Ptn dominant-negative mutation). In these mice, the number and diameter of the seminiferous tubules were markedly reduced, and primary spermatocytes were present in the seminiferous tubules but the number of these germ cells was dramatically reduced and the primary spermatocytes themselves were degenerate with dark, pyknotic nuclei and occasional karyorrhexis (Fig. 4D). The cytoplasm of the primary spermatocytes was variable and stained eosinophilic to slightly basophilic. Some of the primary spermatocytes formed large multinucleated cells. Staining with Apop Tag Plus-Peroxidase identified striking clusters of apoptotic germ cells in the sterile males as compared with the extent of cell death known to occur in normal spermatogenesis (Fig. 4 E and F). In some of the tubules, spermatogonia and spermatocytes showed vacuolation and ballooning of the cytoplasm and focal loss of all germ cells. Neither identifiable secondary spermatocytes nor mature sperm were observed in any of the seminiferous tubules examined. However, the interstitial (Leydig) cells appeared normal.
We also compared 10 sterile chimeric males carrying the Ptn dominant-negative mutation with their normal counterparts between 6 and 8 months of age. These mice had the same abnormalities in the seminiferous tubules as did 2-month-old males. In addition, the number of seminiferous tubules was markedly reduced; the seminiferous tubules that were present were lined with a single layer of degenerating spermatogonia with pale nuclei, large nucleoli, and indistinct nuclear membranes. The cytoplasm was basophilic, slightly granular, and contained numerous small clear vacuoles or large single vesicles (Fig. 4G). Spermatocytes could not be definitively identified and mature sperm were not observed. In contrast to the atrophied seminiferous tubules, the interstitial cells had proliferated dramatically (Fig. 4H). None of these abnormalities were found in any of the highly chimeric mice with the Ptn dominant-negative mutation that were examined at 2 weeks of age (data not shown). We examined the remaining organs and tissues of the sterile animals; no abnormalities were found in brain, pituitary, lung, heart, gastrointestinal tract, bone mar- row, skeletal muscle, accessory sex glands, thymus, spleen, lymph nodes, adrenal glands, urinary bladder, or kidney (data not shown).
DISCUSSION
PTN is a weak mitogen for endothelial (3, 14, 16), epithelial (3, 13), and fibroblastic cells (1, 2). PTN also induces differentiation of oligodendrocyte progenitors and neurite outgrowth (2, 3, 23; H.-J.Y. and T.F.D., unpublished data). The results of this study suggest that PTN is important for spermatogenesis in the mouse. In this work, we demonstrate that interruption of PTN signaling by the introduction of the dominant-negative mutant into germ line of mouse leads to apoptosis of spermatogonia in all stages of development. Growth factor withdrawal from dependent cell lines is known to lead to apoptosis in transformed mammalian cell lines (23, 24). However, platelet-derived growth factor-stimulated fibroblasts become apoptotic when serum is withdrawn but not in the presence of insulin, suggesting that growth factors, in this case PTN, and other cytokines are needed to suppress apoptosis in specific settings.
PTN is widely expressed in different cell types and at different times in neuroectoderm and mesoderm during embryogenesis. However, in adult tissues, PTN is expressed only at low levels in some cells in the brain, bones, gut, uterus, and ovary, but, in testis, PTN expression levels are significantly higher (3, 5, 23). It is possible that the process of continuous self-renewal and differentiation in developing spermatogonia make them uniquely susceptible to loss of PTN signaling that results from this mutation and that other growth factors functionally compensate in other tissues.
The PTN dominant-negative mutant gene encodes a polypeptide of 40 residues (after cleavage of the signal peptide) that forms inactive heterodimers (18) with the wt PTN associated with loss of PTN signaling. However, it is possible the PTN 1–40 forms heterodimers with the wt product of the MK gene (14, 25, 26), a protein that shares nearly 50% amino acid identity and perfect conservation of cysteines with PTN (2), and has similar functional activities as PTN as well as being highly expressed during development. PTN 1–40 heterodimerizes with MK itself, because there is complete conservation of 3 cysteine residues in the first 40 aa of both proteins. However, it is equally possible that PTN 1–40 interferes with the normal interactions of wt PTN and a second protein of vital importance in spermatogenesis. This report demonstrate the unique importance of a single growth factor in the regulation of spermatogenesis in mice. Because abnormal spermatogenesis is a feature of many cases of infertility in human males (27, 28), the mice generated in this study may be useful models to study infertility in human males.
Acknowledgments
This work was supported by National Institutes of Health Grants DK53557, CA66029, HL31102, and CA49712.
Footnotes
Abbreviations: PTN, pleiotrophin; MK, midkine; ES, embryonic stem.
References
- 1.Milner P G, Li Y S, Hoffman R M, Kodmor C M, Siegel N R, Deuel T F. Biochem Biophys Res Commun. 1989;165:1096–1103. doi: 10.1016/0006-291x(89)92715-0. [DOI] [PubMed] [Google Scholar]
- 2.Li Y S, Milner P G, Chauhan A K, Watson M A, Hoffman, Kodner C M, Deuel T F. Science. 1990;250:1690–1694. doi: 10.1126/science.2270483. [DOI] [PubMed] [Google Scholar]
- 3.Rauvala H. EMBO J. 1989;8:2933–2941. doi: 10.1002/j.1460-2075.1989.tb08443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Merenmies J, Rauvala H. J Biol Chem. 1990;265:16721–16724. [PubMed] [Google Scholar]
- 5.Van der Winden J, Mailleux P, Schiffmann S N, Vanderhaeghen J J. Anat Embryol. 1992;186:387–406. doi: 10.1007/BF00185989. [DOI] [PubMed] [Google Scholar]
- 6.Silos-Santiago I, Yeh H J, Gerrieri M A, Guillman R p, Li Y S, Wolf L, Deuel T F. J Neurobiol. 1996;32:283–296. doi: 10.1002/(SICI)1097-4695(199611)31:3<283::AID-NEU2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 7.Yeh H J, He Y Y, Xu J, Hsu C Y, Deuel T F. J Neurosci. 1998;18:3699–3707. doi: 10.1523/JNEUROSCI.18-10-03699.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y S, Gurrieri M, Deuel T F. Biochem Biophys Res Commun. 1992;184:427–432. doi: 10.1016/0006-291x(92)91211-8. [DOI] [PubMed] [Google Scholar]
- 9.Laaroubi K, Delbe J, Vachorote F, Desgranges P, Tardieu M, Jaye M, Courty J. Growth Factors. 1994;10:89–98. doi: 10.3109/08977199409010982. [DOI] [PubMed] [Google Scholar]
- 10.Fang W, Hartmann N, Chow D T, Reigel A T, Wellstein A. J Biol Chem. 1992;267:25889–25897. [PubMed] [Google Scholar]
- 11.Tezuka K, Takeshita S, Hadeda Y, Kumrgawa M, Kikuno R, Hashimoto-Gotoh T. Biochem Biophys Res Commun. 1990;173:246–251. doi: 10.1016/s0006-291x(05)81048-4. [DOI] [PubMed] [Google Scholar]
- 12.Takamatsu H, Itoh M, Kimura M, Gospodarowicz D, Amann E. Biochem Biophys Res Commun. 1992;185:224–230. doi: 10.1016/s0006-291x(05)80979-9. [DOI] [PubMed] [Google Scholar]
- 13.Zhou H-Z, Ohnuma Y, Takita H, Fujisawa R, Mizuno M, Kuboki Y. Biochem Biophys Res Commun. 1992;186:1288–1293. doi: 10.1016/s0006-291x(05)81545-1. [DOI] [PubMed] [Google Scholar]
- 14.Kadomatsu K, Tomomura M, Muramatsu T. Biochem Biophys Res Commun. 1988;151:1312–1318. doi: 10.1016/s0006-291x(88)80505-9. [DOI] [PubMed] [Google Scholar]
- 15.Muramatsu H, Shirahama H, Yonezawa S, Maruta H, Muranatsu T. Dev Biol. 1993;159:392–402. doi: 10.1006/dbio.1993.1250. [DOI] [PubMed] [Google Scholar]
- 16.Chauhan A K, Li Y S, Deuel T F. Proc Natl Acad Sci USA. 1993;90:679–682. doi: 10.1073/pnas.90.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsutsui J, Kadomatsu K, Matsubara A, Hamanoue S, Takao S, Shimazu H, Ohi Y, Muramatsu T. Cancer Res. 1993;53:1281–1285. [PubMed] [Google Scholar]
- 18.Zhang N, Zhong R, Wang Z Y, Deuel T F. J Biol Chem. 1997;272:16730–16736. doi: 10.1074/jbc.272.27.16733. [DOI] [PubMed] [Google Scholar]
- 19.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 20.Schmitz G. Anal Biochem. 1991;192:222–231. doi: 10.1016/0003-2697(91)90212-c. [DOI] [PubMed] [Google Scholar]
- 21.Yeh H-J, Ruit K G, Wang Y X, Parks W C, Snider W D, Deuel T F. Cell. 1991;64:209–216. doi: 10.1016/0092-8674(91)90222-k. [DOI] [PubMed] [Google Scholar]
- 22.Soriano P, Montgomery C, Geske R, Bradley A. Cell. 1991;64:693–702. doi: 10.1016/0092-8674(91)90499-o. [DOI] [PubMed] [Google Scholar]
- 23.Harrington E A, Bennett M R, Fanidi A, Evan G I. EMBO J. 1994;13:3286–3295. doi: 10.1002/j.1460-2075.1994.tb06630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaganti R S K, German J. Am J Hum Genet. 1979;31:634–664. [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H G, Milan J A, Cox A D, Der C J, Rapp U R, Beck T, Zha H, Reed J C. J Cell Biol. 1995;129:1103–1114. doi: 10.1083/jcb.129.4.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muramatsu T. Dev Growth Differ. 1994;36:1–8. doi: 10.1111/j.1440-169X.1994.00001.x. [DOI] [PubMed] [Google Scholar]
- 27.Fernandez-Sarabia M J, Bischoff J R. Nature (London) 1994;366:274–275. doi: 10.1038/366274a0. [DOI] [PubMed] [Google Scholar]
- 28.Micic M, Nikolis J, Micic S. Hum Reprod. 1992;7:1118–1120. doi: 10.1093/oxfordjournals.humrep.a137804. [DOI] [PubMed] [Google Scholar]