What is it about movement in cells that commands our attention? What biologist has not enjoyed turning a microscope on a cell, almost any cell, really, and watching all the commotion? When vesicles, mitochondria, chloroplasts, nuclei, or chromosomes move, we are being treated to an elegant, easily observable manifestation of molecular events. Decades of effort to understand intracellular movement have given rise to two of the great thrusts of modern cell biology: the study of which things go where, usually referred to as intracellular trafficking; and the identification of the protein machines that generate movement, the molecular motors. But we have an incomplete picture of how the cell's array of motor proteins gives rise to the variety of journeys that their cargoes make. The kinesin motors that generate movement along microtubule tracks are a case in point (Vale and Fletterick 1997). As with other motor proteins, the dyneins or myosins, the study of kinesin by the methods of molecular genetics has demonstrated that kinesins are a large family of related motor proteins present across all eukaryotic phyla, and numbering 30–40 members in humans and mice (Kim and Endow 2000; http://www.blocks.fhcrc.org/kinesin). Most analysis of this diversity thus far indicates that different kinesins serve to move different cargoes in the cell (Manning and Snyder 2000). So, where does the trafficking information for the motor proteins reside? What is the cargo “receptor” for kinesin, and what specific protein–protein interactions govern this important matchmaking in the cell? Most work on this question has focused on the first kinesin family member to be discovered, so-called “conventional” kinesin, or kinesin-I. In the case of kinesin-I, the ER membrane protein kinectin has been proposed to be a cargo receptor, but its restricted cellular and phylogenetic distributions (Toyoshima and Sheetz 1996; Goldstein and Gunwardena 2000) have prompted some investigators to look further.
This search has recently borne fruit: two groups have reported that kinesin-I binds to cargoes via a set of proteins involved in intracellular signaling (Bowman et al. 2000; Verhey et al. 2001). The proteins, JIP-1, JIP-2, and JIP-3, are thought to serve as scaffolding proteins for the c-Jun NH2-terminal kinase (JNK) signaling pathway (Davis 2000). The high affinity and specificity of kinesin binding to the JIP proteins indicates that the complex pairing of motors and cargoes will soon be on the same footing with other protein–protein interactions essential to membrane traffic. Perhaps more exciting, these results connect the organization of organelle traffic with that of cell signaling: whereas the JIP proteins themselves apparently function to hold enzymes of the JNK pathway in proximity to each other, their interaction with kinesin-I may also determine the collective spatial organization of the signaling pathway within the cell.
One potential kinesin receptor was identified by Bowman et al. 2000 when they screened Drosophila melanogaster mutants for elements of the machinery of movement other than the motor proteins themselves. They examined larvae with potential axonal transport phenotypes previously seen in kinesin-I mutants (Hurd and Saxton 1996; Gindhart et al. 1998), and identified a Drosophila homologue of the proposed mammalian JNK scaffolding protein, JIP-3. It was clearly essential for transport in Drosophila, as mutant larvae had accumulations of vesicles along the axons of their segmental nerves. GFP-tagged JIP-3 protein was expressed in CV-1 cells, where it colocalized with kinesin-I and with Golgi and early secretory vesicles, but not with mitochondria or the ER-to-Golgi intermediate compartment. When they probed the interaction of kinesin-I and JIP-3 by yeast two-hybrid analysis and coprecipitation methods, they found that the NH2-terminal domain of JIP-3 bound a region of the kinesin light chain (LC) that contains six tetratricopeptide repeat (TPR) motifs (Blatch and Lasle 1999). They propose that JIP-3 (which they named Sunday Driver) is an organelle membrane protein whose interaction with kinesin is required for transport.
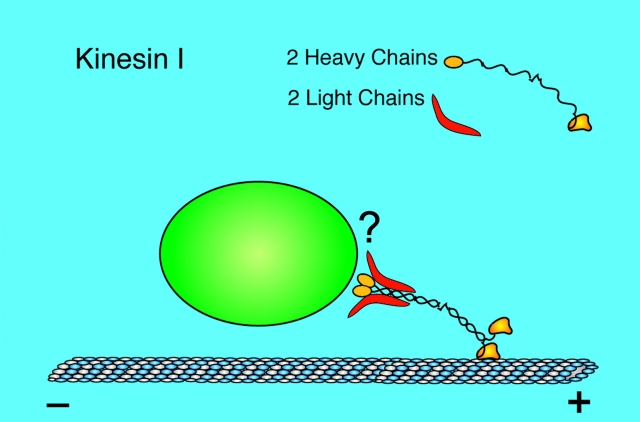
In retrospect, it is not surprising that potential cargo receptors specifically bind the TPR domain of the kinesin LC. Kinesin-I is a heterotetramer comprised of 2 LCs and two heavy chains (HCs; see Fig. 1). It has been thought for some time that the kinesin tail region binds to the motor's cargo (Vale and Fletterick 1997). Both the HCs and LCs occupy this region of the tetramer, but the preponderance of genetic and biochemical data indicate that the LCs are important or even essential for cargo binding (Yu et al. 1992; Stenoien and Brady 1997; Gindhart et al. 1998; Tsai et al. 2000). Furthermore, the TPR domain of the LC stands out specifically as a likely binding site because antibodies directed against it disrupt kinesin–cargo interactions (Stenoien and Brady 1997), and it has well characterized, specific protein binding properties (Blatch and Lasle 1999).
Figure 1.
Kinesin-I is a heterotetramer formed by a coiled-coil interaction between two heavy chains (HCs) and the binding of a LC to the COOH-terminal region of each HC. Each HC has an NH2-terminal catalytic motor domain that interacts with a microtubule during movement. The heterotetramer binds to its cargo at its tail, as shown. However, the exact nature of the interaction, what kinesin is binding to, and how, has not been clear. The LCs have been thought to be essential for this interaction, and the two recent papers discussed here (Bowman et al. 2000; Verhey et al. 2001) identify the TPR domain of the LC as a site of cargo binding. For illustrative purposes, the kinesin molecule is drawn here approximately three times larger than true scale relative to the microtubule and vesicle. (Figure courtesy of W.M. Saxton)
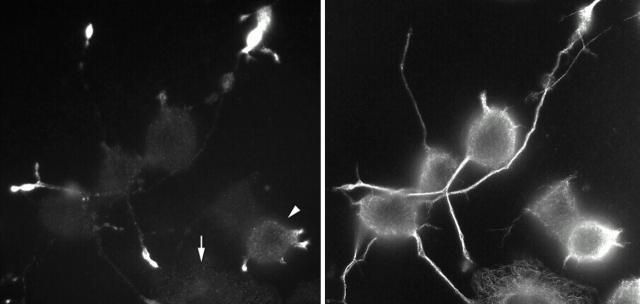
The study by Verhey et al. 2001(this issue) used this view of the LC as a point of departure. They employed the kinesin LC TPR domain as bait in a yeast two-hybrid screen of a mouse brain cDNA library and fished out three binding partners for kinesin: not only JIP-3, but also JIP-1 and JIP-2, which are unrelated to JIP-3, but very similar to each other. Closer examination of kinesin–JIP binding confirmed that the LC TPR domain binds the NH2-terminal region of JIP-3. But JIP-1 and JIP-2 resembled more closely other TPR-binding proteins, in that they interacted with kinesin via their COOH termini. Verhey et al. 2001 found that the mutation of a single tyrosine three residues from the COOH terminus eliminated JIP-1 binding to kinesin. This surprising result invites comparisons between motor–cargo binding and tyrosine-based sorting signals for protein traffic (Bonifacino and Dell'Angelica 1999). So kinesin binds, but does it deliver? To address this, they examined the distribution JIP-1 in neuronal cell lines (Fig. 2) and found that the kinesin-I/JIP-1 interaction was necessary for JIP-1 to accumulate in the tips of the neurites. They propose that the transport of JIP-1 to the neurite tip by kinesin-I is important in neuronal development.
Figure 2.
Immunofluorescent staining of differentiating CAD cells shows the expression and localization of endogenous JIP-1 (left) and tubulin (right) proteins. In cells that have not yet begun to extend neurites (arrow at left), JIP-1 expression and localization are not apparent. But, as soon as this neuron-like cell line has established neurites, JIP-1 is localized to their tips via kinesin-I. The cell denoted by an arrowhead at left is just beginning to produce neurites, whereas the two cells near the center of the field have longer neurites and bright JIP-1 staining at the distal ends. (Image courtesy of K.J. Verhey)
But is this really to do with signaling pathways, or is JIP-1 doing double duty as a kinesin receptor? When Verhey et al. 2001 examined whether kinesin also carries any of the signaling proteins that are thought to bind to the JIP-1 scaffold, they identified one kinase in the kinesin/JIP-1 complex that functions upstream of the JNK pathway. Also present was ApoER2, a membrane receptor that may serve as the link between a kinesin/JIP-1 complex and the cargo membrane. They suggest that the link between kinase scaffolding proteins and kinesin motors serves not only to localize membrane proteins and conventional cargoes, but to provide motor-driven spatial regulation of cytoplasmic signaling pathways.
So, is it time to start drawing seminar slides of kinesin-based transport centered on these two classes of receptors, JIP-1/2 and JIP-3? Of course not! Already another cargo receptor for kinesin-I has been identified: the amyloid precursor protein, a well-known membrane protein that also interacts with the LC TPR domain (Kamal et al. 2000). In addition, there is good evidence that some cargoes bind the kinesin LC, via other receptors, outside the TPR domain. For example, although several classes of organelles are thought to be moved by kinesin-I, Verhey et al. 2001 found that blocking the binding of the kinesin LC TPR domain to other proteins did not disrupt the organelle distribution in CAD cells. Also, antibody disruption of binding to the LC only displaces about one-third of the kinesin-I from vesicles (Yu et al. 1992), even when the antibodies are directed specifically against the TPR domain (Stenoien and Brady 1997). And it remains possible that some cargoes bind not the LCs, but the HCs, of kinesin-I at least in some organisms. Not only can the HCs of heterotetrameric kinesin-I bind vesicles in vitro (Skoufias et al. 1994), but Neurospora crassa kinesin, which lacks LCs completely, binds its cargo via a site on the HC that is highly conserved among members of the kinesin-I family (Seiler et al. 2000).
The existence of many motor protein receptors, with or without signaling functions, seems not only likely but essential, given the plethora of motors and cargoes in the cell. Indeed, receptors or signaling molecules that bind to other kinesin family members have been reported already (Nagata et al. 1998; Nakagawa et al. 2000; Setou et al. 2000). If you would like to take part in the construction of the phylogenetic tree of motor protein receptors, you had better order your PCR primers soon.
Acknowledgments
I am grateful to L.S.B. Goldstein and K.J. Verhey for discussions and to an anonymous reviewer for helpful suggestions for the manuscript.
Work in the author's laboratory is supported by a grant from the National Institutes of Health (NS27073).
Footnotes
Abbreviations used in this paper: JNK, c-Jun NH2-terminal kinase; LC, light chain; TPR, tetratricopeptide repeat;
References
- Blatch G.L., Lasle M. The tetratricopeptide repeata structural motif mediating protein–protein interactions. Bioessays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Bonifacino J.S., Dell'Angelica E.C. Molecular bases for the recognition of tyrosine-based sorting signals. J. Cell Biol. 1999;145:923–926. doi: 10.1083/jcb.145.5.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman A.B., Kamal A., Ritchings B.W., Philp A.V., McGrail M., Gindhart J.G., Goldstein L.S.B. Kinesin-dependent axonal transport is mediated by the Sunday driver (SYD) protein. Cell. 2000;103:583–594. doi: 10.1016/s0092-8674(00)00162-8. [DOI] [PubMed] [Google Scholar]
- Davis R.J. Signal transduction by the JNK kinase group of MAP kinases. Cell. 2000;103:239–252. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- Gindhart J.G., Desai C.J., Beushausen S., Zinn K., Goldstein L.S.B. Kinesin light chains are essential for axonal transport in Drosophila . J. Cell Biol. 1998;141:443–454. doi: 10.1083/jcb.141.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein L.S.B., Gunwardena S. Flying through the Drosophila cytoskeletal genome. J. Cell Biol. 2000;150:F63–F68. doi: 10.1083/jcb.150.2.f63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd D.D., Saxton W.M. Kinesin mutations cause motor neuron disease phenotypes by disrupting fast axonal transport in Drosophila . Genetics. 1996;144:1075–1085. doi: 10.1093/genetics/144.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal A., Stokin G.B., Yang Z.H., Xia C.H., Goldstein L.S.B. Axonal transport of amyloid precursor protein is mediated by direct binding to the kinesin light chain subunit of kinesin-I. Neuron. 2000;28:449–459. doi: 10.1016/s0896-6273(00)00124-0. [DOI] [PubMed] [Google Scholar]
- Kim A.J., Endow S.A. A kinesin family tree. J. Cell Sci. 2000;113:3681–3682. doi: 10.1242/jcs.113.21.3681. [DOI] [PubMed] [Google Scholar]
- Manning B.D., Snyder M. Drivers and passengers wanted! The role of kinesin-associated proteins. Trends Cell Biol. 2000;10:281–289. doi: 10.1016/s0962-8924(00)01774-8. [DOI] [PubMed] [Google Scholar]
- Nagata K., Puls A., Futter C., Aspenstrom P., Schaefer E., Nakata T., Hirokawa N., Hall A. The MAP kinase kinase kinase MLK2 colocalizes with activated JNK along microtubules and associates with kinesin superfamily member KIF3. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:149–158. doi: 10.1093/emboj/17.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Setou M., Seog D.H., Ogasawara K., Dohmae N., Takio K., Hirokawa N. A novel motor, KIF13A, transports mannose-6-phosphate receptor to plasma membrane through direct interaction with AP-1 complex. Cell. 2000;103:569–581. doi: 10.1016/s0092-8674(00)00161-6. [DOI] [PubMed] [Google Scholar]
- Seiler S., Kirchner J., Horn C., Kallipolitou A., Woehlke G., Schliwa M. Cargo binding and regulatory sites in the tail of fungal conventional kinesin. Nat. Cell Biol. 2000;2:333–338. doi: 10.1038/35014022. [DOI] [PubMed] [Google Scholar]
- Setou M., Nakagawa T., Seog D.H., Hirokawa N. Kinesin superfamily motor protein KIF17 and mLin-10 in NMDA receptor-containing vesicle transport. Science. 2000;288:1796–1802. doi: 10.1126/science.288.5472.1796. [DOI] [PubMed] [Google Scholar]
- Skoufias D.A., Cole D.G., Wedaman K.P., Scholey J.M. The carboxy-terminal domain of kinesin heavy chain is important for membrane binding. J. Biol. Chem. 1994;269:1477–1485. [PubMed] [Google Scholar]
- Stenoien D.L., Brady S.T. Immunochemical analysis of kinesin light chain function. Mol. Biol. Cell. 1997;8:675–689. doi: 10.1091/mbc.8.4.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima I., Sheetz M.P. Kinectin distribution in chicken nervous system. Neurosci. Letters. 1996;211:171–174. doi: 10.1016/0304-3940(96)12752-x. [DOI] [PubMed] [Google Scholar]
- Tsai M.Y., Morfini G., Szebenyi G., Brady S.T. Release of kinesin from vesicles by hsc70 and regulation of fast axonal transport. Mol. Biol. Cell. 2000;11:2161–2173. doi: 10.1091/mbc.11.6.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale R.D., Fletterick R.J. The design plan of kinesin motors. Annu. Rev. Cell Dev. Biol. 1997;13:745–777. doi: 10.1146/annurev.cellbio.13.1.745. [DOI] [PubMed] [Google Scholar]
- Verhey K.J., Meyer D., Deehan R., Blenis J., Schnapp B.J., Rapoport T.A., Margolis B. Cargo of kinesin identified as JIP scaffolding proteins and associated signaling molecules. J. Cell Biol. 2001;152:959–970. doi: 10.1083/jcb.152.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Toyoshima I., Steuer E.R., Sheetz M.P. Kinesin and cytoplasmic dynein binding to brain microsomes. J. Biol. Chem. 1992;267:20457–20464. [PubMed] [Google Scholar]