Abstract
Proline-rich tyrosine kinase 2 (PYK2), a tyrosine kinase structurally related to focal adhesion kinase (FAK), is implicated in regulating cytoskeletal organization. However, mechanisms by which PYK2 participates in and regulates cytoskeletal organization remain largely unknown. Here we report identification of PSGAP, a novel protein that interacts with PYK2 and FAK and contains multiple domains including a pleckstrin homology domain, a rhoGTPase-activating protein domain, and a Src homology 3 domain. PYK2 interacts with PSGAP Src homology 3 domain via the carboxyl-terminal proline-rich sequence. PSGAP is able to increase GTPase activity of CDC42 and RhoA in vitro and in vivo. Remarkably, PYK2, but not FAK, can activate CDC42 via inhibition of PSGAP-mediated GTP hydrolysis of CDC42. Moreover, PSGAP is localized at cell periphery in fibroblasts in a pleckstrin homology domain–dependent manner. Over expression of PSGAP in fibroblasts results in reorganization of cytoskeletal structures and changes of cellular morphology, which requires rhoGTPase-activating activity. Taken together, our results suggest that PSGAP is a signaling protein essential for PYK2 regulation of cytoskeletal organization via Rho family GTPases.
Keywords: proline-rich tyrosine kinase 2, focal adhesion kinase, CDC42, rhoGAP, PSGAP
Introduction
Integrin signaling triggered by the interaction of extracellular matrix and cell-surface integrin is essential in regulating cell migration, cell growth, and cell survival (Hynes 1992; Schwartz et al. 1995; Parsons 1996). The potential mediators of integrin signaling include protein kinases [focal adhesion kinase (FAK; Schaller et al. 1992), c-Src (Kaplan et al. 1994), COOH-terminal Src kinase (Bergman et al. 1995), protein kinase C (Jaken et al. 1989; Barry and Critchley 1994), and phosphatidylinositol 3-kinase (Chen and Guan 1994; Guinebault et al. 1995)], adapter proteins [Grb2 (Schlaepfer et al. 1994), Crk (Birge et al. 1993), p130Cas (Polte and Hanks 1995; Harte et al. 1996), and paxillin (Burridge et al. 1992)], and small GTP-protein guanine nucleotide exchange factors (C3G and Sos; Tanaka et al. 1994). Although many components involved in integrin signaling have been identified, the signaling events that regulate cytoskeletal organization and cell migration remain to be elucidated. Two major classes of signaling molecules (i.e., small GTP-binding protein Rho subfamily members and protein tyrosine kinases) have been found to participate in and regulate integrin signaling.
Small GTP-binding proteins of the Rho subfamily, particularly CDC42, Rac1, and RhoA, play important roles in diverse cellular events including actin cytoskeletal organization, membrane trafficking, transcriptional regulation, and cell growth control (Van Aelst and D'Souza-Schorey 1997; Hall 1998). In fibroblasts, CDC42, Rac1, and RhoA function to organize actin cytoskeleton, including filopodium extension, lamellipodium formation, and generation of actin stress fibers and focal adhesions, respectively (Ridley and Hall 1992; Nobes and Hall 1995; Van Aelst and D'Souza-Schorey 1997; Hall 1998). The ability of Rho family GTPases to participate in signaling events is determined by the ratio of GTP/GDP-bound forms in the cell. The cycling of GTPases between active GTP-bound states and inactive GDP-bound states is regulated by the opposing effects of guanine nucleotide exchange factors, which enhance the exchange of bound GDP for GTP, and the GTPase-activating proteins (GAPs), which increase hydrolysis of bound GTP. In addition, GTPases are regulated by guanine nucleotide dissociation inhibitors, which can inhibit both the exchange of GTP and the hydrolysis of bound GTP.
In addition to small GTP-binding protein Rho family members, tyrosine kinases are essential in integrin signaling. A major family of tyrosine kinases regulating integrin signaling and localized at focal adhesions is the FAK family (Guan and Shalloway 1992; Schaller et al. 1992). FAK family tyrosine kinases include FAK and proline-rich tyrosine kinase 2 (PYK2), also known as cellular adhesion kinase β, related adhesion focal tyrosine kinase, and calcium-dependent tyrosine kinase (Avraham et al. 1995; Lev et al. 1995; Sasaki et al. 1995; Li and Earp 1997). FAK and PYK2 are highly homologous to each other in structure. Both contain a central catalytic domain and large amino (NH2) and carboxyl (COOH)-terminal noncatalytic regions (Avraham et al. 1995; Lev et al. 1995; Sasaki et al. 1995; Li and Earp 1997). The COOH-terminal region has two proline-rich sequences and a focal adhesion-targeting (FAT) domain. The proline-rich sequences provide the binding motifs for Src homology 3 (SH3) domains of p130Cas (Crk-associated substrate), Graf (GTPase regulator associated with FAK), and Paps [pleckstrin homology (PH) and SH3 domain containing ArfGAP proteins] (Polte and Hanks 1995; Hildebrand et al. 1996; Astier et al. 1997; Manie et al. 1997; Ohba et al. 1998; Xiong et al. 1998; Andreev et al. 1999). The FAT domain that is both necessary and sufficient for the targeting of FAK/PYK2 to focal adhesions is critical for the association with the cytoskeletal proteins paxillin (Hildebrand et al. 1993; Tachibana et al. 1995; Salgia et al. 1996; Li and Earp 1997; Xiong et al. 1998). In addition to these common binding partners, PYK2 also interacts with proteins that do not bind to FAK. For example, PYK2, but not FAK, interacts with Nirs (mammalian homologues of Drosophila retinal degeneration B) (Lev et al. 1999).
While FAK plays an important role in regulating integrin-mediated cell migration and cell spreading (Illc et al. 1995; Carey et al. 1996; Richardson et al. 1997), few insights were available regarding the role of PYK2 in regulating integrin signaling. PYK2 is implicated in diverse cellular events including regulation of neurotransmission or neuroplasticity (Lev et al. 1995), osteoclastic bone resorption, protein secretion (Fuortes et al. 1999), and cell growth control (Xiong and Parsons 1997; Zhao et al. 2000). We have found that FAK and PYK2 mediate different effects on cytoskeletal organization. Overexpression of PYK2, but not FAK, in fibroblasts induces reorganization of cytoskeletal structures, and PYK2-induced cytoskeletal reorganization can be rescued by FAK. In addition, expression of PYK2, but not FAK, in fibroblasts induces apoptosis (Xiong and Parsons 1997). The mechanisms underlying PYK2-induced cytoskeletal reorganization and the differential functions between PYK2 and FAK are largely unknown.
In an attempt to study the mechanisms involved in PYK2-mediated cytoskeletal reorganization, we used the yeast two-hybrid cloning system to identify proteins that interact with PYK2. Here we report the identification of a novel PYK2-interacting protein, PSGAP (PH and SH3 domain containing rhoGAP protein). PSGAP stimulates GTPase activity of CDC42 and RhoA both in vitro and in vivo. PYK2 bound to PSGAP SH3 domain, inhibited the effect of PSGAP on CDC42, and activated CDC42. Our results suggest that PSGAP is a mediator for PYK2 to regulate cytoskeletal organization via small G proteins.
Materials and Methods
Reagents
Rabbit antisera against PSGAP were raised using a glutathione-S-transferase (GST) fusion protein containing the COOH-terminal 60 amino acids of PSGAP SH3 domain (amino acid residues 731–786). The anti–PSGAP antibodies were purified by affinity column using PSGAP antigen. Monoclonal antibodies were purchased from Santa Cruz Biotechnology, Inc. (anti–Myc), Sigma-Aldrich (anti–Flag), or Transduction Laboratories (anti–PYK2). Mouse brain mRNAs and primers for RACE-PCR were purchased from CLONTECH Laboratories, Inc.
Yeast Two-Hybrid Studies
The PYK2 COOH terminus (amino acid residues 781–1009) was generated by PCR, subcloned into pGBT10 (pGBT10-PYK2Δ1-781; provided by Dr. Ian Macara, University of Virginia, Charlottesville, VA), and used as bait to screen a mouse brain cDNA library fused to the GAL4 transcriptional activation domain (provided by Dr. Jeff Chamberlain, University of Michigan, East Lansing, MI). The Y190 yeast strain was first transformed with pGBT10-PYK2Δ1-781 and subsequently with the mouse brain cDNA library. Positive transformants were selected by growth without tryptophan (Trp−), leucine (Leu−), and histidine (His−), and by β-galactosidase (β-Gal) activity (Huang et al. 2000). Plasmid DNA was purified from His+ β-Gal+ colonies, and retransformed into yeast with different bait vectors to determine specificity.
To characterize binding between PYK2 and PSGAP, plasmids encoding PYK2 and PSGAP mutants were cotransformed into yeast Y190. Interactions were characterized by growth of transformed yeasts on plates without leucine, tryptophan, or histidine and by β-Gal activity. Binding affinity of PSGAP with PYK2 COOH terminus, and PYK2 proline mutants was also characterized by a liquid β-Gal assay as described previously (Huang et al. 2000).
Cloning of Full-Length cDNA of PSGAP
RACE-PCR was employed to obtain entire open reading frame using the Marathon cDNA Amplification Kit and Advantage cDNA Polymerase (CLONTECH Laboratories, Inc.) following the manufacturer's instructions. Two specific primers designed from mouse PSGAP sequences and two primers directed toward the cDNA adapters were used for RACE-PCR. The first primer, 5′-GACCCAAGTGGACCCAAATGGGGCAGGCC-3′, was complementary to bp 851–878 of PSGAP cDNA, whereas the second primer, 5′-GCCTCGCCATCCCCCAGTTTCCCCCCAGATCTGTG-3′, was complementary to bp 936–971 of PSGAP cDNA. PCR products were purified and subcloned into the TA vector (Invitrogen), and positive clones were sequenced. The Blast program from NCBI and the Smart program from EMBL were used for sequence analysis (Schultz et al. 1998).
Expression Vectors
To generate GST-fusion proteins, cDNAs encoding PSGAP PH/GAP (amino acid residues 265–590), PSGAP-PH (amino acid residues 265–453), and PSGAP-SH3 (amino acid residues 731–786) were amplified by PCR and subcloned into pGEX-2T vector using BamHI and EcoRI sites. Full-length FAK and PYK2 were subcloned into mammalian expression vectors downstream of a Myc epitope (MEQKLISEEDL) under the control of the cytomegalovirus (CMV) promoter (pCMV-c-Myc) (Xiong and Parsons 1997). cDNAs encoding PSGAP or its mutants were subcloned into mammalian expression vectors downstream of a Flag epitope (MDYKDDDDKGP) under the control of the CMV promoter. Point mutations in PYK2 or PSGAP were generated by site-directed mutagenesis using Quick Change kit (Stratagene). The authenticity of all mutants was verified by DNA sequencing.
GST Pull-Down Assays
Transiently transfected HEK293 cells were lysed in modified RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM sodium chloride, 1% NP-40, 0.25% sodium-deoxycholate, and proteinase inhibitors). Cell lysates were precleared with GST immobilized on glutathione-sepharose 4B (Amersham Pharmacia Biotech), and then incubated with GST fusion proteins (2–5 μg) immobilized on glutathione-sepharose beads at 4°C for 1 h with constant rocking. Beads were washed three times with modified RIPA buffer and assayed for bound proteins by immunoblotting.
Cell Culture and Transfection
HEK293 cells and 10T1/2 fibroblasts were maintained in DMEM supplemented with 10% fetal calf serum, 100 μg/ml penicillin G, and 100 μg/ml streptomycin (GIBCO BRL). For HEK293 cell transfection experiments, cells were plated for 12 h at a density of 106 cells/10-cm culture dish, and were transfected using the calcium phosphate precipitation method. 24–36 h after transfection, cells were lysed in the modified RIPA buffer. Lysates were subjected to immunoprecipitation or immunoblotting. For 10T1/2 cell transfection, cells were transfected with 30–40 μl of lipofectamine (GIBCO BRL) or superinfect (Invitrogen) with various constructs (3–4 μg DNA) in DMEM serum-free medium. After incubation for 5 h, serum-containing medium was added. 24 h later, cells were trypsinized, replated on fibronectin-coated cover slips, and fixed with 4% paraformaldehyde for immunostaining.
Immunoprecipitation and Immunoblotting
Immunoprecipitation of individual proteins was carried out as previously described (Xiong et al. 1998). In brief, cell lysates (1 mg protein) were incubated with antibodies (1–10 μg) at 4°C for 1 h in a final volume of 1 ml modified RIPA buffer with constant rocking. After the addition of protein A-agarose beads, reactions were incubated at 4°C for 1 h. Immune complexes were resolved by SDS-PAGE and subjected to immunoblotting.
Northern Blotting
To study mRNA distribution in different tissues, an RNA filter comprising poly(A)-selected RNAs of multiple mouse tissues (CLONTECH Laboratories, Inc.) was hybridized with specific 32P-labeled cDNAs as described previously (Xiong et al. 1994). In brief, mouse PSGAP probe (bp 796–1358) or human Graf probe (bp 1–717 in KIAA 0621) was radiolabeled with [α-32P]CTP by nick translation using random primers. Probes (∼4 × 108 cpm/μg) were hybridized with the RNA filter overnight at 42°C and washed twice (30 min) in 0.2× SSC–0.1% SDS at room temperature, and at 45°C. Autoradiograms were obtained by exposure of blots to XAR film (Eastman Kodak Co.) for 2–24 h.
Immunocytochemistry
Cells were fixed with 4% paraformaldehyde for 20 min, blocked with 10% BSA, and incubated with antibodies against PSGAP (rabbit polyclonal), PYK2 (goat polyclonal; Santa Cruz Biotechnology, Inc.), or paxillin (monoclonal; Transduction Laboratories). Double-label immunostaining was done with appropriate fluorochrome-conjugated secondary antibodies. Fluorescent images of cells were captured on a CCD camera (Sony) mounted on a E600 microscope (Nikon) using Photoshop imaging software.
RhoGAP Assays
In vitro RhoGAP assay was carried out as described previously (Hildebrand et al. 1996). In brief, RhoA, Rac1, Cdc42 (gifts from Alan Hall, University College, London, UK), Ran, and PSGAP GAP domain were expressed as GST fusion proteins in Escherichia coli, purified, and cleaved from GST. RhoA, Rac1, Cdc42, and Ran (40 ng each) were incubated with 15 μCi of [γ-32P] GTP (6,000 Ci/mmol; Dupont) in loading buffer (50 mM Tris-HCl, pH 7.5, 50 mM NaCl, 5 mM EDTA, 1 mg BSA per ml, 1 mM DTT) for 15 min at room temperature in a total volume of 30 μl. After MgCl2 was added to a final concentration of 10 mM, and the GTPase mixture was placed on ice. The GTP-loaded GTPase (final concentration of 5 nM) was incubated with or without 1, 5, 10, or 20 nM PSGAP GAP domain for 15 min at room temperature. The reaction was stopped with 1 ml wash buffer (50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM DTT) and the mixture was filtered through nitrocellulose membranes. Membranes were washed with 10 ml of wash buffer three times, air dried, and counted to determine the amount of [γ-32P]GTP remaining.
To test RhoGAP activity in vivo, GTP-bound CDC42, Rac1, or RhoA was determined by specific binding to the p21-binding domain of PAK1 (GST-PBD) or rhotekin (GST-RBD) (Fujisawa et al. 1998; del Pozo et al. 2000). In brief, cell lysates expressing Myc-tagged wild-type small GTPases (CDC42, Rac1, or RhoA) with or without Flag-tagged PSGAP were incubated with 5 μg of recombinant GST-PBD, or GST-RBD conjugated with glutathione-agarose beads (Amersham Pharmacia Biotech) for 60 min at 4°C, washed with lysis buffer, and eluted with SDS sample buffer. Bound CDC42, Rac, or Rho was analyzed by Western blotting using anti–Myc antibodies (Santa Cruz Biotechnology, Inc.). Whole-cell lysates were also analyzed for the presence of expressed CDC42, Rac, RhoA, and PSGAP for normalization.
Nucleotide Sequence Accession Number
The nucleotide sequences of the PSGAP cDNAs have been given the GenBank accessions numbers AF297029 (PSGAP-s) and AF297030 (PSGAP-m).
Results
Identification and Molecular Cloning of a Novel PYK2 Interacting RhoGAP Protein, PSGAP
To identify proteins that bind to PYK2, we used the yeast two-hybrid system. Using the PYK2 COOH terminus (amino acids 781–1009) as bait, a positive clone was isolated from a mouse brain cDNA library. Sequence analysis revealed that this clone contained an insert of ∼2.5 kb with a predicted open reading frame of 526 amino acids contiguous with the GAL4 activation domain (Gal4AD), missing an apparent NH2 terminus. RACE-PCR was used to isolate 5′cDNA from a mouse brain cDNA. Two splice variants that overlapped with the original cDNA clone were isolated. Both share a common COOH-terminal coding sequence, but vary in the positions of their starting codons and in the sequences of their respective 5′-untranslated regions. Whereas one isoform encoded a protein of 786 amino acids, the other splice variant appeared use methionine 104 as the translational starting codon (indicated by an arrow), resulting in an open reading frame for a protein of 683 amino acids (Fig. 1 A). By searching the human EST databases, we were able to obtain a partial human cDNA clone that corresponded to the residues of 508–786 of the mouse clone with 84% identity (Fig. 1 B).
Figure 1.
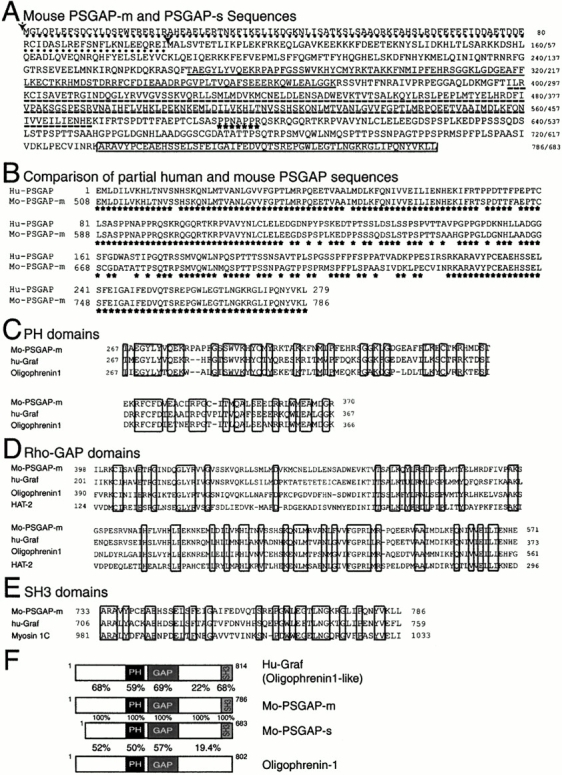
Cloning of PSGAP, a novel rhoGAP-containing protein that interacts with PYK2. (A) Deduced amino acid sequences of mouse PSGAP-m and PSGAP-s. Filled arrows indicate the two methionines as the translational starting codons for the splice variants, PSGAP-m (786 amino acids) and PSGAP-s (683 amino acids), respectively. PSGAP-m contains a different amino terminus (∼103 amino acids) that is underlined with dots. The PH domain (amino acids 267–370 in PSGAP-m) is underlined, rhoGAP domain (amino acids 398–571 in PSGAP-m) is underlined with dashed lines, the proline rich motif is indicated by stars, and the SH3 domain (amino acids 733–786 in PSGAP-m) is boxed. The original clone isolated by yeast two-hybrid screen contains amino acids 265–786. (B) Comparison of the partial human and mouse PSGAP protein sequences. Partial human PSGAP (hu-PSGAP) (279 amino acids) were obtained by searching human EST databases, which corresponded to the residues of 508–786 of mouse PSGAP-m (mo-PSGAP-m) with 84% identity. Stars indicate identical residues. (C) Sequence alignment of PH domains of mouse PSGAP-m, human Graf, and oligophrenin-1. (D) Sequence alignment of the RhoGAP domains of mo-PSGAP-m, hu-Graf, Oligophrenin-1, HAT-2. (E) Sequence alignment of the SH3 domains of mo-PSGAP-m, hu-Graf, and myosin 1C. Alignments were determined using the program pileup from the Genetics Computer Group. (C, D, and E) Identical residues are boxed. (F) Comparison of domain structures of mo-PSGAP-m, mo-PSGAP-s, hu-Graf, and oligophrenin-1. The PH, rhoGAP, and SH3 domains are indicated. The amino acid identities (%) within a specific domain are shown.
Protein structural analysis using SMART program (Schultz et al. 1998) revealed that this novel protein contained multiple domains, including a PH domain, a RhoGAP domain, and a SH3 domain in the COOH-terminal region (Fig. 1A and Fig. F). Therefore, we named this protein PSGAP. The PH domain of PSGAP located from amino acid 267 to 370 showed homology to the PH domain of centaurin α (a PH domain containing ArfGAP protein) (Hammonds-Odie et al. 1996). Interestingly, Graf, a previously identified FAK and PYK2-interacting rhoGAP protein (Hildebrand et al. 1996), and oligophrenin 1, a protein implicated in mental retardation (Billuart et al. 1998), also contained PH domains that were highly related to that of PSGAP (Fig. 1 C). The centrally located RhoGAP domain (residues 398–571) was highly homologous to rhoGAP domains of Graf, oligophrenin 1, and HAT-2 (George and Clayton 1992) (Fig. 1 D). The SH3 domain (residues 733–786) shared homology with SH3 domains of several proteins including Graf and myosin 1C (Fig. 1 E). The overall structure of PSGAP resembled that of Graf, and PSGAP showed high homology to Graf with 68% identity in amino terminal regions, 59% in PH domains, 69% in rhoGAP domains, and 68% in SH3 domains (Fig. 1 F). PSGAP was also highly homologous to oligophrenin 1 in the amino terminus (52% identity), PH domain (50% identity), and rhoGAP domain (57% identity) (Fig. 1 F). These results indicate that PSGAP and Graf comprise a novel family of rhoGAP proteins that interact with PYK2 and FAK.
Differential Distribution between PSGAP and Graf
We compared mRNA expression patterns of PSGAP and Graf by Northern blot analysis. Three RNA transcripts of PSGAP were detected at ∼8, 6, and 3.8 kb, suggesting that there may be splice variants of PSGAP mRNAs as revealed by cDNA cloning. Of the three transcripts, the 3.8 kb appeared most abundant and was expressed at high levels in the testis, heart, lung, skeletal muscle, kidney, and brain, with a low level of expression in the spleen (Fig. 2). In contrast, expression of Graf was different from that of PSGAP. First, its transcript was detected as a single band at 9 kb. Second, Graf was highly expressed in the brain and heart, at low levels in the lung and skeletal muscle, but undetectable in the liver and skeletal muscle (Fig. 2).
Figure 2.
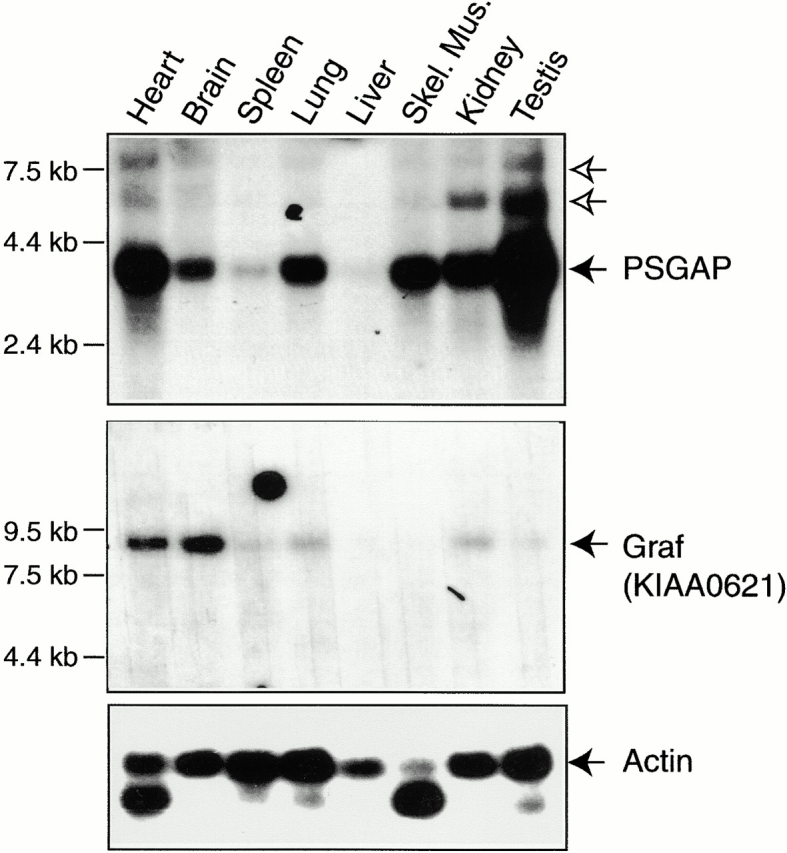
Expression of PSGAP and Graf mRNAs in various tissues. Northern blot analyses were carried out using a multiple tissue blot (CLONTECH Laboratories, Inc.). Mouse cDNA fragments (796–1,358 bp for PSGAP and 1–717 bp of KIAA0621 for Graf) were labeled with [32P-α]dCTP by the random prime method and hybridized with the blot. A major transcript at 3.8 kb was detected for PSGAP (top, solid arrow). In addition, two more PSGAP transcripts, at 6 and 8 kb, respectively, were detected (top, open arrows). Graf transcript was detected at ∼9 kb (bottom, arrow). Molecular weight markers are indicated (left).
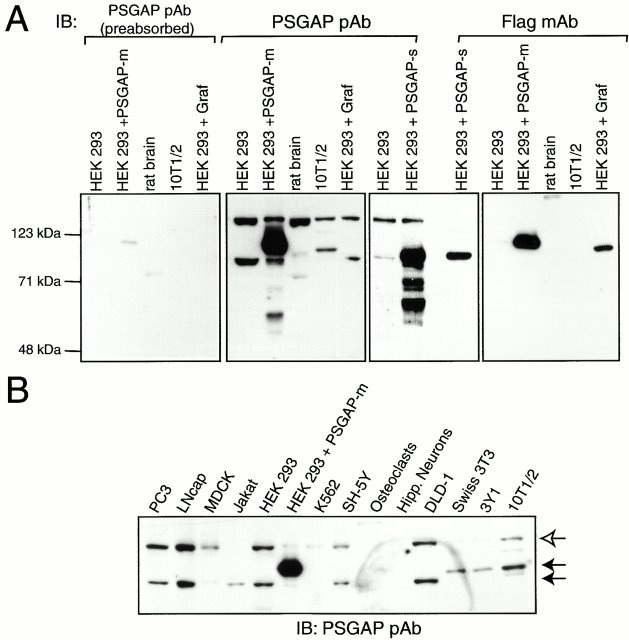
To characterize PSGAP protein distribution, specific antibodies were raised in rabbits using a recombinant PSGAP protein (amino acids 731–786) as an antigen. Western blot analysis detected three protein bands of ∼140, 95, and 85 kD (Fig. 3 A). These bands were recognized by the antibodies, but not preimmune sera (data not shown). Preabsorption of antibodies with the antigen inhibited their ability to detect all three bands (Fig. 3 A). Furthermore, although Graf and PSGAP share a high homology of sequences, the PSGAP antibodies did not cross react with Graf, whose expression was evident by immunoblotting with anti–Flag antibodies (Fig. 3 A). These results suggest that the three bands represent specifically three variants of PSGAP, which may be generated by mRNA splicing as revealed by Northern blot analysis. The 95- and 85-kD proteins appeared to be encoded by the two identified splice variants, PSGAP-m (786 amino acids) and PSGAP-s (683 amino acids), respectively, since expression of both transcripts yielded proteins of similar molecular masses (Fig. 3 A). In addition, the three PSGAP isoforms exhibited a unique expression pattern. The 95-kD protein (PSGAP-m) was expressed in fibroblastic cell lines, including 10T1/2, 3Y1, and Swiss 3T3 fibroblasts, whereas the 140- and 85-kD (PSGAP-s) proteins appeared to be expressed ubiquitously with a relatively high level in tumor cell lines including prostate (PC3 and LnCap) and colon (DLD-1) cancer cells (Fig. 3 B).
Figure 3.
Expression of PSGAP in various cells. (A) Characterization of anti–PSGAP antibody. A GST fusion protein containing the mouse PSGAP SH3 domain (amino acids 731–786) was used to generate rabbit polyclonal antibodies as described in Materials and Methods. Lysates (10 μg protein) of 293 cells expressing Flag-tagged PSGAP (PSGAP-m and PSGAP-s) and Graf, and other indicated cell lysates (20 μg protein) were resolved on SDS-PAGE and subjected to immunoblotting with anti–PSGAP antibodies, antibodies preabsorbed with the GST-PSGAP antigen, or an anti–Flag antibody. (B) Western blot analysis of PSGAP expression. Lysates (20 μg protein) of various cells were resolved on SDS-PAGE and subjected to immunoblotting with anti–PSGAP antibodies (6 μg protein of HEK293 cells expressing PSGAP was loaded). (Solid arrows) Transfected PSGAP and endogenously expressed PSGAP. Molecular weight markers are indicated (left).
Dependence of PSGAP Binding to PYK2 on the PSGAP's SH3 Domain and PYK2's Proline-rich Region
To understand the mechanism of the PSGAP–PYK2 interaction, we mapped the domain in PSGAP required for interaction in yeast two-hybrid assays. As shown in Fig. 4 A, deletion of the SH3 domain (PSGAPΔSH3) inhibited almost completely PSGAP binding to PYK2. Moreover, PSGAP SH3 domain (PSGAP-SH3) alone was able to bind to PYK2. These data indicated that the SH3 domain of PSGAP was required and sufficient for the binding to PYK2 in yeast two-hybrid system.
Figure 4.
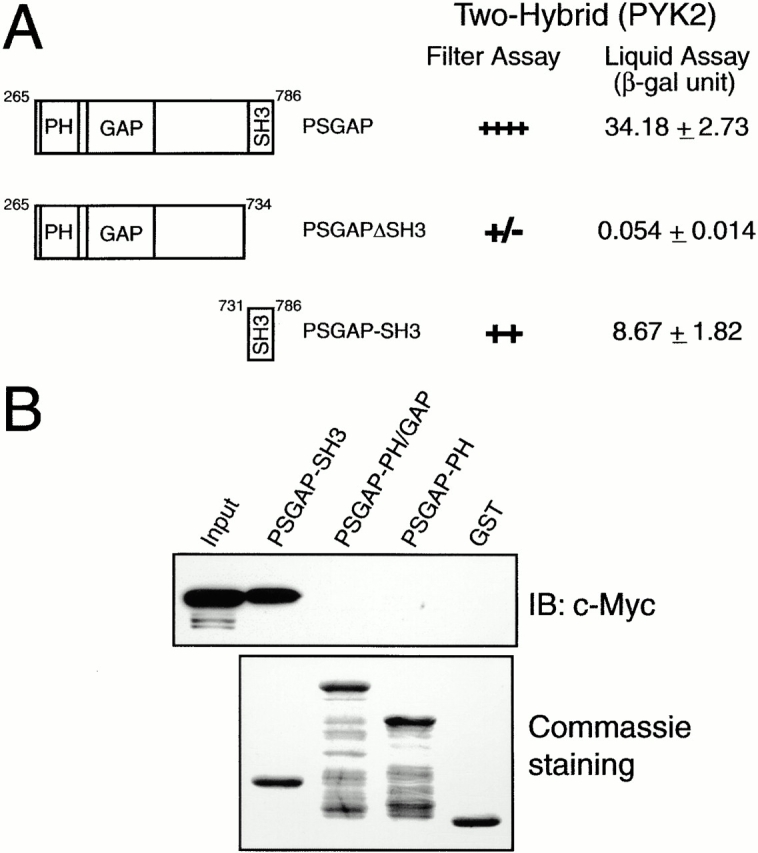
SH3 domain of PSGAP is essential for the interaction with PYK2. (A) Mapping of the binding region in PSGAP by yeast two-hybrid assays. Yeast cells were cotransformed with a vector encoding the Gal4 binding domain fused to different PSGAP constructs and Gal4AD fused to PYK2. Transformed yeast cells were seeded in Leu−, Trp−, and His− plates and assayed for β-Gal activity. Transformed yeast cells were also grown in Leu− and Trp− medium for liquid β-Gal assays. (B) Analysis of PSGAP binding to PYK2 by in vitro GST pull-down assays. HEK293 cells were transfected with Myc-tagged PYK2. Lysates of transfected cells were incubated with various GST-PSGAP fusion proteins (SH3, amino acids 731–786; PH-GAP, amino acids 265–590; and PH, amino acids 265–454) immobilized on beads. Lysate input and bound proteins were resolved on SDS-PAGE and subjected to immunoblotting with an anti–Myc antibody (top). An equal amount of GST fusion proteins was used as revealed by Coomassie staining (bottom).
We next examined the ability of PSGAP SH3 domain to bind to PYK2 in pull-down assays. Lysates from cells transfected with Myc-tagged PYK2 were incubated with different GST fusion proteins immobilized on beads (Fig. 4 B). Consistent with results from yeast two-hybrid assays, GST-PSGAP-SH3 fusion protein was able to precipitate PYK2, whereas little PYK2 was pulled down by GST alone, or GST-PSGAP fusion proteins without SH3 domain (GST-PSGAP-PH/GAP and GST-PSGAP-PH) (Fig. 4 B), further indicating the requirement of and sufficiency for the PSGAP SH3 domain to mediate the interaction with PYK2.
The dependence of the PSGAP–PYK2 interaction on PSGAP's SH3 domain suggests the importance of proline-rich sequences in PYK2. PYK2 COOH-terminal domain contains two proline-rich regions that may function as ligands for SH3 domains. The first proline-rich motif spans residues 712–719 (PPPKPSR). However, this proline-rich sequence of PYK2 appeared dispensable in the PSGAP–PYK2 interaction, since mutations of this motif either by deletion or a site-directed mutagenesis had no apparent effect on PSGAP binding to PYK2 in yeast two-hybrid assays (Fig. 5). The second proline-rich motif contains the sequence PPQKPPR (residues 855–861). This proline-rich motif appeared to be critical for the interaction, since deletion of the region (amino acids 838–869) containing this motif abolished the interaction (Fig. 5 A). The failure of PYK2Δ1-869 or PYK2-Nterm to interact with PSGAP was not due to improper expression of these constructs. As shown in Fig. 5 A, PYK2Δ1-869 and PYK2-Nterm were expressed and were able to interact with Hic5 (a paxillin-related protein) or m-rdgB (the mouse homologue of Drosophila rdgB), respectively. To further ascertain the role of this proline-rich sequence in PYK2-PSGAP interaction, we mutated prolines to alanines and characterized the binding of PSGAP to PYK2 and its mutants. As shown in Fig. 5 B, a point mutation in proline 859 or double mutations in prolines 717 and 859 significantly reduced PSGAP binding to PYK2, suggesting that this second proline-rich sequence was critical for PYK2 binding to PSGAP. In contrast, PYK2 binding to the PSGAP-related protein Graf required both the first and second proline-rich motifs (Fig. 5 B). While mutation in either proline 717 or 859 reduced the binding significantly, only the double mutant was able to nearly abolish the interaction of Graf to PYK2 (Fig. 5 B).
Figure 5.
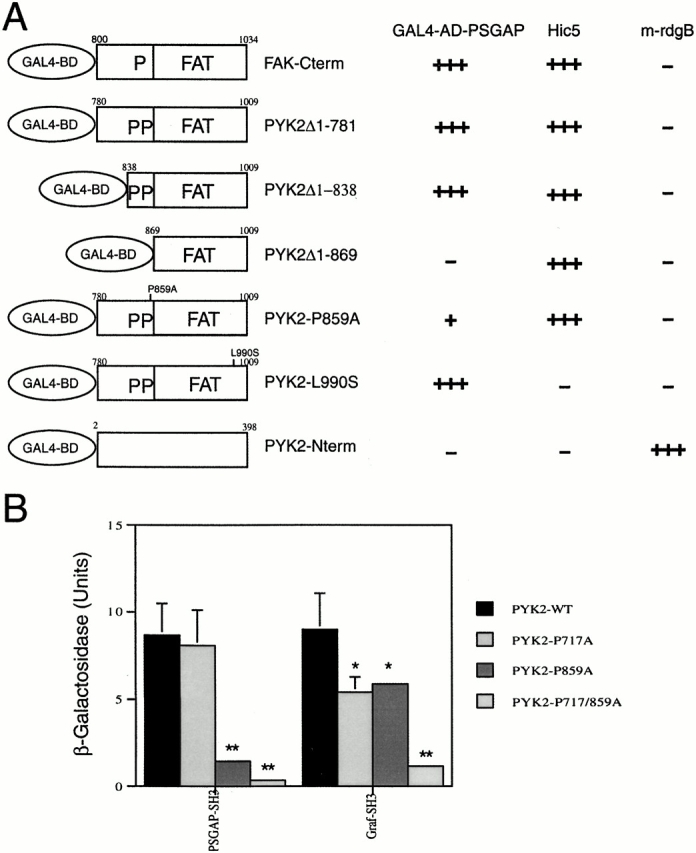
Proline 859 in PYK2 is required for PSGAP binding. (A) Analyses of PYK2 domains essential for binding to PSGAP by yeast two-hybrid filter assays. Yeast cells were cotransformed with a vector encoding the Gal4 binding domain (Gal4-BD) fused to different constructs of PYK2, or FAK and Gal4AD/PSGAP. Gal4AD/Hic5 and Gal4AD/mrdgB were used as controls. Transformed yeast cells were seeded in Leu−, Trp−, and His− plates and scored for growth and β-Gal activity (blue color). (−) No blue color in the yeast after 24 h in reaction, (+) blue reaction after 4 h, (+++) blue reaction after 30 min. (B) Reduction of the ability of P859A mutant to bind PSGAP in yeast two-hybrid liquid assays. Yeast cells were cotransformed with a vector encoding the Gal4 DB fused to SH3 domain of PSGAP or Graf and wild-type or mutant PYK2 in Gal4AD. Transformed yeast cells were grown in His−, Leu−, and Trp− medium for liquid β-Gal assays. *P < 0.05 and **P < 0.01 in comparison with PYK2-WT, Student's t test.
Interaction of PYK2 and FAK with PSGAP In Vivo
To determine whether PYK2 and FAK interact with PSGAP in vivo, we examined their association in HEK293 cells transfected with Flag-tagged PSGAP with Myc-tagged PYK2 or FAK. Lysates of transfected cells were incubated with anti–Flag antibodies immobilized on beads. PSGAP-interacting proteins were purified by immunoprecipitation followed by immunoblotting with anti–Myc antibodies. PYK2 and FAK were detected in immunoprecipitates of PSGAP (Fig. 6 A), suggesting that PYK2 and FAK associated with PSGAP in mammalian cells. Again, the interaction of PYK2 with PSGAP required the SH3 domain of PSGAP because a mutant with deletion of the SH3 domain (PSGAPΔSH3) failed to interact with PYK2 (Fig. 6 A). This interaction was unaffected by the addition of 103 amino acid residues in the NH2 terminus of PSGAP-m isoform, since similar amounts of PSGAP-m and PSGAP-s were observed in PYK2 immunoprecipitates (data not shown). To further assess the ability of endogenous PYK2 to bind PSGAP, the in vivo coimmunoprecipitation between PSGAP and PYK2 was carried out in 10T1/2 fibroblasts and in HEK293 cells, where both PYK2 and PSGAP were expressed endogenously. As observed with transfected cells, 1–5% PYK2 was recovered in PSGAP immunocomplexes, but not in preimmune complexes (Fig. 6 B), indicating that PSGAP associates with PYK2 in vivo.
Figure 6.
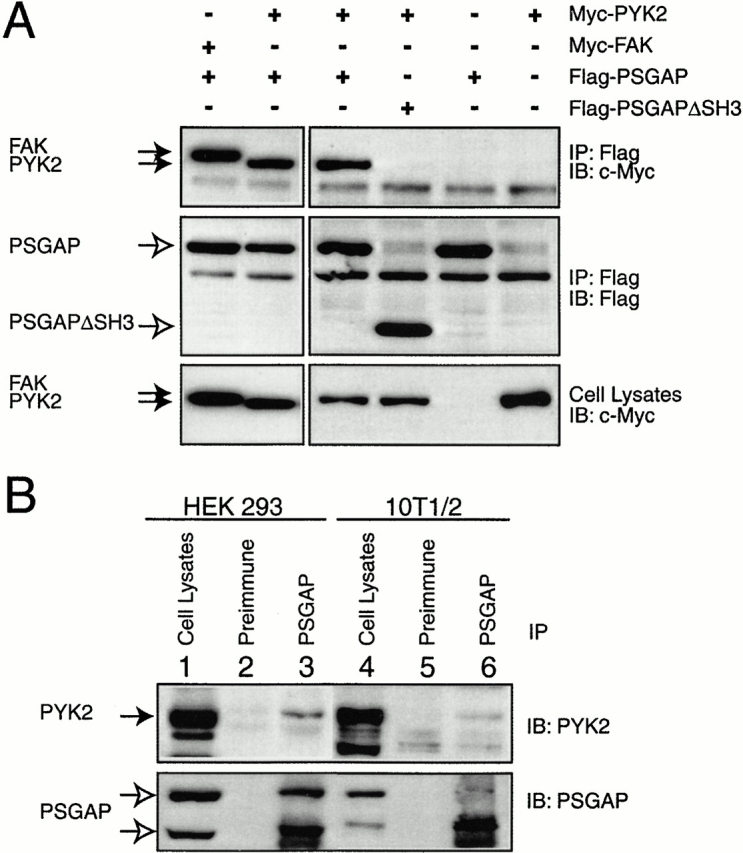
Interaction of PSGAP with PYK2 and FAK in vivo. (A) Coimmunoprecipitation of PSGAP with PYK2 or FAK in mammalian-expressing cells. HEK293 cells cotransfected Myc-tagged PYK2 or FAK with Flag-tagged PSGAP or PSGAPΔSH3 were lysed. Immunoprecipitations (IP) with Flag antibody was revealed by immunoblotting (IB) with anti–Myc (top) or anti–Flag (middle) antibodies. Expression of PYK2 and FAK was demonstrated at bottom. (B) Coimmunoprecipitation of PSGAP with PYK2 in vivo. Cell lysates from HEK293 or 10T1/2 cells were immunoprecipitated with PSGAP antibodies and immunoblotted with PYK2 antibodies. (Solid arrow) PYK2; (open arrows) PSGAP.
Stimulation of GTPase Activity of CDC42 and RhoA by PSGAP
To assess the GTPase-activating activity of PSGAP, we constructed a GST fusion protein containing the entire GAP domain (residues 390–590) and assayed its ability to activate GTPase activity of GTP-bound CDC42, Rac1, and RhoA. Purified small G proteins (RhoA, Rac1, CDC42, and Ran) were loaded with [γ-32P]GTP and incubated with different concentrations of PSGAP GAP domain. The GTPase-activating activity was determined by measuring the ratio of GTPase-bound radioactivity without GAP domain versus that with the GAP domain. As shown in Fig. 7 A, the PSGAP GAP domain was able to stimulate the intrinsic GTPase activity of RhoA and CDC42 in a concentration-dependent manner. However, the effects of the PSGAP GAP domain on the GTPase activity of Rac1 and Ran were undetectable. These results indicate that CDC42 and RhoA may be preferred substrates for PSGAP in the cell-free system.
Figure 7.
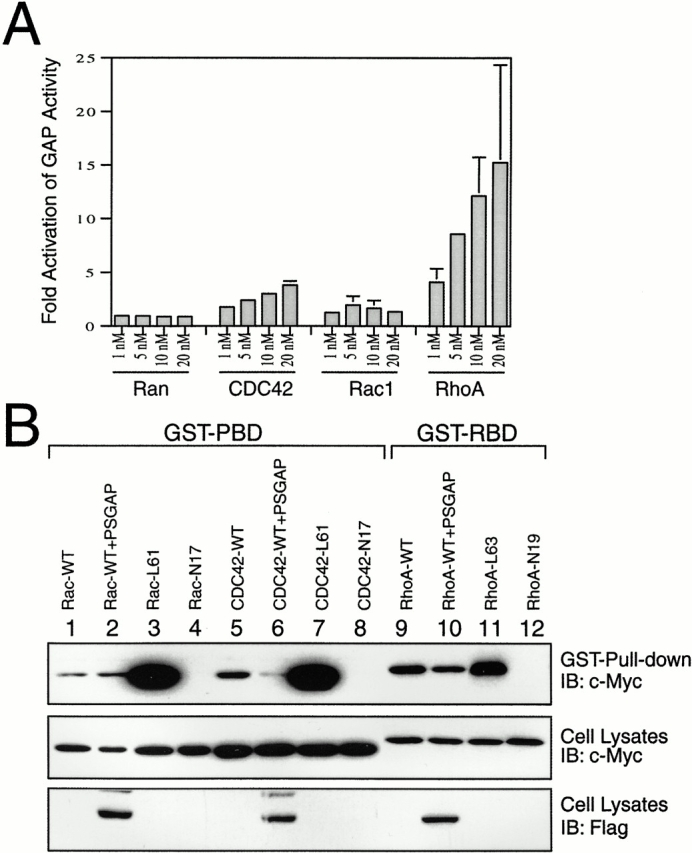
GTPase-activating activity of PSGAP. (A) Characterization of GTPase-activating activity of PSGAP in vitro. Recombinant proteins of RhoA, Rac1, CDC42, and Ran GTPases were loaded with [γ-32P] GTP and incubated without or with increasing concentrations of PSGAP GAP domain. Hydrolyzed and GTPase-bound radioactive GTP was determined by nitrocellulose filtration assay as described in Materials and Methods. Fold activation represents the ratio of radioactivity remaining after incubation in the absence of GAP domain versus the amount of radioactivity remaining after incubation in the presence of GAP domain. (B) Analysis of the effects of PSGAP on GST-PBD or GST-RBD binding to transfected small G proteins. HEK293 cells were transfected with wild-type, active, or inactive mutants of Rac1, CDC42, or RhoA (all Myc-tagged) with or without Flag-tagged PSGAP. Cell lysates were incubated with GST-PBD or GST-RBD immobilized on beads. Bound small G proteins were resolved on SDS-PAGE and detected by immunoblotting (top). Equal amounts of small G proteins were expressed as shown in the middle. (Bottom) Expression of PSGAP.
The GTP-bound Rho family GTPases bind to downstream proteins specifically and with high affinity. For example, activated CDC42 or Rac binds to PAK1, while activated RhoA binds to rhotekin specifically (Reid et al. 1996; Van Aelst and D'Souza-Schorey 1997; Hall 1998). To assess the GTPase-activating activity of PSGAP in vivo, GST-PBD or GST-RBD was used to isolate activated small G proteins from cell lysates coexpressing wild-type CDC42, Rac, or RhoA and PSGAP (Reid et al. 1996; del Pozo et al. 2000). As shown in Fig. 7 B, while a similar amount of CDC42 and RhoA was expressed in transfected cells, the amount of precipitated active CDC42 and RhoA was significantly lower in cells expressing PSGAP, indicating that PSGAP-stimulated GTPase activities of CDC42 and RhoA in vivo (Fig. 7 B). However, the GTPase-activating activity of PSGAP towards Rac1 was undetectable in this assay (Fig. 7 B). These results suggest that CDC42 and RhoA may be substrates for PSGAP in vivo.
Differential Regulation of PSGAP and CDC42 by PYK2 and FAK
While both PYK2 and FAK interacted with PSGAP, effects of PYK2 and/or FAK on PSGAP were unknown. To address this question, we first examined whether PSGAP could be tyrosine phosphorylated by PYK2 or FAK. PSGAP was cotransfected in HEK293 cells with wild-type, mutant PYK2, or FAK. Tyrosine phosphorylation was revealed by immunoprecipitation and immunoblotting using antibodies against phosphotyrosine. As shown in Fig. 8 A, PSGAP was tyrosine phosphorylated in cells coexpressing PYK2. Tyrosine phosphorylation of PSGAP was diminished in cells coexpressing PYK2-KD (a catalytically inactive PYK2) or PYK2-Y402F (an autophosphorylation mutant), suggesting the involvement of PYK2 kinase activity and autophosphorylation in this event. In contrast, PSGAP was not tyrosine phosphorylated by FAK (Fig. 8 A). These results suggest that the regulation of PSGAP by PYK2 may be different from that by FAK. To further address the differential effects of PYK2 and FAK on PSGAP, we examined whether PYK2 and FAK affected PSGAP's regulation of GTPase activity of CDC42. GST-PBD binding to CDC42 in cell lysates coexpressing PSGAP and PYK2 or FAK was carried out. While PSGAP exhibited significant reduction of the precipitated GTP-bound CDC42 (Fig. 8 B), coexpressing PYK2 increased the GTP-bound CDC42, suggesting that PSGAP's negative effect on CDC42-GTP loading was abolished when PYK2 was coexpressed (Fig. 8 B). PYK2-mediated inhibition of PSGAP depended on PYK2 binding to PSGAP, since mutations in PYK2 (PYK2-P859A) or PSGAP (PSGAPΔSH3) that block the interaction of PYK2 with PSGAP failed to exhibit such inhibitory effect (Fig. 8 B). Interestingly, although FAK was able to interact with PSGAP, it seemed unable to regulate PSGAP (Fig. 8 B). These results further indicated that PYK2 and FAK regulated PSGAP differently.
Figure 8.
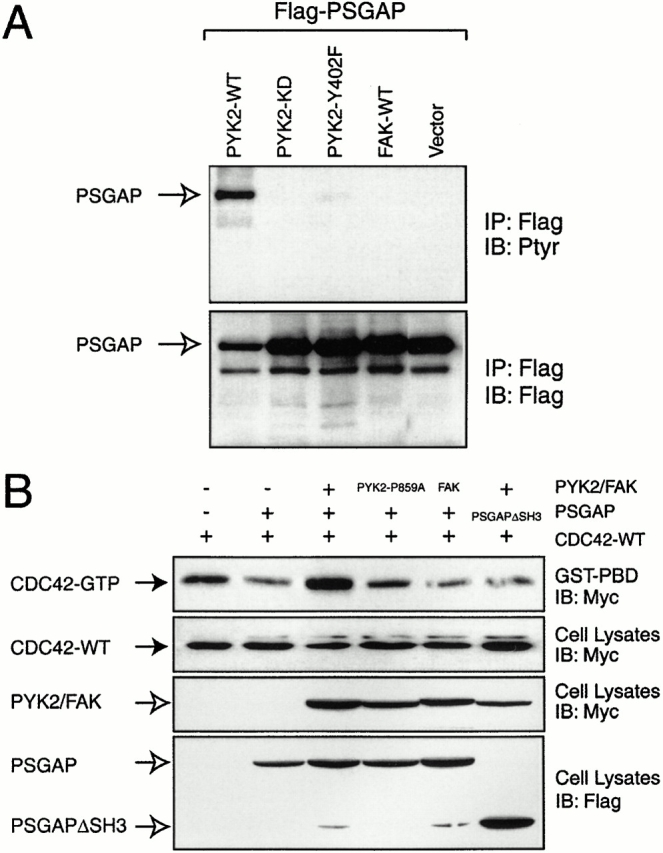
Regulation of PSGAP by PYK2, but not FAK. (A) Tyrosine phosphorylation of PSGAP by PYK2, but not FAK. HEK293 cells were transfected with indicated constructs. Cell lysates were immunoprecipitated with Flag antibodies (for PSGAP) and immunoblotted with antibodies against phosphotyrosine (Ptyr) and Flag. Open arrows indicate PSGAP. (B) Inhibition of PSGAP's effect on CDC42 by PYK2, but not FAK. HEK293 cells were transfected with indicated constructs. Cell lysates were incubated with GST-PBD immobilized on beads. Bound active CDC42 was resolved on SDS-PAGE and detected by immunoblotting. Equal amounts of CDC42 were expressed as indicated. The expression of PYK2, FAK, and PSGAP are also shown. (Solid arrows) CDC42; (open arrows) PYK2 or PSGAP.
PYK2 inhibition of PSGAP's effect on CDC42 suggested that PYK2 might activate CDC42. To test this hypothesis, we examined whether PYK2 increases the GTP loading of CDC42 when PYK2 was coexpressed with small GTP-binding proteins. As we expected, expressing PYK2 indeed increased the GTP-bound CDC42, but not Rac (Fig. 9 A). Interestingly, PYK2-induced activation of CDC42 appeared to require PYK2 catalytic activity and binding to PSGAP, since the catalytic inactive mutant (PYK2-KD) and mutant that did not bind PSGAP (PYK2-P859A) failed to increase GTP-bound CDC42 (Fig. 9 B). Remarkably, GTP-bound CDC42 was increased only in cells expressing PYK2, but not FAK (Fig. 9 B), suggesting again the differential regulation of CDC42 between PYK2 and FAK.
Figure 9.
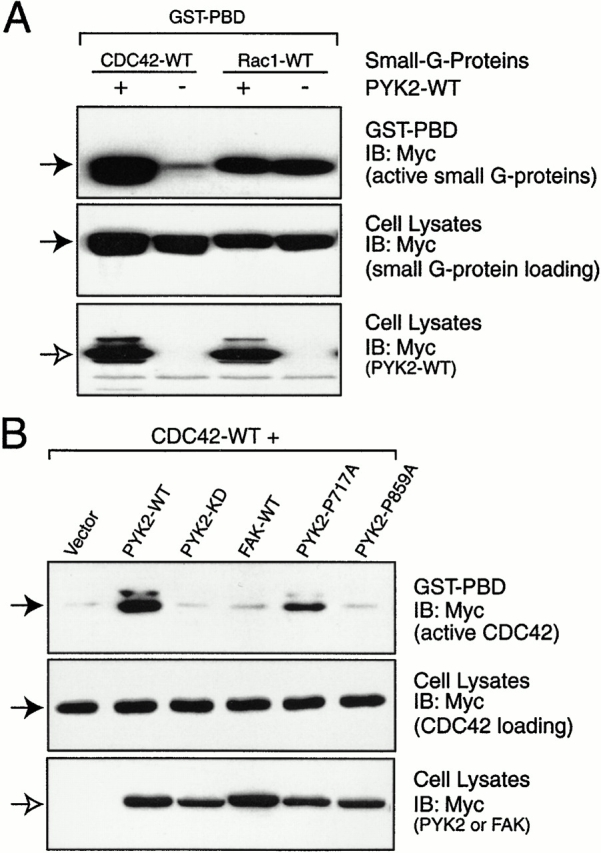
Activation of CDC42 by PYK2, but not FAK. (A) PYK2 activation of CDC42, but not Rac. (B) PYK2, but not FAK, activation of CDC42 requires PYK2 catalytic activity and its binding region to PSGAP. (A and B) HEK293 cells were transfected with indicated constructs. Cell lysates were incubated with GST-PBD immobilized on beads. Bound active CDC42 or Rac was resolved on SDS-PAGE and detected by immunoblotting. Equal amounts of small G proteins were expressed as indicated. The expression of PYK2 and FAK are also shown. (Solid arrows) Small G proteins; (open arrows) PYK2 or FAK.
Subcellular Localization of PSGAP in Fibroblasts
Subcellular localization of PSGAP was characterized in 10T1/2 fibroblasts that expressed PSGAP, PYK2, and FAK endogenously. 10T1/2 cells were plated on fibronectin-coated cover slips, fixed, and immunostained using antibodies against PSGAP (polyclonal) and paxillin (monoclonal). As shown in Fig. 10, PSGAP was distributed in perinuclear regions as well as in peripheral regions as punctuate aggregates where paxillin was also localized. The staining was specific, since it could be blocked by the PSGAP antigen (Fig. 10 A). Interestingly, the punctuate cell periphery staining of PSGAP appeared to be a transitory event. It only appeared in cells grown on fibronectin-coated cover slips within 90 min (data not shown). Remarkably, PYK2 was also localized at the cell periphery in addition to the perinuclear regions as well as focal contacts (Fig. 10 B). To confirm the subcellular localization of PSGAP, we examined the distribution of the transfected PSGAP in 10T1/2 cells. As described in the following section, when PSGAP was overexpressed, it caused abnormal morphology. Thus, the subcellular localization of transfected PSGAP was studied in cells with a low level of transfected PSGAP. Consistent with endogenous PSGAP, transfected PSGAP was distributed in perinuclear and cell-periphery regions (Fig. 10 C).
Figure 10.
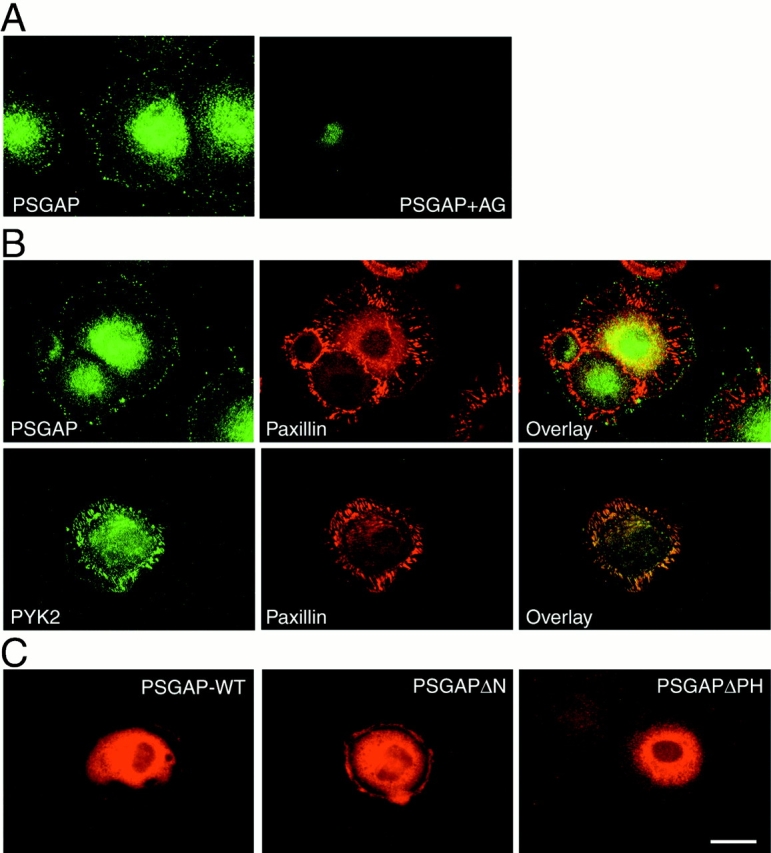
Expression of PSGAP at the cell periphery in 10T1/2 fibroblasts. 10T1/2 cells were fixed with 4% paraformaldehyde for 20 min, blocked with 10% BSA, and immunostained with indicated antibodies. (A) Immunostaining using antibodies against PSGAP or antibodies preabsorbed with the GST-PSGAP antigen. (B) Coimmunostaining with anti–PSGAP or anti–PYK2 and antipaxillin antibodies. PSGAP and PYK2 were visualized by FITC-conjugated secondary antibodies, whereas paxillin appeared in red with rhodamine-conjugated secondary antibodies. (C) Immunostaining of PSGAP wild-type (PSGAP-WT), NH2-terminal–deleted PSGAP (PSGAPΔN), and PH-domain–deleted PSGAP (PSGAPΔPH) in transfected 10T1/2 fibroblasts. Cells were transfected with the indicated constructs and stained with anti–PSGAP antibodies. Bar, 50 μm.
To determine which domains of PSGAP were required for its cell periphery localization, we generated mutants with deletions of either NH2 terminus (PSGAPΔN, aa1-265 deleted) or the NH2 terminus and PH domain (PSGAPΔPH, amino acids 1–390 deleted). While PSGAPΔN exhibited a similar localization pattern as the wild type, there was little, if any, staining of PSGAPΔPH at the cell periphery (Fig. 10 C). These results indicated that the PH domain was critical for PSGAP cell periphery localization.
Cytoskeletal Reorganization in Fibroblasts Expressing PSGAP
To study potential roles of PSGAP in regulating cytoskeletal organization, we examined the cytoskeletal organization and cellular morphology in 10T1/2 fibroblasts expressing PSGAP or its mutants. Expression of wild-type PSGAP disrupted focal adhesions and cell morphology in approximately half of transfected cells (Fig. 11). The number (73–75%) of cells with disorganized cytoskeleton or abnormal morphology increased in cells expressing PSGAPΔN or PSGAPΔPH, indicating that the NH2 terminus and PH domains were not required for these events. To examine whether rhoGAP activity was required for PSGAP-mediated disorganization of cytoskeleton, we generated a rhoGAP-inactive mutant (PSGAPΔN-GD) containing an arginine 424-to-glutamine mutation in the GAP domain (Fig. 11 B). Arginine 424 in PSGAP is conserved with arginine 85 in p50rhoGAP, a critical residue for the direct contacts with CDC42 (Barrett et al. 1997; Rittinger et al. 1997). Mutation of this residue rendered many rhoGAP proteins enzymatically inactive (Barrett et al. 1997; Rittinger et al. 1997; Taylor et al. 1999). Indeed, PSGAPΔN-GD was unable to stimulate CDC42 or RhoA GTPase activity in vitro (data not shown). Remarkably, a majority of cells expressing PSGAPΔN-GD exhibited normal focal adhesions and cellular morphology (Fig. 11), indicating that the rhoGAP activity was required for PSGAP-mediated cytoskeletal reorganization.
Figure 11.
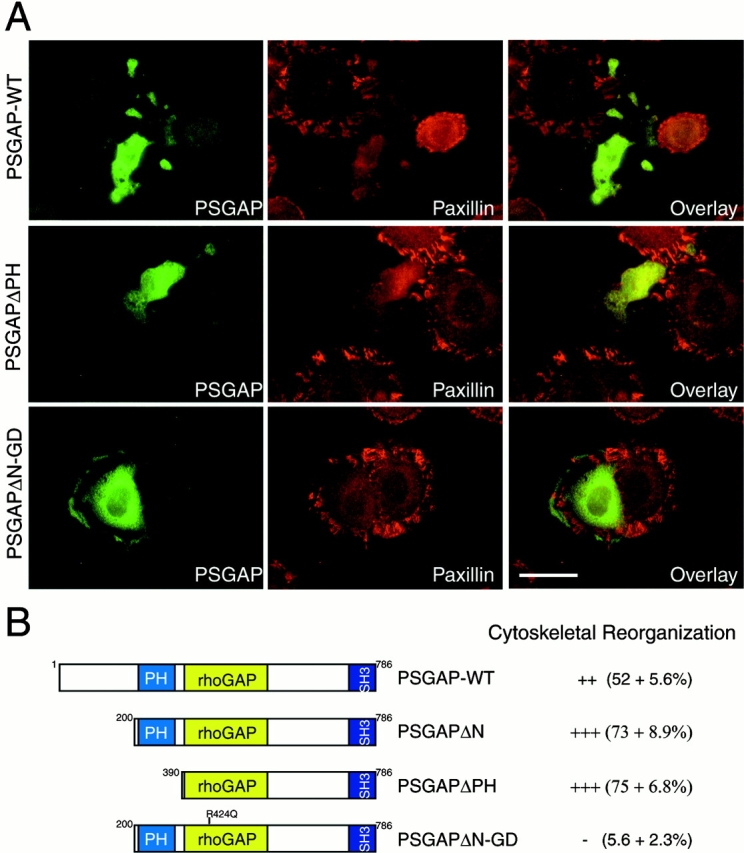
Cytoskeletal reorganization in fibroblasts expressing PSGAP. (A) 10T1/2 cells were transfected with the indicated constructs and stained with anti–PSGAP (green) and anti–paxillin (red) antibodies. Bar, 50 μm. (B) Schematic representation of PSGAP and PSGAP mutants. The numbers represent the amino acid residues deleted from PSGAP-m. The PH domain, rhoGAP domain, and SH3 domain are indicated. Cytoskeletal reorganization index (mean ± SEM) was determined by counting the morphologically altered PSGAP-expressing cells divided by the total number of PSGAP-expressing cells, and listed at right.
Discussion
This paper described identification and characterization of PSGAP, a novel PYK2-interacting protein that may play a pivotal role in PYK2 regulation of cytoskeleton. PSGAP interacts with PYK2 via the COOH-terminal SH3 domain and PYK2's proline-rich region. In addition to the SH3 domain, PSGAP has a PH domain and a RhoGAP domain. These domains appear to be of functional importance. The PH domain is essential for subcellular localization, whereas the rhoGAP domain stimulates CDC42 and RhoA GTPase activity and is required for PSGAP-mediated cytoskeletal reorganization. Furthermore, we provide evidence that the interaction of PYK2 with PSGAP regulates the activation of CDC42 GTPase. These results suggest that PSGAP is an important mediator for PYK2 to regulate Rho-family GTPases, providing insight into mechanisms by which PYK2 regulates cytoskeletal organization.
SH3 domains are important in mediating protein–protein interactions or directing cell compartmentalization (Koch et al. 1991; Bar-Sagi et al. 1993). PSGAP SH3 domain alone, but not PSGAP with the deletion of SH3 domain, interacts with PYK2, demonstrating that the SH3 domain mediates the protein–protein interaction. The ligand for PSGAP SH3 domain appears to be the second proline-rich sequence (residues 874–880), PPQKPPR, at the COOH terminus of PYK2. This proline-rich motif belongs to class II ligands of SH3 domains (Feng et al. 1994) and is conserved in FAK. Indeed, FAK can interact with PSGAP in vitro and in vivo. On the other hand, this motif also binds to other SH3 domain containing GAP proteins including Graf and Paps, a PH and SH3 domain containing ArfGAP protein (Andreev et al. 1999). We speculate that these binding proteins may compete with PSGAP to interact with PYK2 or FAK, and that PYK2 and FAK may regulate different GAP proteins in different subcellular compartments.
PSGAP is localized at the cell periphery, which appears to correspond to the cell membrane. Interestingly, such localization occurs in a PH domain–dependent manner. PH domains are small protein modules of ∼120 amino acids found in many proteins involved in cell signaling and cytoskeletal rearrangement (Blomberg et al. 1999). PH domains function as membrane targeting signals and bind specifically to phosphoinositides (Blomberg et al. 1999). The exact ligand of the PH domain of PSGAP is not yet known. It is possible that phosphoinositides regulated by integrin stimulation may be important for PSGAP localization and rhoGAP activity. The transient cell periphery localization of PSGAP during integrin-mediated cell spreading (data not shown) suggests that function of the PSGAP PH domain may be regulated by integrin stimulation.
The presence of a RhoGAP domain in PSGAP suggests to us that PSGAP contains an intrinsic GTPase activating activity for Rho family proteins. Indeed, the recombinant RhoGAP domain exhibited GTPase activating activity towards several small G proteins, including RhoA and CDC42 in vitro and in vivo. It is worth noting, however, that the substrate profile of PSGAP's GTPase-activating activity is different between in vitro and in vivo assays. While it activates RhoA more than CDC42 in vitro, the GTPase-activating activity for RhoA is weakly detected when PSGAP is expressed in HEK293 cells. The discrepancy between in vitro and in vivo GTPase-activating activities of PSGAP may result from the difference of the assays, or different affinities of the GST-PBD and GST-RBD to the active small G proteins. This discrepancy has been reported previously for other GAP proteins. For example, the substrate spectrum of p50rhoGAP in vitro encompasses CDC42, Rac, and Rho, whereas it appears to be restricted to Rho in vivo (Ridley et al. 1993). The apparent difference of PSGAP's GTPase-activating activities between in vitro and in vivo assays suggest that PSGAP rhoGAP activity may be regulated by phospholipids and/or subcellular localization.
Although both PYK2 and FAK bind to PSGAP, only PYK2, but not FAK, negatively regulates the GTPase-activating activity of PSGAP. This finding is interesting since it provides an explanation of different functions of PYK2 and FAK, and implicates a possibility of activation of CDC42 by PYK2-inactivating PSGAP. Indeed, CDC42 was activated by PYK2 in HEK293 cells. Whether PYK2 activates CDC42 in vivo and how PYK2 regulates PSGAP activity is unclear. PSGAP became tyrosine phosphorylated in cells overexpressing PYK2, but not FAK, implying that PYK2-directed PSGAP phosphorylation may be a mechanism for the regulation of PSGAP activity by PYK2. The requirement of the PYK2 catalytic activity for the inhibition of PSGAP supports this speculation. It will be interesting to further investigate how phosphorylation of PSGAP regulates its GAP activity.
Although PSGAP is highly related to Graf in structure, they are encoded by different genes. Partial cDNA encoding Graf is initially cloned from chicken as a FAK-binding protein (Hildebrand et al. 1996). Graf orthologs have been identified in humans, including oligophrenin 1-like or KIAA0621, which exhibit an overall sequence identity of >90%, suggesting that Graf is evolutionarily conserved. Interestingly, PSGAP is also evolutionarily conserved. In addition to human orthologs of PSGAP, analysis of EST database reveals clones in rat or Xenopus laevis that are highly homologous to PSGAP, with sequence identity of 85–95%. However, human and mouse PSGAP show ∼50% identities with chicken and human Graf, suggesting that both Graf and PSGAP are derived from two different biologically conserved genes. Moreover, PSGAP exhibits a tissue expression pattern different from Graf. Graf is expressed in a relatively restricted pattern, with mRNA detected in tissues including heart and brain. In contrast, PSGAP is expressed in a wide variety of tissues and cell lines, including testis, heart, skeletal muscles, kidney, lung, and brain. Remarkably, PSGAP, but not Graf, is expressed in fibroblasts. Thus, in fibroblasts, PSGAP, but not Graf, may be an important mediator for PYK2 or FAK to regulate cytoskeletal organization. In terms of substrate specificity, PSGAP and Graf are also different. While Rho is the preferred in vivo substrate for Graf (Taylor et al. 1999), PSGAP has high GTPase-activating activity towards CDC42 in vivo.
In summary, we have identified a novel PH and SH3 domain-containing rhoGAP protein, PSGAP, which interacts with both PYK2 and FAK in a SH3 domain-dependent manner. PSGAP, a CDC42/Rho GTPase-activating protein, may be an important mediator for PYK2 to regulate cytoskeletal organization via Rho family GTPases.
Acknowledgments
We are grateful to Dr. Jeff Chamberlain for the cDNA library, Dr. John Collard for GST-fusion proteins, and Dr. Nagaso for providing KIAA0621 cDNA clones.
This study was supported in part by a start-up fund from the School of Medicine, University of Alabama at Birmingham and grants from the American Heart Association Southeastern Affiliate Grand In Aid 051566B (to W.-C. Xiong) and the National Institute of Neurological Disorders and Stroke (NS40480 and NS34062) (to L. Mei).
Footnotes
Abbreviations used in this paper: AD, activation domain; β-Gal, β-galactosidase; FAK, focal adhesion kinase; GAP, GTPase-activating protein; GST, glutathione-S-transferase; PBD, p21-binding domain of PAK1; PH, pleckstrin homology; PYK2, proline-rich tyrosine kinase 2; RBD, p21-binding domain of rhotekin; SH3, Src homology 3.
References
- Andreev J., Simon J.P., Sabatini D.D., Kam J., Plowman G., Randazzo P.A., Schlessinger J. Identification of a new PYK2 target protein with Arf-GAP activity. Mol. Cell. Biol. 1999;19:2338–2350. doi: 10.1128/mcb.19.3.2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astier A., Avraham H., Manie S.N., Groopmen J., Canty T., Avraham S., Freedman A.S. The related adhesion focal tyrosine kinase is tyrosine-phosphorylated after β1-integrin stimulation in B cells and binds to p130Cas . J. Biol. Chem. 1997;272:228–232. doi: 10.1074/jbc.272.1.228. [DOI] [PubMed] [Google Scholar]
- Avraham S., London R., Fu Y., Ota S., Hiregowdara D., Li J., Jiang S., Pasztor L.M., White R.A., Groopman J.E., Avraham H. Identification and characterization of a novel related adhesion focal tyrosine kinase (RAFTK) from megakaryocytes and brain. J. Biol. Chem. 1995;270:27742–27751. doi: 10.1074/jbc.270.46.27742. [DOI] [PubMed] [Google Scholar]
- Bar-Sagi D., Rotin D., Batzer A., Mandiyan V., Schlessinger J. SH3 domains direct cellular localization of signaling molecules. Cell. 1993;74:83–91. doi: 10.1016/0092-8674(93)90296-3. [DOI] [PubMed] [Google Scholar]
- Barrett T., Xiao B., Dodson E.J., Dodson G., Ludbrook S.B., Nurmahomed K., Gamblin S.J., Musacchio A., Smerdon S.J., Eccleston J.F. The structure of the GTPase-activating domain from p50rhoGAP. Nature. 1997;385:458–461. doi: 10.1038/385458a0. [DOI] [PubMed] [Google Scholar]
- Barry S.T., Critchley D.R. The RhoA-dependent assembly of focal adhesions in Swiss 3T3 cells is associated with increased tyrosine phosphorylation and the recruitment of both pp125FAK and protein kinase C-δ to focal adhesions. J. Cell Sci. 1994;107:2033–2045. doi: 10.1242/jcs.107.7.2033. [DOI] [PubMed] [Google Scholar]
- Bergman M.V., Vladimir V., Virtanen I., Alitalo K. Overexpression Csk tyrosine kinase is localized in focal adhesions, causes reorganization of αvβ5 integrin, and interferes with cell spreading. Mol. Cell. Biol. 1995;15:711–722. doi: 10.1128/mcb.15.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billuart P., Bienvenu T., Ronce N., des Portes V., Vinet M.C., Zemni R., Roest Crollius H., Carrie A., Fauchereau F., Cherry M. Oligophrenin-1 encodes a rhoGAP protein involved in X-linked mental retardation. Nature. 1998;392:923–926. doi: 10.1038/31940. [DOI] [PubMed] [Google Scholar]
- Birge R.B., Fajardo J.E., Reichman C., Shoelson S.E., Songyang Z., Cantley L.C., Hanafusa H. Identification and characterization of a high-affinity interaction between v-Crk and tyrosine-phosphorylated paxillin in CT10-transformed fibroblasts. Mol. Cell. Biol. 1993;13:4648–4656. doi: 10.1128/mcb.13.8.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomberg N., Baraldi E., Nilges M., Saraste M. The PH superfolda structural scaffold for multiple functions. TIBS. 1999;24:441–445. doi: 10.1016/s0968-0004(99)01472-3. [DOI] [PubMed] [Google Scholar]
- Burridge K., Turner C.E., Romer L.H. Tyrosine phosphorylation of paxillin and pp125FAK accompanies cell adhesion to extracellular matrixa role in cytoskeletal assembly. J. Cell Biol. 1992;119:893–903. doi: 10.1083/jcb.119.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey L.A., Chang J.F., Guan J.L. Stimulation of cell migration by overexpression of focal adhesion kinase and its association with Src and Fyn. J. Cell Sci. 1996;109:1787–1794. doi: 10.1242/jcs.109.7.1787. [DOI] [PubMed] [Google Scholar]
- Chen H.-C., Guan J.L. Association of focal adhesion kinase with its potential substrate phosphatidylinositol 3-kinase. Proc. Natl. Acad. Sci. USA. 1994;91:10148–10152. doi: 10.1073/pnas.91.21.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo M.A., Price L.S., Alderson N.B., Ren X.D., Schwartz M.A. Adhesion to the extracellular matrix regulates the coupling of the small GTPase Rac to its effector PAK. EMBO (Eur. Mol. Biol. Organ.) J. 2000;19:2008–2014. doi: 10.1093/emboj/19.9.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Chen J.K., Yu H., Simon A., Schreiber S.L. Two binding orientations for peptides to the Src SH3 domaindevelopment of a general model for SH3-ligand interactions. Science. 1994;266:1241–1247. doi: 10.1126/science.7526465. [DOI] [PubMed] [Google Scholar]
- Fujisawa K., Madaule P., Ishizaki T., Watanabe G., Bito H., Saito Y., Hall A., Narumiya S. Different regions of Rho determine Rho-selective binding of different classes of rho target molecules. J. Biol. Chem. 1998;273:18943–18949. doi: 10.1074/jbc.273.30.18943. [DOI] [PubMed] [Google Scholar]
- Fuortes M., Melchior M., Han H., Lyon G.J., Nathan C. Role of the tyrosine kinase PYK2 in the integrin-dependent activation of human neutrophils by TNF. J. Clin. Invest. 1999;104:327–335. doi: 10.1172/JCI6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George J.M., Clayton D.F. Differential regulation in the avian song control circuit of an mRNA predicting a highly conserved protein related to protein kinase C and the bcr oncongene. Brain Res. Mol. Brain Res. 1992;12:323–329. doi: 10.1016/0169-328x(92)90134-w. [DOI] [PubMed] [Google Scholar]
- Guan J.L., Shalloway D. Regulation of pp125FAK both by cellular adhesion and by oncogenic transformation. Nature. 1992;358:690–692. doi: 10.1038/358690a0. [DOI] [PubMed] [Google Scholar]
- Guinebault C., Payratre B., Racaud-Sultan C., Mazarguil H., Breton M., Mauco G., Plantavid M., Chap H. Integrin-dependent translocation of phosphoinositide 3-kinase to the cytoskeleton of thrombin-activated platelets involves specific interactions of p85α with actin filaments and focal adhesion kinase. J. Cell Biol. 1995;129:831–842. doi: 10.1083/jcb.129.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. G proteins and small GTPasesdistant relatives keep in touch. Science. 1998;279:509–514. [Google Scholar]
- Hammonds-Odie L.P., Jackson T.R., Profit A.A., Blader I.J., Turck C.W., Prestwish G.D., Theibert A.B. Identification and cloning of centaurin-alpha. A novel phosphatidylinositol 3,4,5-trisphosphate-binding protein from rat brain. J. Biol. Chem. 1996;271:18859–18868. doi: 10.1074/jbc.271.31.18859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harte M.T., Hildebrand J.D., Burnham M.R., Bouton A.H., Parsons J.T. p130Cas, a substrate associated with v-Src and c-Crk, localizes to focal adhesions and binds to focal adhesion kinase. J. Biol. Chem. 1996;271:13649–13655. doi: 10.1074/jbc.271.23.13649. [DOI] [PubMed] [Google Scholar]
- Hildebrand J.D., Schaller M.D., Parsons J.T. Identification of sequences required for the efficient localization of the focal adhesion kinase, pp125FAK to cellular focal adhesions. J. Cell Biol. 1993;123:993–1005. doi: 10.1083/jcb.123.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand J.D., Taylor J.M., Parsons J.T. An SH3 domain-containing GTPase-activating protein for Rho and Cdc42 associates with focal adhesion kinase. Mol. Cell. Biol. 1996;16:3169–3178. doi: 10.1128/mcb.16.6.3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.Z., Won S., Ali D.W., Wang Q., Tanowitz M., Du Q.S., Pelkey K.A., Yang D.J., Xiong W.C., Salter M.W., Mei L. Regulation of neuregulin signaling by PSD95 interacting with ErbB4 at CNS synapses. Neuron. 2000;26:443–455. doi: 10.1016/s0896-6273(00)81176-9. [DOI] [PubMed] [Google Scholar]
- Hynes R.O. Integrinsversatility, modulation, and signaling in cell adhesion. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- Illc D., Furuta Y., Kanazawa S., Takeda N., Sobue K., Nakatsuji N., Nomura S., Fujimoto J., Okada M., Yamamoto T., Alzawa S. Reduced cell motility and enhanced focal adhesion contact formation in cells from FAK-deficient mice. Nature. 1995;377:539–544. doi: 10.1038/377539a0. [DOI] [PubMed] [Google Scholar]
- Jaken S., Leach K., Klauck T. Association of type 3 protein kinase C with focal adhesions in rat embryo fibroblasts. J. Cell Biol. 1989;109:697–704. doi: 10.1083/jcb.109.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan K.B., Bibbins K.B., Swedlow J.R., Arnaud M., Morgan D.O., Varmus H.E. Association of the amino-terminal half of c-Src with focal adhesions alters their properties and is regulated by phosphorylation of tyrosine 527. EMBO (Eur. Mol. Biol. Organ.) J. 1994;13:4747–4756. doi: 10.1002/j.1460-2075.1994.tb06800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C., Anderson D., Moran M., Ellis C., Pawson T. SH2 and SH3 domainselements that control interactions of cytoplasmic signaling proteins. Science. 1991;252:668–674. doi: 10.1126/science.1708916. [DOI] [PubMed] [Google Scholar]
- Lev S., Hernandez J., Martinez R., Chen A., Plowman G., Schlessinger J. Identification of a novel family of targets of PYK2 related to Drosophila retinal degeneration B (rdgB) protein. Mol. Cell. Biol. 1999;19:2278–2288. doi: 10.1128/mcb.19.3.2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev S., Moreno H., Martinez R., Canoll P., Peles E., Musacchio J.M., Plowman G.D., Rudy B., Schlessinger J. Protein tyrosine kinase PYK2 involved in Ca2+-induced regulation of ion channel and MAP kinase functions. Nature. 1995;376:737–745. doi: 10.1038/376737a0. [DOI] [PubMed] [Google Scholar]
- Li X., Earp H.S. Paxillin is tyrosine-phosphorylated by and preferentially associates with the calcium-dependent tyrosine kinase in rat liver epithelial cells. J. Biol. Chem. 1997;272:14341–14348. doi: 10.1074/jbc.272.22.14341. [DOI] [PubMed] [Google Scholar]
- Manie S.N., Beck A.R.P., Astier A., Law S.F., Canty T., Hirai H., Druker B.J., Avraham H., Haghayeghi N., Sattler M. Involvement of p130Cas and p105HEF1, a novel Cas-like docking protein, in a cytoskeleton-dependent signaling pathway initiated by ligation of integrin or antigen receptor on human B cells. J. Biol. Chem. 1997;272:4230–4236. doi: 10.1074/jbc.272.7.4230. [DOI] [PubMed] [Google Scholar]
- Nobes C.D., Hall A. Rho, Rac, CDC42, GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- Ohba T., Ishino M., Aoto H., Sasaki T. Interaction of two proline-rich sequences of cell adhesion kinase b with SH3 domain of p130Cas-related proteins and a GTPase-activating protein Graf. Biochem. J. 1998;330:1249–1254. doi: 10.1042/bj3301249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons J.T. Integrin-mediated signalingregulation by protein tyrosine kinases and small GTP-binding proteins. Curr. Opin. Cell Biol. 1996;8:146–152. doi: 10.1016/s0955-0674(96)80059-7. [DOI] [PubMed] [Google Scholar]
- Polte T.R., Hanks S.K. Interaction between focal adhesion kinase and Crk-associated tyrosine kinase substrate p130Cas . Proc. Natl. Acad. Sci. USA. 1995;92:10678–10682. doi: 10.1073/pnas.92.23.10678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid T., Furuyashiki T., Ishizaki T., Watanabe G., Watanabe N., Fujisawa K., Morii N., Madaule P., Narumiya S. Rhotekin, a new putative target for Rho bearing homology to a serine/threonine kinase, PKN, and rhophilin in the rho-binding domain. J. Biol. Chem. 1996;271:13556–13560. doi: 10.1074/jbc.271.23.13556. [DOI] [PubMed] [Google Scholar]
- Richardson A., Malik R.K., Hildebrand J.D., Parsons J.T. Inhibition of cell spreading by expression of the C-terminal domain of focal adhesion kinase (FAK) is rescued by coexpression of Src or catalytically inactive FAKa role for paxillin tyrosine phosphorylation. Mol. Cell. Biol. 1997;17:6906–6914. doi: 10.1128/mcb.17.12.6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley A.J., Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- Ridley A.J., Self A.J., Kasmi F., Paterson H.F., Hall A., Marshall C.J., Ellis C. Rho family GTPase activating proteins p190, bcr and rhogap show distinct specificities in vitro and in vivo. EMBO (Eur. Mol. Biol. Organ.) J. 1993;12:5151–5160. doi: 10.1002/j.1460-2075.1993.tb06210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittinger K., Walker P.A., Eccleston J.F., Nurmahomed K., Owen D., Laue E., Gamblin S.J., Smerdon S.J. Crystal structure of a small G protein in complex with the GTPase-activating protein rhoGAP. Nature. 1997;388:693–697. doi: 10.1038/41805. [DOI] [PubMed] [Google Scholar]
- Salgia R., Avraham S., Pisick E., Li J.-L., Raja S., Greenfield E.A., Sattler M., Avraham H., Griffin J.D. The related adhesion focal tyrosine kinase forms a complex with paxillin in hematopoietic cells. J. Biol. Chem. 1996;271:31222–31226. doi: 10.1074/jbc.271.49.31222. [DOI] [PubMed] [Google Scholar]
- Sasaki H., Nagura K., Ishino M., Tobioka H., Kotani K., Sasaki T. Cloning and characterization of cell adhesion kinase b, a novel protein-tyrosine kinase of the focal adhesion kinase subfamily. J. Biol. Chem. 1995;270:21206–21219. doi: 10.1074/jbc.270.36.21206. [DOI] [PubMed] [Google Scholar]
- Schaller M.D., Borgman C.A., Cobb B.S., Vines R.R., Reynolds A.B., Parsons J.T. pp125FAK, a structurally distinctive protein-tyrosine kinase associated with focal adhesions. Proc. Natl. Acad. Sci. USA. 1992;89:5192–5196. doi: 10.1073/pnas.89.11.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaepfer D.D., Hanks S.K., Hunter T., van der Geer P. Integrin-mediated signal transduction linked to Ras pathway by Grb2 binding to focal adhesion kinase. Nature. 1994;372:786–791. doi: 10.1038/372786a0. [DOI] [PubMed] [Google Scholar]
- Schultz J., Milpetz F., Bork P., Ponting C.P. SMART, a simple modular architecture research toolidentification of signaling domains. Proc. Natl. Acad. Sci. USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M., Schaller M., Ginsberg M.H. Integrinsemerging paradigms of signal transduction. Annu. Rev. Cell. Dev. Biol. 1995;11:549–599. doi: 10.1146/annurev.cb.11.110195.003001. [DOI] [PubMed] [Google Scholar]
- Tachibana K., Sato T., D'Avirro N., Morimoto C. Direct association of pp125FAK with paxillin, the focal adhesion-targeting mechanism of pp125FAK . J. Exp. Med. 1995;182:1089–1100. doi: 10.1084/jem.182.4.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Morishita T., Hashimoto Y., Hattori S., Nakamura M., Shibuya K., Matuoka T., Takenawa T., Kurata K., Nagachima K., Matsuda M. C3G, a guanine nucleotide-releasing protein expressed ubiquitously, binds to the Src homology 3 domains of CRK and Grb2/ASH proteins. Proc. Natl. Acad. Sci. USA. 1994;91:3443–3447. doi: 10.1073/pnas.91.8.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J.M., Macklem M., Parsons J.T. Cytoskeletal changes induced by Graf, the GTPase regulator associated with focal adhesion kinase, are mediated by Rho. J. Cell Sci. 1999;112:231–242. doi: 10.1242/jcs.112.2.231. [DOI] [PubMed] [Google Scholar]
- Van Aelst L., D'Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- Xiong W.C., Macklem M., Parsons J.T. Expression and characterization of splice variants of PYK2, a focal adhesion kinase-related protein. J. Cell Sci. 1998;111:1981–1991. doi: 10.1242/jcs.111.14.1981. [DOI] [PubMed] [Google Scholar]
- Xiong W.C., Okano H., Patel N.H., Blendy J.A., Montell C. Repo encodes a glial-specific homeo domain protein required in the Drosophila nervous system. Genes Dev. 1994;8:981–994. doi: 10.1101/gad.8.8.981. [DOI] [PubMed] [Google Scholar]
- Xiong W.C., Parsons J.T. Induction of apoptosis after expression of PYK2, a tyrosine kinase structurally related to focal adhesion kinase. J. Cell Biol. 1997;139:529–539. doi: 10.1083/jcb.139.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zheng C., Guan J. PYK2 and FAK differentially regulate progression of the cell cycle. J. Cell Sci. 2000;113:3063–3072. doi: 10.1242/jcs.113.17.3063. [DOI] [PubMed] [Google Scholar]