Abstract
Myosin V is a double-headed unconventional myosin that has been implicated in organelle transport. To perform this role, myosin V may have a high duty cycle. To test this hypothesis and understand the properties of this molecule at the molecular level, we used the laser trap and in vitro motility assay to characterize the mechanics of heavy meromyosin–like fragments of myosin V (M5HMM) expressed in the Baculovirus system. The relationship between actin filament velocity and the number of interacting M5HMM molecules indicates a duty cycle of ≥50%. This high duty cycle would allow actin filament translocation and thus organelle transport by a few M5HMM molecules. Single molecule displacement data showed predominantly single step events of 20 nm and an occasional second step to 37 nm. The 20-nm unitary step represents the myosin V working stroke and is independent of the mode of M5HMM attachment to the motility surface or light chain content. The large M5HMM working stroke is consistent with the myosin V neck acting as a mechanical lever. The second step is characterized by an increased displacement variance, suggesting a model for how the two heads of myosin V function in processive motion.
Keywords: myosin V; in vitro motility; laser trap; single molecule; mechanics
Introduction
Myosin V, like all members of the myosin super family of motor proteins, is characterized by a globular motor domain that interacts with actin to generate force and motion—processes that are powered by the hydrolysis of ATP (Mermall et al., 1998; Hodge and Cope, 2000). Although myosin family members share these structural and enzymatic properties, their physiological roles encompass a wide range of cellular motile functions including organelle transport, cell division, and muscle contraction (for review see Baker and Titus, 1998). These specialized functions are likely due to structural and enzymatic variations that exist among the various classes of myosin.
The phenotype of myosin V mutations in yeast, mice, and humans suggests that myosin V may function in organelle transport (Jenkins et al., 1981; Johnston et al., 1991; Hill et al., 1996; Provance et al., 1996; Pastural et al., 1997). To serve in this capacity and assure long range transport along actin filaments without the vesicle diffusing away from its actin track, myosin V molecules must remain attached to actin for a large fraction of their mechanochemical cycle (i.e., a high duty cycle). This is necessary, since the low density of myosin V molecules on the vesicle surface allows only a few myosin V molecules to interact simultaneously with the actin filament. Consistent with this prediction, dilution studies indicate that a single tissue-purified myosin V molecule can undergo several mechanochemical cycles (i.e., multiple steps) along an actin filament without diffusing away (Mehta et al., 1999; Rief et al., 2000) and thus has been termed a “processive motor.” In contrast, myosin II, the motor that powers muscle contraction, is characterized by a low duty cycle where myosin II is attached to actin for only a small percentage (i.e., <5%) of its mechanochemical cycle (Uyeda et al., 1990; Harris and Warshaw, 1993). The low duty cycle is compatible with muscle function given that there are hundreds of myosin molecules per thick filament and the sarcomere provides a structure that prevents actin filaments from diffusing away.
Myosin V is believed to produce force and motion in a way analogous to myosin II. The prevailing model for the mechanism by which myosin II displaces actin filaments is based on the atomic structure of chicken skeletal myosin II (Rayment et al., 1993) and numerous biophysical studies (Huxley and Kress, 1985; Thomas et al., 1995; Baker et al., 1998; Irving et al., 2000). In this model, the “neck,” an ∼8.5-nm α-helix that extends from the motor domain, acts as a rigid lever to amplify small movements originating in the head to generate a 5–15-nm working stroke. The lever arm model is supported by indirect measurements of actin filament velocity (Anson et al., 1996; Uyeda et al., 1996) and the direct measurements of single molecule unitary displacements generated by engineered myosin II molecules with necks having different lengths (Warshaw et al., 2000; Ruff et al., 2001).
Myosin V has a particularly long neck that contains six IQ motifs, each IQ motif having the ability to bind either calmodulin or a tissue-specific light chain (Espindola et al., 2000). Compared with myosin II with its two IQs, the long neck of myosin V provides a naturally occurring test of the lever arm model. Based on this model, one would expect that the working stroke of myosin V could be as much as threefold greater than that of conventional myosin II.
In this study, we characterized the duty cycle and working stroke size of a Baculovirus-expressed heavy meromyosin (HMM)*–like fragment of the murine myosin V heavy chain through the use of the in vitro motility assay and optical trap, respectively. Myosin V constructs translocated actin filaments at very low surface densities, which is characteristic of a ≥50% duty cycle. In addition, myosin V supports actin filament motility at ionic strengths >300 mM potassium chloride, which provides evidence that myosin V has high actin affinity.
Single molecule optical trapping studies show predominantly single step displacement events of 20 nm with an occasional displacement event that consists of two consecutive 20-nm steps. These laser trap data support the hypothesis that the longer myosin V neck acts as a lever that is threefold longer than in myosin II, whereas the double step displacements suggest a model for the coordination of myosin V's two heads in processive motion.
The large working stroke, the high affinity for actin, and high duty cycle are properties that provide myosin V the capacity to act as an organelle transporter under conditions where only a small number of myosin V molecules are available to interact with an actin filament.
Results
Purification of the expressed myosin V HMM constructs
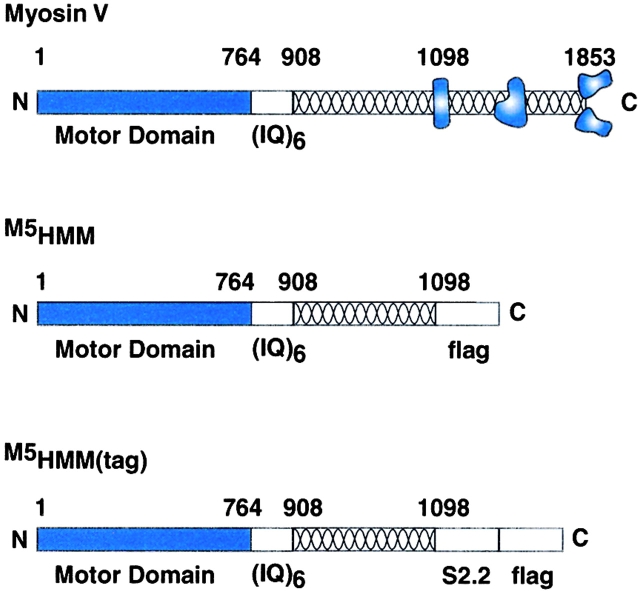
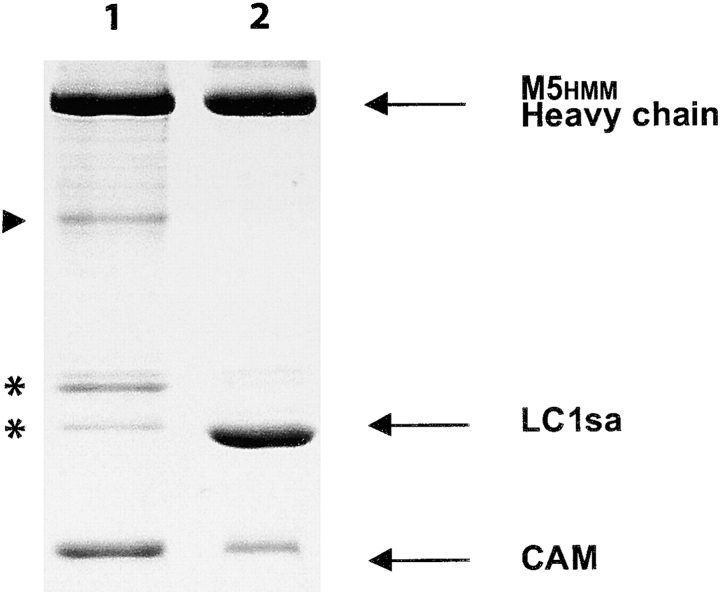
A double-headed HMM-like fragment of the murine myosin V heavy chain was expressed using the Baculovirus/insect cell system (Fig. 1). The heavy chain was coexpressed with calmodulin, which was identified in the purified myosin V HMM (M5HMM) by its characteristic calcium-dependent shift in gel migration rate. This preparation contains three minor bands in the light chain region (molecular weight ∼20–30 kD), which react with anti-FLAG monoclonal antibody by Western blotting, confirming that they are derived from the COOH terminus of the myosin V heavy chain (Fig. 2) and are not essential light chains. The minor band (Fig. 2, arrowhead), which does not react with anti-FLAG antibody, is likely to be the complementary NH2-terminal piece of the heavy chain. In the mechanical assays, exogenous calmodulin was added (see Materials and methods) to the assay solution to assure full occupancy of the six IQ motifs.
Figure 1.
Schematic representation comparing the sequences of the expressed myosin V constructs to wild-type myosin V. We constructed two COOH terminally truncated myosin V constructs, M5HMM and M5HMM(tag), where amino acids 1099–1853 were removed. Note that removal of the residues, which include the tail region of the molecule, results in a M5HMM protein that resembles the proteolytic HMM fragment of vertebrate smooth and skeletal muscle. M5HMM and M5HMM(tag) are identical except that M5HMM(tag) contains the S2.2 epitope for specific attachment to the motility surface (described in Materials and methods).
Figure 2.
SDS-PAGE of the expressed constructs used in this study. (Lane 1) The M5HMM construct expressed with calmodulin only. (Lane 2) The M5HMM construct expressed with calmodulin and LC1sa. Asterisks indicate several minor proteolytic cleavage products that react with anti-FLAG antibody, indicating that they are derived from the COOH terminus of the M5HMM heavy chain. An arrowhead indicates the complementary proteolytic cleavage product that does not react with the anti-FLAG antibody. Note that these cleavages are suppressed in the presence of the tighter binding LC1sa light chain.
An alternative way of assuring full occupancy of the IQ motifs was to coexpress M5HMM with both calmodulin and the nonmuscle essential light chain isoform (LC1sa), which has a higher affinity for the heavy chain than calmodulin (De La Cruz et al., 1999). This construct, although not physiologically relevant (Wang et al., 2000), allowed us to test if the binding of a specific light chain affects the mechanical properties of M5HMM. As expected, the isolated protein predominantly contained LC1sa, confirming that LC1sa has a much higher affinity for the heavy chain than calmodulin (Fig. 2). Note that the tighter binding suppressed the small amount of cleavage in the neck region that was observed when calmodulin was the sole light chain.
In vitro motility of the expressed constructs
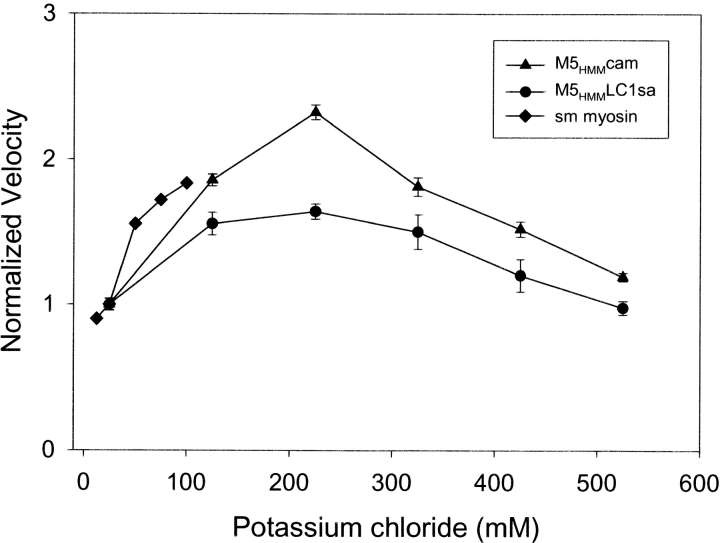
To determine if light chain composition and/or surface attachment of M5HMM is associated with functional alterations, each myosin construct was analyzed for its ability to move actin in the in vitro motility assay over a range of ionic strengths. The results from these experiments (Table I and Fig. 3) show that M5HMM, containing only calmodulin, translocated actin at slightly higher rates than that of M5HMM containing LC1sa. This result was obtained regardless of surface attachment strategy (see Materials and methods; Table I) and at ionic strengths between 25 and 525 mM potassium chloride in the absence of methylcellulose (Fig. 3). The velocity difference is surprising, since both the unitary displacement and displacement duration were independent of light chain content (see below; Table I), suggesting that the slower filament velocity for the M5HMM is not due to a change in the working stroke size or ADP release rate, respectively (see below).
Table I. Summary of in vitro motility and laser trap data.
| Heavy chain construct
|
Light chain composition
|
Velocity
|
Unitary displacement
|
Displacement duration
|
|---|---|---|---|---|
| μm s−1 | nm | ms | ||
| M5HMM | Calmodulin | 0.41 ± 0.06a | 21 ± 2 (6) | 120 ± 12 (6) |
| M5HMM | Calmodulin/LC1sa | 0.30 ± 0.07 | 18 ± 2 (5) | 111 ± 31 (5) |
| M5HMM(tag) | Calmodulin | 0.46 ± 0.04a | 18 ± 2 (5) | 100 ± 13 (14) |
| M5HMM(tag) | Calmodulin/LC1sa | 0.26 ± 0.06 | 24 ± 2 (6) | 138 ± 21 (5) |
p < 0.01 when the effects of light chain composition were compared for a given heavy chain construct.
The in vitro motility values are means ± SD at 25 mM KCl. The unitary displacement values are means ± SEM. The quantities in parentheses represent the number of MV histograms (i.e., data sets) that contribute to the mean. LC1sa refers to the nonmuscle essential light chain.
Figure 3.
The ionic strength dependence of M5 HMM in vitro motility. Normalized filament velocity (mean ±SEM, n > 10 filaments) as a function of added potassium chloride for M5HMM expressed with LC1sa (•) and calmodulin (▴) (M5HMM loading concentration of 10–25 μg/ml). Normalized filament velocity as a function of added potassium chloride for smooth muscle myosin II (♦) from Harris and Warshaw (1992) for comparison (loading concentration of 100 μg/ml). Note the reduction in actin filament velocity at potassium chloride concentrations <125 mM for both M5HMM and myosin II. Also note the plateau at physiological ionic strengths and greater (between 125 mM and 325 mM potassium chloride) for M5HMM. Velocities were normalized to the velocity at 25 mM potassium chloride. A similar dependence of actin filament velocity was observed regardless of light chain content.
The rate of actin filament movement increases with ionic strength up to ∼300 mM potassium chloride. As the ionic strength was increased, the number of filaments moving on the surface was reduced and actin filament motion became more erratic. A similar dependence of actin filament velocity on potassium chloride concentration was observed for single-headed myosin V fragments in the in vitro motility assay (Wang et al., 2000). For comparison, the velocity of actin filaments propelled by smooth muscle myosin II also increases steadily with ionic strength up to ∼100 mM potassium chloride (Fig. 3; Harris and Warshaw, 1992) but required both a 5–10-fold increase in myosin surface density and the presence of methylcellulose. In stark contrast to myosin V, myosin II no longer supports motility >100 mM potassium chloride because actin filaments diffuse away from the coverslip surface.
Duty cycle estimation
All myosin V constructs regardless of light chain content or surface attachment were capable of translocating actin filaments at very low surface densities (1–25 μg/ml) in the absence of methylcellulose, a high molecular weight polymer that restricts diffusion of actin filaments away from the coverslip surface (Uyeda et al., 1990; Warshaw et al., 1990; Homsher et al., 1992). At similar myosin II concentrations, methylcellulose is required for motility (Uyeda et al., 1990; Harris and Warshaw, 1993). These results suggests that myosin V may have a higher duty cycle than myosin II.
To support actin filament motility at such low surface densities, a myosin V molecule must remain bound for a large fraction of its mechanochemical cycle (i.e., have a high duty cycle). We estimated the M5HMM duty cycle (f ) by measuring actin filament velocity as a function of filament length for motility surface loading concentrations of 1 and 5 μg/ml.
Using solution and motility surface NH4-ATPase assays, we then estimated the number of myosin heads (N) available to interact with a given length of actin filament. The NH4-ATPase activity depended on the amount of myosin present with a rate of 1.6 nmol phosphate/min/μg for M5HMM and a rate of 6.6 nmol phosphate/min/μg for smooth muscle myosin II (Harris and Warshaw, 1993). Assuming that the NH4-ATPase activity of myosin attached to the motility surface was the same as when in solution, the amount of myosin on the surface was calculated. Using these surface density estimates and the geometry of the myosin molecule, we then estimated the number of heads (N) available to interact per micrometer of actin filament (see Materials and methods; Table II).
Table II. Summary of surface density estimates.
| Myosin | ATPase rate | Myosin density | Available heads | |
|---|---|---|---|---|
| μg/ml | nM/min | heads/μm2 | heads/μm actin | |
| M5HMM | 1 | 0.03 | 72 | 3.7 |
| M5HMM | 5 | 0.09 | 215 | 11.1 |
| Myosin IIa | 5 | 0.35 | 270 | 7.0 |
| Myosin IIa | 10 | 0.67 | 514 | 13 |
Smooth muscle myosin II data taken from Harris and Warshaw, 1993.
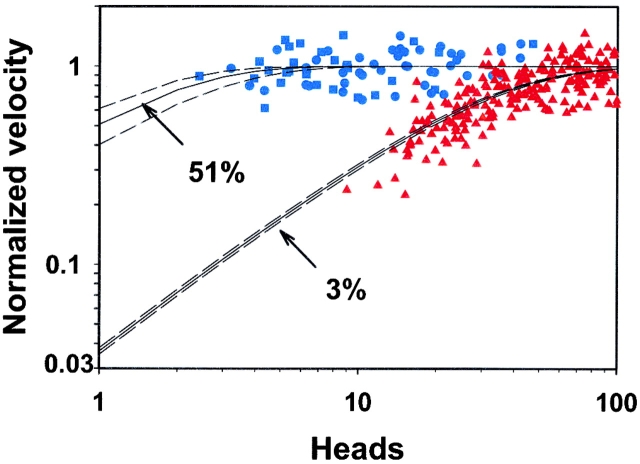
The actin filament velocity was plotted as a function of N (Fig. 4) and the best fit (f ± SE) of the data to Eq. 1 resulted in a 51 ± 10% duty cycle estimate (Fig. 4, top solid line). One characteristic of such a high duty cycle is the observation that maximum actin filament velocity was maintained when only a few heads were available to interact with the actin filament. It is worth noting that the duty cycle may in fact be >50%, but the sensitivity of the estimate is extremely low between 50 and 100% (Fig. 4); therefore, interpretation of these data with regards to processive versus nonprocessive motility should be taken with caution. Published data from smooth muscle myosin II (Harris and Warshaw, 1993) with a 3.4 ± 0.1% duty cycle are plotted for comparison (Fig. 4, bottom solid line) to demonstrate the striking difference between the M5HMM and myosin II duty cycles.
Figure 4.
The dependence of actin filament velocity on the number of interacting M5 HMM heads. Normalized filament velocity as a function the number of M5HMM heads available to interact with each filament at a loading concentration of 5 (▪) and 1 μg/ml (•). Note the independence of M5HMM-driven actin filament velocity on the number of interacting heads. The number of available heads was calculated via surface ATPase assays (described in Materials and methods). For comparison, normalized filament velocity as a function the number of smooth muscle myosin heads available to interact with each filament (▴; data from Harris and Warshaw, 1993) at a loading concentration of 10 μg/ml. (Bottom solid line) A least squares fit of the myosin II data to Eq. 1, revealing a 3% duty cycle for the myosin II data. (Top solid line) A least squares fit of the M5HMM data to Eq. 1, revealing a 51% duty cycle. Dashed lines represent the range of the fit determined by standard error of the estimate.
Optical trapping studies
Unitary events
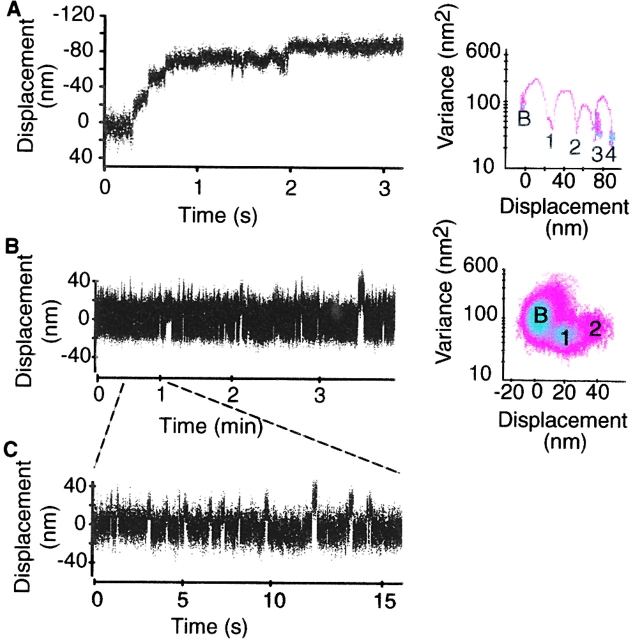
To observe M5HMM displacements at high resolution (<1 nm), we used an optical trapping assay (Dupuis et al., 1997; Guilford et al., 1997). Fig. 5 shows displacement data for M5HMM obtained with a flow cell loading of 1 μg/ml (Fig. 5 A) and 0.1 μg/ml (Fig. 5, B and C). At the higher surface density, displacement traces with frequent staircase events were observed where the actin filament was pulled for several 20-nm step increments (Fig. 5 A). These step increments were discernible as individual displacement event populations offset by ∼20 nm in the mean variance (MV) histogram (Fig. 5 A) and at a lower bead position variance compared with baseline, which is characteristic of a stiffer system because of myosin's attachment to actin. Interestingly, the step size within the staircases appears to decrease with distance from trap center, suggesting that load may influence the size of the working stroke.
Figure 5.
Laser trap data time series of single M5 HMM molecules. (A) Representative time series for M5HMM at a flow cell loading of 1 μg/ml. For clarity, a 3-s portion of the total ∼140-s time series is shown. The corresponding MV histogram for this 3-s time series is shown with the baseline population indicated by a “B.” The event populations are numbered consecutively and are separated from the baseline by an increase in mean displacement and a reduction in system variance. (B) Representative 220-s time series for M5HMM at a flow cell loading of 0.1 μg/ml. The corresponding MV histogram for the entire time series is also shown. Note that the variance of the second event population is between baseline and the first event population. (C) To illustrate event size and duration more clearly, a 15-s portion of the time series in B is also shown.
At comparable flow cell loadings, myosin II molecules generate primarily unitary displacements. However, with increased density (2–5 μg/ml) applied to the coverslip surface, myosin II can also produce staircases with step increments of ∼10 nm (Table III; see Fig. 8 A), which presumably reflect the interaction of multiple myosin molecules with actin (see below). Based on system geometry and the surface ATPase assays, we estimate that at 1 μg/ml there are on average 1.7 M5HMM molecules on the surface of the bead platform that can interact with the actin filament. This surface density coupled to the high duty cycle of M5HMM increases the probability that the staircase events observed at the 1 μg/ml loading (Fig. 5 A) result from the interaction of more than one M5HMM molecule with actin.
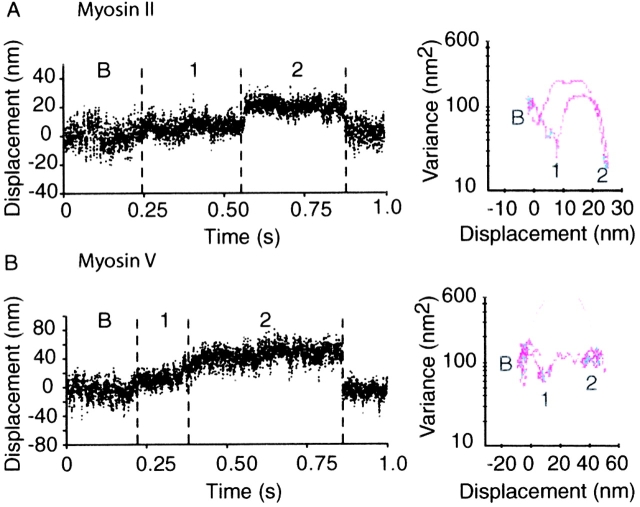
Figure 8.
Double step events. (A) Representative “double step” event for smooth muscle myosin II taken at higher myosin surface densities where presumably more than one molecule is available to interact with the actin filament. Note that the second step is characterized by a similar duration and lower variance than the first step. (B) Representative “double step” event for myosin V, which gives rise to the intermediate variance 37-nm steps (summarized in Table III). Note the higher variance and longer duration of the second step. “B” denotes the baseline population, and event populations are numbered to highlight at the respective mean and variance for each particular population.
To increase the likelihood of sampling single M5HMM molecules, we decreased the M5HMM concentration by an order of magnitude (i.e., 0.17 myosin molecules capable of interacting with the actin filament). Consistent with these experiments being performed in the single molecule regime, we observed myosin activity on <9% of the surface platforms. A sample data trace for M5HMM at 0.1 μg/ml is shown in Fig. 5 B. Staircase events were no longer observed, but rather unitary displacements of 20 nm were seen (Fig. 5 C and Table I). If staircase events were prevalent at this limiting myosin concentration, this could have been interpreted as evidence for M5HMM behaving in a processive manner as reported for tissue-purified myosin V (Mehta et al., 1999; Rief et al., 2000). However, this was not the case for this expressed construct.
The 20-nm step size for M5HMM shown in Fig. 5 is approximately half that reported for tissue-purified chicken myosin V (Mehta et al., 1999; Rief et al., 2000). Here, we address the possibility that this is an underestimate due to (a) the mode of attachment of the construct to the motility surface, (b) the light chain content, and (c) the presence of compliant elements within the experimental setup.
To address the concern that the surface attachment may impede the full mechanical expression of myosin V's working stroke (Anson et al., 1996) or render one head unavailable, we measured unitary displacements from the M5HMM(tag) construct, which was attached to the nitrocellulose surface via an antibody (see Materials and methods). This attachment would elevate the myosin V molecule above the surface and allow unimpeded motion generation. In addition, both the M5HMM and M5HMM(tag) constructs were studied with calmodulin or both calmodulin and LC1sa bound to the neck (see Materials and methods). Table I shows that there were no significant differences in step size regardless of the mode of attachment and light chain composition.
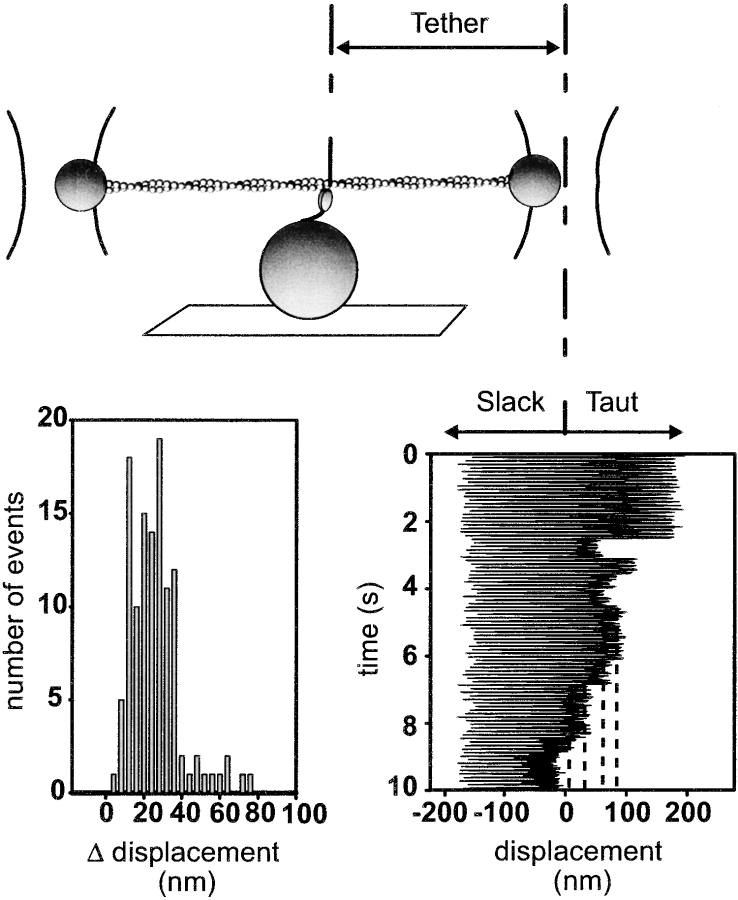
To control for any effect of compliance within the experimental setup on our estimate of the working stroke, we pulled any compliant linkages taught by oscillating one of the trapped beads with a ∼300-nm sinusoidal displacement (Mehta et al., 1999). In these experiments, it was necessary to use sufficient myosin concentrations to assure staircase events (1 μg/ml). Representative data are shown in Fig. 6. In the absence of an attached M5HMM molecule, the bead followed the sinusoidal driving function. Upon attachment of an M5HMM molecule, clipping of the bead oscillation was observed. As other M5HMM molecules bind successively to the actin filament and undergo their working strokes, the tether length becomes progressively shorter, and the difference between the clipped levels was recorded as a measure of the true M5HMM working stroke. These data give a broad step size distribution that centers at 23 nm, thus indicating an ∼15% compliance in our system. This compliance estimate is consistent with our previous estimates (Dupuis et al., 1997; Tyska et al., 1999). In summary, the difference between murine M5HMM and the tissue-purified chicken myosin V step size cannot be accounted for by an underestimate due to surface attachment, light chain content, or system compliance.
Figure 6.
Compliance-corrected step measurements. The experimental configuration for the oscillation experiments (described in Materials and methods). An example trace for M5HMM at 100 μM ATP is also shown (bottom right). Note that when myosin binds the sinusoidal oscillation of the trapped bead is clipped. The difference between clipped levels (dashed lines) provides a measure of the compliance-corrected displacement. The histogram of step increments indicates a displacement of ∼23 nm (bottom left).
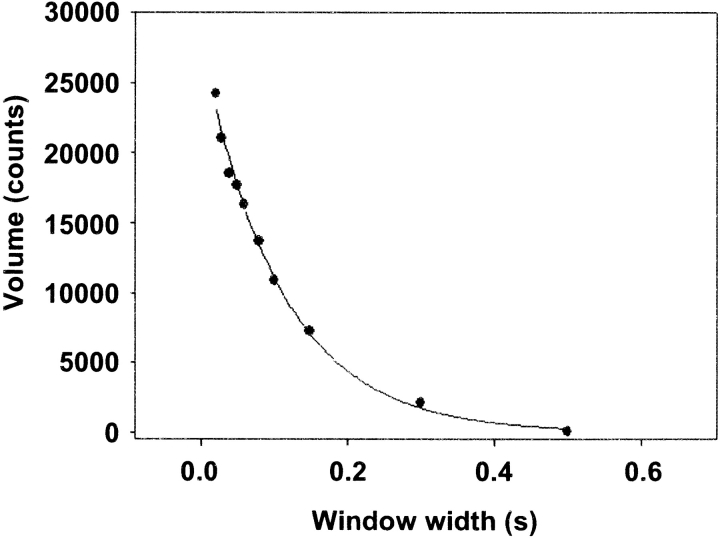
For segments of the data where predominately unitary events were present, we used MV analysis to estimate event durations (see Materials and methods). Fig. 7 shows a representative plot of event volume versus widow width for M5HMM. Because event volume decays exponentially with increasing window width (Fig. 7), it is possible to estimate the average event duration by fitting these data to Eq. 2. The average t on for myosin V at 500 μM ATP was determined for each construct studied and presented in Table I. Values for t on ranged from 100 to 140 ms, independent of surface attachment or light chain content (Table I).
Figure 7.
Determination of event duration. Plot of MV histogram event volumes versus window width for a 127-s M5HMM(tag) displacement time series. A fit of Eq. 2 to the data yields an average event duration of 104 ms.
Double step events
In displacement records obtained at 0.1 μg/ml where presumably only one myosin V molecule can interact with the actin filament, there were occasional events that gave rise to a second displacement population at 40 nm. This second population was also characterized by an increased bead position variance relative to the first step (Fig. 5 B, Fig. 8 B, and Table III). Although MV analysis readily reveals the existence of this second population, determining its relationship to other events requires a closer inspection of the data time series.
Table III.
Summary of double step events b
| Myosin | Mean displacement | Displacement duration | Displacement variance | Normalized variance | |
|---|---|---|---|---|---|
| nm | ms | nm2 | |||
| M5HMM | Step 1 | 19 ± 1 | 213 ± 38 | 55 ± 3 | 0.58 ± 0.02 |
| Step 2 | 18 ± 1 | 687 ± 132a | 64 ± 4a | 0.68 ± 0.03a | |
| Unitary steps | 18 ± 1 | 192 ± 17 | 59 ± 1 | 0.61 ± 0.01 | |
| Step 1 | 12 ± 1 | 468 ± 134 | 26 ± 2 | 0.58 ± 0.04 | |
| Step 2 | 9 ± 2 | 400 ± 95 | 17 ± 1a | 0.38 ± 0.01a |
Values are means ± SEM. M5HMM double steps, n = 59 events; unitary steps, n = 357 events. Normalized variance is the ratio of event variance to baseline variance for that experiment.
Significant differences between step 1 and 2 of each myosin type was determined by Student's t test (p < 0.01).
The myosin II used was a baculovirus-expressed (–)insert isoform of smooth muscle myosin HMM (Lauzon et al., 1998) with experiments done at an ATP concentration of 10 μM (n = 25 events). Displacement amplitude, duration, and variance were calculated from hand-tallied events.
Direct analysis of displacement events from the 33 datasets in this study indicates that double step events are found in ∼50% of the datasets irrespective of light chain content and mode of surface attachment (where a dataset is a record containing on average ≥100 events from a single actin filament over a single surface platform). In addition, the high variance second population at 40 nm was the result of two successive 20-nm steps (Fig. 8 B). In these double step events, the first step yields a displacement of 19 ± 1 nm (mean ± SEM) with an average event duration of 213 ± 38 ms. The displacement event magnitude, duration, and variance associated with the first step are in the same range as the values obtained for individual single step events (Table III). The second step provides an additional 18 nm to yield a second displacement step at 37 ± 1 nm, which was characterized by a significantly increased bead position variance and longer duration (Table III and Fig. 8 B).
Similar double step transitions have been observed for other myosins, although at much higher surface densities than those mentioned above (Lauzon et al., 1998; Tyska et al., 1999). Analysis of double step events from smooth muscle myosin II are also presented in Table III and Fig. 8 A for comparison to the myosin V double steps. In contrast to the myosin V data, the displacement duration of the second step is similar to the duration of the first step. Furthermore, the variance level associated with the second step in the myosin II data is significantly lower than the variance of the first step. The progressive decrease in variance indicates that the second step in myosin II data records is most likely due to additional myosin molecules attaching to the actin filament, leading to increased stiffness and thus lower position variance of the bead-actin-bead system.
Although it is also possible that the M5HMM double step events result from more than one myosin V molecule interacting with actin, the data in Table III indicate that this is unlikely due to the unique high variance signature of the second step of myosin V, which was not the case for myosin II (Table III). For the variance of the second step to increase, the stiffness of any structural elements attached to the actin filament must be reduced. This argues strongly that fewer rather than more myosin heads are attached during the second step compared with the first step.
Discussion
In this study, we determined the in vitro properties of expressed fragments of murine myosin V. The inherent mechanical and biochemical properties of the expressed myosin V are consistent with its proposed role as an organelle transport motor. The ensemble motility experiments indicate that one adaptation appears to be an increase in the fractional time that myosin V remains bound to its actin track during its mechanochemical cycle. This becomes critical when the organelle surface has only a small number of myosin V molecules available to interact with the actin filament (Miller and Sheetz, 2000). In addition, the single myosin molecule mechanical data suggest that the longer neck of myosin V results in a larger unitary displacement as predicted by the lever arm model for myosin motion generation.
Myosin V: a high duty cycle motor
To avoid diffusion away from its actin track, myosin V molecules must have a high duty cycle (i.e., spend most of their cycle time attached to actin). We estimated the myosin V duty cycle to be at a minimum 50%, which is more than an order of magnitude greater than that for muscle myosin II (Harris and Warshaw, 1993; Fig. 3). Is it possible to determine which biochemical and mechanical states within its ATPase cycle give rise to myosin V's high duty cycle?
Regardless of function, all myosins hydrolyze ATP in a cycle that can be broadly divided into weakly and strongly bound states. The kinetics of myosin V are distinct from those of myosin II (De La Cruz et al., 1999; Trybus et al., 1999), and these differences provide a basis for their respective duty cycles. The rate-limiting step for the myosin II actin-activated ATPase cycle is either ATP hydrolysis (White et al., 1997) or phosphate release (Ma and Taylor, 1994); therefore, the predominant state would be bound weakly or detached from actin. In contrast, the rate-limiting step for the myosin V actin-activated ATPase is ADP release (De La Cruz et al., 1999; De La Cruz et al., 2000a,b). Therefore, myosin V would spend the majority of its steady state cycle strongly bound to actin and thus would have a higher duty cycle. The high duty cycle of myosin V may also explain its ability to translocate actin filaments at ionic strengths much higher than physiological levels in the absence of methylcellulose (Fig. 3).
Can the rate of ADP release be determined at the single molecule level? We have previously modeled t on measured in the laser trap as the time waiting for both ADP to be released from, followed by the time waiting for ATP binding to, the active site (Lauzon et al., 1998; Warshaw et al., 1998; Palmiter et al., 1999). Based on biochemically determined second order rate constants, we estimate the time for ATP binding and subsequent myosin V detachment to be 2–4 ms at saturating ATP (De La Cruz et al., 1999; Trybus et al., 1999). Therefore, at saturating ATP t on should be dominated by the time to release ADP from the active site. This is supported by the similarity in our estimates of t on (100–140 ms; Table I) and those calculated from ADP release rates measured in solution (65 –125 ms; De La Cruz et al., 1999; Trybus et al., 1999; Wang et al., 2000). Rief et al. (2000) also reported similar event durations in tissue-purified myosin V at saturating ATP and based on their relationship between event duration and ATP concentration concluded that ADP release must be rate limiting for the observed step durations. Therefore, the laser trap studies provide an important model to study single myosin molecule enzymology.
Myosin V exhibits a large unitary displacement
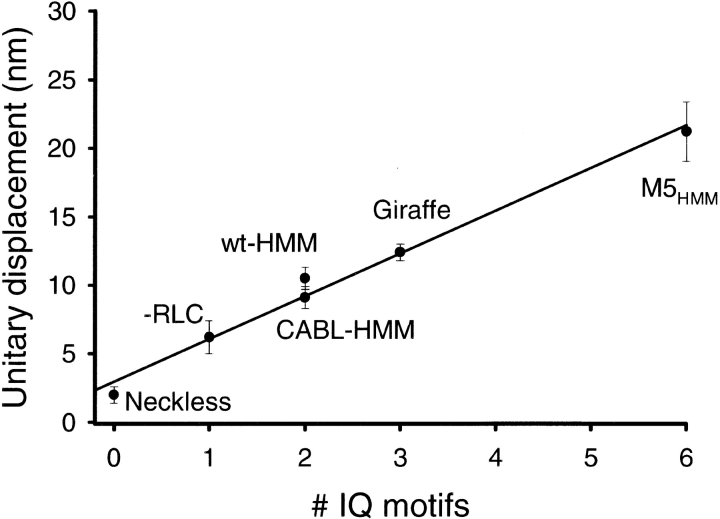
If the myosin neck acts as a lever then myosin V, with a neck that contains six IQ motifs, should generate larger unitary displacements than myosins with shorter necks. In fact, the 20-nm step size for M5HMM obtained from unitary single step events (Fig. 5 B) is greater than any reported step size measurement for myosins with a shorter neck (Finer et al., 1994; Molloy et al., 1995; Guilford et al., 1997; Mehta et al., 1997). If the M5HMM step size reflects the working stroke and originates from a lever arm mechanism as proposed for myosin II, then these data can be related to data obtained in a previous study (Warshaw et al., 2000) where unitary displacements were obtained from mutant smooth muscle myosins with shorter and longer necks created by either deleting or adding light chain binding sites, respectively. In Fig. 9, the previously determined relationship between the unitary displacement amplitude and the number of IQ motifs for smooth muscle myosin (Warshaw et al., 2000) is plotted with the addition of the M5HMM data obtained in this study. The M5HMM data appear to be predicted by the extrapolation of the linear regression through the smooth muscle myosin data. This suggests that under lightly loaded conditions the myosin V neck acts as a rigid lever and the 20-nm step size reflects the working stroke for the M5HMM construct. It is worth noting that the preliminary data of Yanagida and coworkers (Tanaka, H., K. Kitamura, A.H. Iwane, and T. Yanagida. 2000. Biophys. J. 78:A3) challenges the lever arm model by showing that mutant myosins without a neck are capable of generating displacements of the same magnitude produced by the wild- type myosin. Given our previous results (Warshaw et al., 2000) and with the addition of the M5HMM data, we gain further confidence that at least one major role for the neck region is to serve as a lever.
Figure 9.
Unitary step size is a linear function of relative lever arm length. Unitary displacements for smooth muscle myosin II–HMM neck length mutants and M5HMM are plotted as mean ± SEM against the number of IQ motifs. These data were obtained from Warshaw et al. (2000) except for the addition of the M5HMM data, which is the mean of all the M5HMM constructs used in this study (Table I). WT-HMM or CABL-HMM contain two IQ motifs. Zero, one, three, and six IQ motifs were assigned to Neckless, -RLC, Giraffe, and the M5HMM, respectively. The linear regression for the data suggest that the neck region of myosin acts as a rigid lever arm up to 6IQ motifs. Furthermore, the regression does not pass through zero but intersects the y-axis, suggesting that lever may extend into the motor domain.
Correlation between the processive walking stride and working stroke
Recent single molecule data strongly support the ability for full-length tissue-purified myosin V to move processively along an actin filament (Mehta et al., 1999; Rief et al., 2000). When doing so, myosin steps along the actin filament in 40-nm increments. Although the M5HMM construct does not appear to be highly processive, it does serve as a model for the underlying mechanism for processive motion.
As mentioned above, the 20-nm unitary displacement for M5HMM observed in this study is approximately half that reported for tissue-purified myosin V (Mehta et al., 1999; Rief et al., 2000). This is not an underestimate due to compliance within the system (Fig. 6), nonspecific surface attachment, or light chain content (Table I). How this 20-nm displacement relates to the 40-nm step in tissue-purified myosin V may be understood based on electron microscopic images of expressed myosin V binding to actin (Walker et al., 2000). In these images, the two heads of myosin V were observed binding to actin 36 nm apart equal to that of the actin repeat. Furthermore, Walker et al. (2000) showed that the difference in angle between lead and rear heads (∼75°), presumably representing the beginning and end of the working stroke, respectively, would result in a displacement of ∼26 nm, which is similar to the compliance corrected 23-nm unitary step observed here and in preliminary data of Veigel et al. (Veigel C., F. Wang, M.L. Bartoo, J. Hammer, J.R. Sellers, and J.E. Molloy. 2000. Biophys. J. 78:A242 [Abstr.]) In contrast, the staircases with a 40-nm step increment observed for the processive tissue-purified myosin V (Rief et al., 2000) represent the successive walking strides between actin-binding sites.
Although the M5HMM data exhibit primarily unitary displacements of 20 nm, there were double step events in which a second 20-nm step to 40 nm was observed, with the second step characterized by an increased bead position variance (Fig. 8 B and Table III). Since these data are most likely the result of a single myosin molecule (see above), they may represent an aborted attempt by the M5HMM construct to begin processive motion. The higher variance steps are reminiscent of the “half-steps” reported for tissue-purified myosin V at high loads (Rief et al., 2000; Mehta, 2001) and may offer insight to the relationship between the M5HMM working stroke and the processive walking stride observed with the tissue-purified myosin V.
We propose a model for the double step event data observed in this study and the potential explanation for the higher variance second step as illustrated in Fig. 10. In this model, one of M5HMM heads binds to actin and undergoes its working stroke (Fig. 10, states 1 and 2). Rapidly after the initial displacement and within the time resolution of our recording system (∼2 ms), the second head binds to the next available binding site (Fig. 10, state 3). At this time, both heads are attached and account for the low position variance observed for the first step in the double step event. At this point in the cycle, each myosin V head may have ADP in the active site with the leading head negatively strained (Rief et al., 2000; Walker et al., 2000; Mehta, 2001). To assure forward progression, either ADP release from the lead head must be delayed or ADP release from the rear head must be accelerated. Since the dwell period of the first step is comparable to that predicted by the ADP release rate in solution (De La Cruz et al., 1999; Trybus et al., 1999; Wang et al., 2000), ADP release from the lead head is most likely delayed. When the rear head releases ADP, binds ATP, and detaches from actin, intramolecular strain is released, bringing the rear head toward the next actin-binding site (Walker et al., 2000). During processive movement, myosin V would then bind to the next actin-binding site (Fig. 10, states 3–5) thus giving rise to the 40-nm walking stride. However, the expressed M5HMM did not demonstrate processivity. Instead, it stalled during its second step at an intermediate displacement level having high position variance (Fig. 10, state 4).
Figure 10.
The relationship between the double step events and hypothetical states in the walking cycle of a double-headed myosin V molecule. In this model, a myosin V molecule walks from left to right along an actin filament (solid lines). The helical repeat of actin is indicated by tick marks, and dashed lines indicate 20- and 40-nm displacements from the site of initial attachment. After attachment, the myosin V molecule undergoes its working stroke (states 1–2) with the attachment of the second head following very rapidly (state 3). The working stroke and double-headed attachment results in the low variance first step. After the second head's powerstroke, release of the rear head discharges the intramolecular strain, bringing the rear head toward the next actin-binding site. Under normal processive movement, myosin V would then bind to the next actin-binding site (states 3–5), thus giving rise to the 40-nm walking stride. However, the expressed M5HMM did not demonstrate processivity but stalled during its second step at an intermediate displacement level, having high position variance (state 4) resulting from single-headed attachment.
We propose two potential explanations for this high variance intermediate position step. In both cases, one head is strongly bound at all times, whereas the second head either is detached (Fig. 10, state 4) or fluctuating rapidly between attaching to the actin site that is proximal (Fig. 10, state 3) or distal (Fig. 10, state 5) to the strongly bound head. In either case, the high variance results from the reduction in system stiffness due to the effective detachment of one of the M5HMM heads compared with the first step where both heads are attached (Fig. 9, state 3). Given the current data, it is impossible to distinguish between these two possibilities.
The high variance step is also characterized by a longer duration than the first step. Given the load of the trap (∼1.6 pN at the 40-nm displacement), the single attached head may be strained, thus slowing its ADP release rate (Mehta et al., 1999). This strain may be relieved in the processive myosin due to the binding of the lead head to the next available site. Such a strain-dependent ADP release could function to increase the likelihood of the free head binding to the forward site in the event that myosin V organelle transport is prevented by a cellular structure that acts as an obstacle.
A puzzling observation is that the M5HMM did not move processively as did the tissue-purified preparations (Mehta et al., 1999; Rief et al., 2000). It is possible that truncation of the myosin V molecule to form an HMM-like molecule eliminates a region of the molecule that may be necessary to confer processivity. This would be unexpected given that myosin II HMM is a excellent model system for both the mechanics and biochemistry of whole myosin II. However, examples of long range effects within a myosin molecule have been described in other myosin family members (Horowitz and Trybus, 1992), and a full-length myosin V construct will address this possibility.
An alternative explanation for the lack of processivity is that the load that is placed on the myosin molecule in the laser trap places sufficient strain on the molecule to inhibit processivity for this construct. Assays free of external load (Sakamoto et al., 2000) will be used to test this possibility.
Conclusions
Myosin V possesses several structural and enzymatic adaptations, which make it particularly well suited for its physiological role as an organelle transporter. The combination of an elongated neck and two heads allows myosin V to span the actin helix and thus have a walking stride that equals the actin repeat of 36 nm. This walking stride length is greater than the 23-nm working stroke measured in this study that would be associated with the lever action of the neck. In addition, both myosin V's high affinity for actin and its high duty cycle confers upon myosin V the capacity to reliably transport its organelle cargo over long distances with a relatively small number of motor molecules. Further studies with single-headed constructs will test the extent that coordination between heads is necessary for the high duty cycle and whether or not both heads contribute to the working stroke.
Materials and methods
Protein expression
Double-headed myosin V HMM constructs (Fig. 1) were derived from the murine myosin clone (sequence data available from GenBank/EMBL/DDBJ under accession no. X57377; Mercer et al., 1991), which was a gift from Dr. Nancy Jenkins (National Cancer Institute). The construct, referred to as M5HMM, was composed of amino acids M1 to E1098 followed by the FLAG epitope (DYKDDDDK) to facilitate protein purification. A second construct M5HMM(tag) contained an additional epitope tag (S2.2) between E1098 and the FLAG sequence, which allowed us to use monoclonal antibody S2.2 (Trybus and Henry, 1989) to orient the attachment of the construct to the motility surface. The zero heptad zipper construct described in Trybus et al. (1997) was used as a PCR template for insertion of this epitope.
Recombinant virus was generated with linearized BaculoGold DNA (PharMingen) using conventional protocols (O'Reilly et al., 1992). For expression of M5HMM containing only bound calmodulin, Sf9 cells were coinfected with two recombinant viruses: one coding for the myosin V heavy chain and one coding for calmodulin. To investigate the effect of specific light chain binding to M5HMM, some infections contained a third virus coding for LC1sa (sequence data available from GenBank/EMBL/DDBJ under accession no. P14649). M5HMM isolated from this coinfection contained both LC1sa and calmodulin.
Protein purification
Myosin V constructs.
Myosin V constructs were purified as described previously (Trybus et al., 1999) with the following modifications. Purified calmodulin (25 μg/ml) was added to the lysis buffer and the resuspended 70% ammonium sulfate pellet to help maintain complete subunit binding to all of the IQ motifs. Purification on the anti-FLAG affinity column was conducted at 300–400 mM NaCl to maximize the amount of soluble protein. The purified protein was stored at −20°C in buffer containing 50% glycerol.
Actin
Actin was isolated from chicken pectoralis acetone powder by sequential polymerization and depolymerization as described previously (Pardee and Spudich, 1982). Filamentous actin was labeled with tetramethyl rhodamine isothiocyanate phalloidin overnight before experiments to allow F-actin filament visualization using fluorescence microscopy (Warshaw et al., 1990).
NEM myosin
Skeletal myosin was purified from chicken pectoralis (Warshaw et al., 1990). Sulfhydryl modification of skeletal myosin was achieved with N-ethylmaleimide (NEM) to create a strong binding rigor-like myosin (NEM myosin). Briefly, skeletal myosin is reacted with 1 mM NEM for 40 min at 25°C, pH 7.5. The reaction is stopped by the addition of 25 mM DTT (Warshaw et al., 1990). NEM myosin was used to attach actin filaments to polystyrene beads for the laser trap experiments described below.
In vitro motility assay
The in vitro motility of actin over myosin-coated surfaces was performed in low salt actin buffer at saturating ATP (25 mM potassium chloride, 25 mM imidazole, 1 mM EGTA, 4 mM MgCl2, 10 mM DTT, 1 mM ATP, pH 7.4) essentially as described previously (Warshaw et al., 1990) except the actin buffer was supplemented with 100 μg/ml calmodulin and flow cells were loaded with various concentrations of the M5HMM constructs in the presence of 0.5 mg/ml BSA. Monoclonal antibody S2.2 (Trybus and Henry, 1989) was used for attachment of the M5HMM(tag) construct. Alterations in potassium chloride concentration and the addition of methylcellulose are indicated throughout the text where appropriate. Actin filament velocity was determined by software designed to track actin filament images between successive digitized video images (Work and Warshaw, 1992). The average velocity for a given filament was determined from the distance traveled by the filament between 7 and 10 consecutive video images taken at 2–3-s intervals.
Estimating motor density on the motility surface
The density of myosin V molecules bound to the coverslip surface was estimated by comparing the NH4-ATPase of myosin V on the coverslip surface to the NH4-ATPase of known amounts of myosin V in solution (Harris and Warshaw, 1993).
To determine the number of M5HMM heads available to interact with an actin filament, we assumed that a myosin V molecule 23 nm away was capable of reaching out and attaching to the actin filament. Therefore, for any length of an actin filament a 46 nm wide band of myosin V molecules was assumed to be available to interact with the actin filament. The number of molecules within the band was determined by the surface density estimates (Table II).
Estimating myosin V duty cycle
The duty cycle of the M5HMM was estimated by the method of Uyeda et al. (1990). Briefly, the rationale is as follows: actin filaments will be translocated at maximum velocity (V max) if at least one head is interacting with the filament at all times. The probability of this occurring is related to the number of heads available to interact with actin, N, and the fraction of its cycle time that the myosin molecule remains bound, f (i.e., duty cycle). Assuming that the motors act asynchronously, the probability that at least one out of N heads is interacting with actin at all times is given by 1−(1−f)N. Consequently, at low motor surface densities velocity will decrease as a function of f and N according to the equation:
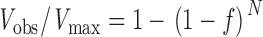 |
(1) |
where V obs is the observed velocity of the actin filament (Uyeda et al., 1990). To estimate the M5HMM duty cycle, we measured actin filament velocity as a function of filament length (i.e., number of heads available to interact with actin; see above) for applied M5HMM concentrations of 1 and 5 μg/ml. Plots of velocity as a function of the number of heads available to interact with a particular filament were then fit by nonlinear regression analysis (SigmaPlot 2000) to Eq. 1 with the duty cycle, f, as the fit parameter.
Laser trap experiments
The laser trap system used in this study has been described previously (Dupuis et al., 1997; Guilford et al., 1997; Lauzon et al., 1998; Palmiter et al., 1999; Tyska et al., 1999). Briefly, flow cells were loaded with various concentrations of the M5HMM constructs in the presence of 0.5 mg/ml BSA and incubated for 2 min. The flow cell was then washed with ATP-free actin buffer to remove any unbound protein. After the washing procedure, 1 mM ATP actin buffer containing 2 nM tetramethyl rhodamine isothiocyanate phalloidin–labeled actin and NEM myosin–coated polystyrenes beads (1 μm NH2 modified; Polysciences) was added. The NEM myosin–coated beads were used to capture an actin filament at each end. After pretensioning to ∼4 pN, the myosin-coated surface was brought into contact with the trapped filament where displacement events were recorded. The single trap stiffness in these experiments was ∼0.02 pN/nm.
For M5HMM(tag), the monoclonal antirod antibody S2.2 was used to ensure a uniform site of attachment to the nitrocellulose surface (Trybus and Henry, 1989; Horowitz and Trybus, 1992). For experiments with M5HMM(tag), the S2.2 antibody was applied at 100 μg/ml. After surface blocking with 0.5 mg/ml BSA, M5HMM(tag) was applied in concentrations ranging from 0.1 to 1 μg/ml in the presence of 0.5 mg/ml BSA and allowed to bind to the antibody for 2 min. The flow cell was then washed with ATP-free actin buffer to remove any unbound protein and treated as described above.
Measurement of event displacement and duration
MV analysis
Myosin V displacement data were analyzed by generating MV histograms, a statistical approach designed originally to analyze single ion channel current records (Patlak, 1993; Guilford et al., 1997). The MV histogram is synthesized by moving a window across the displacement data and calculating a mean and variance at each point. The third dimension in a MV histogram is the number of points at a given mean and variance. MV histograms have well-defined populations associated with regions of data where the mean is relatively constant, whereas the transitions appear as an arch (Fig. 5). Due to the addition of myosin's stiffness upon attachment to the actin filament, the displacement population can also be isolated from the baseline population by its reduction in variance.
We determined the time constant of the displacement events as described by Patlak (1993) and Guilford et al. (1997). Since MV analysis emphasizes events of equal or greater duration than the window width (N), the volume (V mv) in each population is related to the window width (N) used to generate the MV histogram. Assuming that the event durations (t on) are distributed exponentially, the relationship between V mv, N, t on, and the number of events (k) is given by the equation:
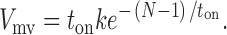 |
(2) |
By plotting V mv versus window width and fitting the data to the above equation, estimates of t on were obtained.
Trap oscillation experiments
The manner in which the actin filament is attached to the beads (Dupuis et al., 1997) and the attachment of the myosin molecule to the motility surface may introduce compliance to the experimental system, which may result in underestimating the myosin-generated displacements. To address this issue and correct for any compliance that is external to the myosin molecule, we attempted to effectively eliminate any compliance by imposing an ∼300 nm peak to peak ∼10-Hz sinusoidal oscillation to one of the trapped beads (Mehta et al., 1999). Compliance-corrected displacement amplitudes were then recorded as the distance between “clipped” levels. Flow cells were prepared as described above except the concentration of ATP was 100 μM and the concentration of M5HMM applied to the flow cell was 1 μg/ml.
Acknowledgments
The authors acknowledge the contributions of the following people: Matt Tyska for collection of the smooth muscle myosin II data; Peter Knight, Josh Baker, and Joe Patlak for helpful discussions; Eric Hayes, Janet Vose, Christine Brosseau, and Guy Kennedy for expert technical assistance; and Steven Work and Don Gaffney for computational assistance. We also thank Yelena Freyzon, Peteranne Joel, and Patricia Fagnant for contributions to this project.
This work was supported by funds from the National Institutes of Health: GM208343 to J.R. Moore, HL38113 to K. Trybus, and AR47906 to K. Trybus and D.M. Warshaw.
Footnotes
Abbreviations used in this paper: HMM, heavy meromyosin; LC1sa, nonmuscle myosin essential light chain isoform; M5HMM, myosin V HMM; MV, mean variance; NEM, N-ethylmaleimide.
References
- Anson, M., M.A. Geeves, S.E. Kurzawa, and D.J. Manstein. 1996. Myosin motors with artificial lever arms. EMBO J. 15:6069–6074. [PMC free article] [PubMed] [Google Scholar]
- Baker, J.E., I. Brust-Mascher, S. Ramachandran, L.E. LaConte, and D.D. Thomas. 1998. A large and distinct rotation of the myosin light chain domain occurs upon muscle contraction. Proc. Natl. Acad. Sci. USA. 95:2944–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, J.P., and M.A. Titus. 1998. Myosins: matching functions with motors. Curr. Opin. Cell Biol. 10:80–86. [DOI] [PubMed] [Google Scholar]
- De La Cruz, E.M., A.L. Wells, S.S. Rosenfeld, E.M. Ostap, and H.L. Sweeney. 1999. The kinetic mechanism of myosin V. Proc. Natl. Acad. Sci. USA. 96:13726–13731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Cruz, E.M., H.L. Sweeney, and E.M. Ostap. 2000. a. ADP inhibition of myosin V ATPase activity. Biophys. J. 79:1524–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Cruz, E.M., A.L. Wells, H.L. Sweeney, and E.M. Ostap. 2000. b. Actin and light chain isoform dependence of myosin V kinetics. Biochemistry. 39:14196–14202. [DOI] [PubMed] [Google Scholar]
- Dupuis, D.E., W.H. Guilford, J. Wu, and D.M. Warshaw. 1997. Actin filament mechanics in the laser trap. J. Muscle Res. Cell Motil. 18:17–30. [DOI] [PubMed] [Google Scholar]
- Espindola, F.S., D.M. Suter, L.B. Partata, T. Cao, J.S. Wolenski, R.E. Cheney, S.M. King, and M.S. Mooseker. 2000. The light chain composition of chicken brain myosin-Va: calmodulin, myosin-II essential light chains, and 8-kDa dynein light chain/PIN. Cell. Motil. Cytoskeleton. 47:269–281. [DOI] [PubMed] [Google Scholar]
- Finer, J.T., R.M. Simmons, and J.A. Spudich. 1994. Single myosin molecule mechanics: piconewton forces and nanometer steps. Nature. 368:113–119. [DOI] [PubMed] [Google Scholar]
- Guilford, W.H., D.E. Dupuis, G. Kennedy, J. Wu, J.B. Patlak, and D.M. Warshaw. 1997. Smooth muscle and skeletal muscle myosins produce similar unitary forces and displacements in the laser trap. Biophys. J. 72:1006–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, D.E., and D.M. Warshaw. 1992. Smooth and skeletal actin are mechanically indistinguishable in the in vitro motility assay. Circ. Res. 72:219–224. [DOI] [PubMed] [Google Scholar]
- Harris, D.E., and D.M. Warshaw. 1993. Smooth and skeletal muscle myosin both exhibit low duty cycles at zero load in vitro. J. Biol. Chem. 268:14764–14768. [PubMed] [Google Scholar]
- Hill, K.L., N.L. Catlett, and L.S. Weisman. 1996. Actin and myosin function in directed vacuole movement during cell division in Saccharomyces cerevisiae. J. Cell Biol. 135:1535–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge, T., and M.J. Cope. 2000. A myosin family tree. J. Cell Sci. 113:3353–3354. [DOI] [PubMed] [Google Scholar]
- Homsher, E., F. Wang, and J.R. Sellers. 1992. Factors affecting movement of F-actin filaments propelled by skeletal muscle heavy meromyosin. Am. J. Physiol. 262:C714–C723. [DOI] [PubMed] [Google Scholar]
- Horowitz, A., and K.M. Trybus. 1992. Inhibition of smooth muscle myosin's activity and assembly by an anti-rod monoclonal antibody. J. Biol. Chem. 267:26091–26096. [PubMed] [Google Scholar]
- Huxley, H.E., and M. Kress. 1985. Crossbridge behavior during muscle contraction. J. Muscle Res. Cell Motil. 6:153–161. [DOI] [PubMed] [Google Scholar]
- Irving, M., G. Piazzesi, L. Lucii, Y.B. Sun, J.J. Harford, I.M. Dobbie, M.A. Ferenczi, M. Reconditi, and V. Lombardi. 2000. Conformation of the myosin motor during force generation in skeletal muscle. Nat. Struct. Biol. 7:482–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins, N.A., N.G. Copeland, B.A. Taylor, and B.K. Lee. 1981. Dilute (d) coat colour mutation of DNA/2J mice is associated with the site of integration of an ecotropic MuLV genome. Nature. 293:370–374. [DOI] [PubMed] [Google Scholar]
- Johnston, G.C., J.A. Prendergast, and R.A. Singer. 1991. The Saccharomyces cerevisiae MYO2 gene encodes an essential myosin for vectorial transport of vesicles. J. Cell Biol. 113:539–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauzon, A.M., M.J. Tyska, A.S. Rovner, Y. Freyzon, D.M. Warshaw, and K.M. Trybus. 1998. A 7-amino acid insert in the heavy chain nucleotide binding loop alters the kinetics of smooth muscle myosin in the laser trap. J. Muscle Res. Cell Motil. 19:825–837. [DOI] [PubMed] [Google Scholar]
- Ma, Y., and E.W. Taylor. 1994. Kinetic mechanism of myofibril ATPase. Biophys. J. 66:1542–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta, A.D. 2001. Myosin learns to walk. J. Cell Sci. 114:1981–1998. [DOI] [PubMed] [Google Scholar]
- Mehta, A.D., J.T. Finer, and J.A. Spudich. 1997. Detection of single-molecule interactions using correlated thermal diffusion. Proc. Natl. Acad. Sci. USA. 94:7927–7931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta, A.D., R.S. Rock, M. Rief, J.A. Spudich, M.S. Mooseker, and R.E. Cheney. 1999. Myosin-V is a processive actin-based motor. Nature. 5:590–593. [DOI] [PubMed] [Google Scholar]
- Mercer, J.A., P.K. Seperack, M.C. Strobel, N.G. Copeland, and N.A. Jenkins. 1991. Novel myosin heavy chain encoded by murine dilute coat colour locus. Nature. 349:709–713. [DOI] [PubMed] [Google Scholar]
- Mermall, V., P.L. Post, and M.S. Mooseker. 1998. Unconventional myosins in cell movement, membrane traffic, and signal transduction. Science. 279:527–533. [DOI] [PubMed] [Google Scholar]
- Miller, K.E., and M.P. Sheetz. 2000. Characterization of myosin V binding to brain vesicles. J. Biol. Chem. 275:2598–2606. [DOI] [PubMed] [Google Scholar]
- Molloy, J.E., J.E. Burns, J. Kendrick-Jones, R.T. Tregear, and D.C. White. 1995. Movement and force produced by a single myosin head. Nature. 378:209–212. [DOI] [PubMed] [Google Scholar]
- O'Reilly, D.R., L.K. Miller, and V.A. Luckow. 1992. Baculovirus Expression Vectors: A Laboratory Manual. L.K. Miller and V.A. Luckow, editors. W.H. Freeman, New York. 347 pp.
- Palmiter, K.A., M.J. Tyska, D.E. Dupuis, N.R. Alpert, and D.M. Warshaw. 1999. Kinetic differences at the single molecule level account for the functional diversity of rabbit cardiac myosin isoforms. J. Physiol. 3:669–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee, J.D., and J.A. Spudich. 1982. Purification of muscle actin. Methods Enzymol. 85:164–181. [DOI] [PubMed] [Google Scholar]
- Pastural, E., F.J. Barrat, R. Dufourcq-Lagelouse, S. Certain, O. Sanal, N. Jabado, R. Seger, C. Griscelli, A. Fischer, and G. de Saint Basile. 1997. Griscelli disease maps to chromosome 15q21 and is associated with mutations in the myosin-Va gene. Nat. Genet. 16:289–292. [DOI] [PubMed] [Google Scholar]
- Patlak, J.B. 1993. Measuring kinetics of complex single ion channel data using mean-variance histograms. Biophys. J. 65:29–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provance, D.W., Jr., M. Wei, V. Ipe, and J.A. Mercer. 1996. Cultured melanocytes from dilute mutant mice exhibit dendritic morphology and altered melanosome distribution. Proc. Natl. Acad. Sci. USA. 93:14554–14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayment, I., H.M. Holden, M. Whittaker, C.B. Yohn, M. Lorenz, K.C. Holmes, and R.A. Milligan. 1993. Structure of the actin-myosin complex and its implications for muscle contraction. Science. 261:58–65. [DOI] [PubMed] [Google Scholar]
- Rief, M., R.S. Rock, A.D. Mehta, M.S. Mooseker, R.E. Cheney, and J.A. Spudich. 2000. Myosin-V stepping kinetics: a molecular model for processivity. Proc. Natl. Acad. Sci. USA. 97:9482–9486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff, C., M. Furch, B. Brenner, D.J. Manstein, and E. Meyhofer. 2001. Single-molecule tracking of myosins with genetically engineered amplifier domains. Nat. Struct. Biol. 8:226–229. [DOI] [PubMed] [Google Scholar]
- Sakamoto, T., I. Amitani, E. Yokota, and T.D. Ando. 2000. Direct observation of processive movement by individual myosin V molecules. Biochem. Biophys. Res. Commun. 272:586–590. [DOI] [PubMed] [Google Scholar]
- Thomas, D.D., S. Ramachandran, O. Roopnarine, D.W. Hayden, and E.M. Ostap. 1995. The mechanism of force generation in myosin: a disorder-to-order transition, coupled to internal structural changes. Biophys. J. 68:135S–141S. [PMC free article] [PubMed] [Google Scholar]
- Trybus, K.M., Y. Freyzon, L.Z. Faust, and H.L. Sweeney. 1997. Spare the rod, spoil the regulation: necessity for a myosin rod. Proc. Natl. Acad. Sci. USA. 94:48–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trybus, K.M., and L. Henry. 1989. Monoclonal antibodies detect and stabilize conformational states of smooth muscle myosin. J. Cell Biol. 109:2879–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trybus, K.M., E. Krementsova, and Y. Freyzon. 1999. Kinetic characterization of a monomeric unconventional myosin V construct. J. Biol. Chem. 274:27448–27456. [DOI] [PubMed] [Google Scholar]
- Tyska, M.J., D.E. Dupuis, W.H. Guilford, J.B. Patlak, G.S. Waller, K.M. Trybus, D.M. Warshaw, and S. Lowey. 1999. Two heads of myosin are better than one for generating force and motion. Proc. Natl. Acad. Sci. USA. 96:4402–4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeda, T.Q., S.J. Kron, and J.A. Spudich. 1990. Myosin step size. Estimation from slow sliding movement of actin over low densities of heavy meromyosin. J. Mol. Biol. 214:699–710. [DOI] [PubMed] [Google Scholar]
- Uyeda, T.Q., P.D. Abramson, and J.A. Spudich. 1996. The neck region of the myosin motor domain behaves as a lever arm to generate movement. Proc. Nat. Acad. Sci. 93:4459–4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, M.L., S.A. Burgess, J.R. Sellers, F. Wang, J.A. Hammer, J. Trinick, and P.J. Knight. 2000. Two-headed binding of a processive myosin to F-actin. Nature. 405:804–807. [DOI] [PubMed] [Google Scholar]
- Wang, F., L. Chen, O. Arcucci, E.V. Harvey, B. Bowers, Y. Xu, J.A. Hammer, and J.R. Sellers. 2000. Effect of ADP and ionic strength on the kinetic and motile properties of recombinant mouse myosin V. J. Biol. Chem. 275:4329–4335. [DOI] [PubMed] [Google Scholar]
- Warshaw, D.M., J.M. Desrosiers, S.S. Work, and K.M. Trybus. 1990. Smooth muscle myosin cross-bridge interactions modulate actin filament sliding velocity in vitro. J. Cell Biol. 111:453–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warshaw, D.M., E. Hayes, D. Gaffney, A.-M., Lauzon, J. Wu, G. Kennedy, K. Trybus, S. Lowey, and C. Berger. 1998. Myosin conformational states determined by single fluorophore polarization. Proc. Natl. Acad. Sci. USA. 95:8034–8039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warshaw, D.M., W.H. Guilford, Y. Freyzon, E. Krementsova, K.A. Palmiter, M.J. Tyska, J.E. Baker, and K.M. Trybus. 2000. The light chain binding domain of expressed smooth muscle heavy meromyosin acts as a mechanical lever. J. Biol. Chem. 275:37167–37172. [DOI] [PubMed] [Google Scholar]
- White, H.D., B. Belknap, and M.R. Webb. 1997. Kinetics of nucleoside triphosphate cleavage and phosphate release steps by associated rabbit skeletal actomyosin, measured using a novel fluorescent probe for phosphate. Biochemistry. 36:11828–11836. [DOI] [PubMed] [Google Scholar]
- Work, S.S., and D.M. Warshaw. 1992. Computer-assisted tracking of actin filament motility. Anal. Biochem. 202:275–285. [DOI] [PubMed] [Google Scholar]