Abstract
The human CC chemokine I-309 is a potent monocyte chemoattractant and inhibits apoptosis in thymic cell lines. Here, we identify a specific human I-309 receptor, and name it CCR8 according to an accepted nomenclature system. The receptor has seven predicted transmembrane domains, is expressed constitutively in monocytes and thymus, and is encoded by a previously reported gene of previously unknown function named, alternatively, CY6, TER1, and CKR-L1. After transfection with the CY6 open reading frame, a mouse pre–B cell line exhibited calcium flux and chemotaxis in response to I-309 (EC50 = 2 nM for each), whereas 20 other chemokines were inactive. Signaling was sensitive to pertussis toxin, suggesting coupling to a Gi-type G protein. These properties parallel those of endogenous I-309 receptors expressed in an HL-60 clone 15 cell line model. The apparent monogamous relationship between I-309 and CCR8 is unusual among known CC chemokines and known CC chemokine receptors. CCR8 may regulate monocyte chemotaxis and thymic cell line apoptosis.
The chemokine superfamily consists of specific leukocyte chemoattractant proteins that can be sorted by structure into four groups, designated C, CX3C, CC, and CXC depending on the number and spacing of conserved cysteines (1–3). The C and CX3C groups each have only one known member, whereas the CC and CXC groups each have many members. CXC chemokines mainly target neutrophils and T cells, and C and CX3C chemokines are specific for T cells. CC chemokines target monocytes, eosinophils, basophils, and T cells with variable selectivity, but, in most cases, they do not target neutrophils.
I-309 is a human CC chemokine first identified by molecular cloning in a search for genes expressed in activated T cell lines (4). Like other CC chemokines, I-309 induces chemotaxis in monocytes (5). Recently, I-309 was purified from CD4+ T cells as a secreted factor that protects murine thymic lymphoma cell lines from dexamethasone-induced apoptosis (6). Other chemokines had little or no anti-apoptotic activity, suggesting a unique signaling pathway.
The first step in chemokine action involves binding to G protein–coupled receptors on the cell surface. Seven CC chemokine receptor subtypes, CCR1-7, have previously been identified by molecular cloning (7–14c). They are all expressed on leukocytes, and together they account for binding sites for most of the known CC chemokines. CCR1, CCR2, CCR3, and CCR5 bind overlapping sets of multiple CC chemokines, whereas only one high affinity ligand has been identified for CCR4, CCR6, and CCR7. None of these receptors has been shown to bind I-309. We have previously reported the genomic DNA and deduced protein sequence of an orphan receptor named CY6, cloned by virtue of its sequence homology to known chemokine receptors (GenBank no. U45983U45983, released April 2, 1996). Subsequently, two groups published the same orphan sequence deduced from genomic clones, and named it TER1 and CKR-L1 (15, 16). Here, we show that CY6/TER1/ CKR-L1 encodes an I-309 receptor.
Materials and Methods
Genomic Cloning and Sequencing.
Genomic DNA from a healthy donor was amplified by PCR using degenerate primers designed from conserved sequences in the predicted third and seventh transmembrane domains of CXCR2 (GenBank no. M73969M73969) and an orphan receptor named 9-6 (GenBank no. U45982U45982). The primer sequences included HincII sites for cloning purposes, and are CC GTC GAC TGC ATI (T/A)(C/G)I GTI GA(C/T) (C/ A)GI TA (primer CY3), and CC GTC GAC AI IGG (A/G)TT IA(A/G) (G/A)CA I(G/C)(A/T) (A/G)TG (primer CY7). The reaction contained 1.3 μg template DNA, 1 μM of each primer, 200 μM dATP, dTTP, dCTP, and dGTP, 10 mM Tris–HCl, pH 8.3, 50 mM KCl, 2.5 mM MgCl2, and 2.5 U of DNA polymerase (Perkin-Elmer Cetus; Norwalk, CT) in 100 μl, and was amplified for 33 cycles (93°C for 1.5 min, 50°C for 2 min, and 72°C for 2 min), then given a final 7-min extension at 72°C. Products were cloned into the HincII site of pUC18 and sequenced (17). A novel sequence named CY6 was identified and used as a probe to isolate a human genomic clone from a λ library (18). A 1.9-kb fragment containing the CY6 ORF was isolated by cutting an EcoRI site in the vector and a genomic XbaI site, subcloned into pUC18, and sequenced completely on both strands. The 5′ end of CY6 RNA was obtained using Clontech Marathon-Ready human thymus cDNA and nested primers from the coding region, named CY6A (CCAGAAGACTGAATACAAACAGGAGGCAA) and CY6B (GTCTGAATAAGTTCCGCATCACAGGGGCTT). The cDNA template was amplified using 10 pmol of CY6A and adaptor primer AP1 (30 cycles of 90°C for 1 min, 60°C for 1min, and 72°C for 2 min). Product from this reaction was reamplified using CY6B and AP2 primers. The 200–250-bp product was gel-purified, digested with NotI and EcoRV (which cuts immediately 5′ of the CY6B primer), cloned into Bluescript, and sequenced.
Mapping.
Fluorescence in situ hybridization (FISH) was carried out as previously described using the CY6 genomic clone as probe (19). Radiation hybrid mapping was performed by PCR using the Stanford G3 panel (Research Genetics, Huntsville, AL) with primers CY6B and CY6 (GCTAGGATTACAGGCATGAGCCACA) to give a 341-bp product.
Creation of Cell Lines Expressing Chemokine Receptors.
The CY6 ORF was first amplified from the 1.9-kb genomic fragment using primers 5′-GCTCTAGATCTGTGACCAGGTCCCGCTGCC (upper strand), which contains an XbaI site (underlined) and nucleotides −4 to −25 relative to the ATG initiator, and 5′-CGG AATTCATATTTAGTCTTCATTGATCCT (lower strand), which contains an XhoI site (underlined) and 21 nucleotides downstream of the stop codon. The PCR product was subcloned into pcDNA3 (Invitrogen, San Diego, CA). Using the same methodology, we created Flag epitope-tagged constructs in pcDNA3 for CCR1, CCR3, and CCR5 using the p4 (GenBank no. L10918L10918), clone 3 (GenBank no. U28694U28694), and 8.5 (GenBank no. U57840U57840) cDNAs, respectively, as templates. The nucleotide sequences were confirmed on both strands. 4DE4 pre-B cell lymphoma cells (gift of L. Staudt, NCI, Bethesda, MD) were grown in RPMI 1640 containing 10% FCS and 50 μM 2-mercaptoethanol. Human embryonic kidney (HEK) 293 cells were grown in DMEM with 10% FCS containing streptomycin 100 μg/ml and penicillin 100 U/ml. 1–1.5 × 107 cells in log phase were electroporated using a GenePulser (Bio-Rad Laboratories, Hercules, CA) with 20 μg of plasmid DNA. HEK 293 cell colonies resistant to 2 g/L G-418 (GIBCO BRL, Gaithersburg, MD) were isolated and expanded in the same media supplemented with 2 g/L G-418. 4DE4 cells were cultured in 1 g/L G-418 and expanded. For CCR1 and CCR3, mixed populations of 4DE4 cells resistant to G-418 were enriched for receptor-expressing cells by chemotaxis in response to appropriate agonists through a ChemoTx chemotactic chamber (Neuroprobe, Inc., Cabin John, MD) with a 5-μm pore size. Clones were obtained by limiting dilution, and receptor expression confirmed by FACS® using the anti-Flag mAb Bio M5 according to the instructions of the manufacturer (Kodak, Rochester, NY).
Cell Culture.
The promyelocytic cell line HL-60 clone 15 (CRL 1964; American Type Culture Collection, Rockville, MD) was maintained and induced to differentiate to eosinophil-like cells by treatment with 0.5 μM butyric acid (Sigma) and 10 ng/ml IL-5 (R&D, Minneapolis, MN), as previously described (20, 21). Human neutrophils and mononuclear cells were purified from the peripheral blood of healthy donors. Mononuclear cells were plated on tissue culture plastic for 18 h and the adherent and nonadherent cells, enriched in monocytes and lymphocytes, respectively, were collected separately for RNA analysis.
RNA Analysis.
Total RNA was prepared using a commercial kit (Stratagene, La Jolla, CA). Blots were prepared and hybridized with 32P-labeled probes as previously described (21).
Intracellular [Ca2+] Measurements.
Cells (107/ml) were incubated in PBS, pH 7.4, and 2.5 μM Fura-2 AM (Molecular Probes, Eugene, OR) for 30–60 min at 37°C in the dark. The cells were subsequently washed twice with HBSS, and resuspended at 1 × 106 cells/ml. 106 cells were stimulated in a total volume of 2 ml in a continuously stirred cuvette at 37°C in a fluorimeter (Photon Technology, Inc., South Brunswick, NJ). Recombinant human chemokine sources: SDF-1β, HCC-1, and I-309, R&D; NAP-2, Bachem (Philadelphia, PA); the BB10010 variant of MIP-1α, a gift from L. Czaplewski (British Biotech, Inc., Oxford, UK); all others, Peprotech (Rocky Hill, NJ). C3a was a gift of C. Hammer. fMLP and recombinant human C5a were from Sigma Chem. Co. (St. Louis, MO). The data were recorded every 200 ms as the relative ratio of fluorescence emitted at 510 nm after sequential excitation at 340 and 380 nm. For some experiments, cells were incubated with 250 ng/ml pertussis toxin, 2 μg/ml cholera toxin, or 2 μM herbimycin A for 4 h before functional assay.
Chemotaxis.
Cells were harvested and washed twice with PBS, then resuspended in serum-free RPMI 1640. Cells were loaded in a total volume of 25 μl into the upper compartment of a microchemotaxis chamber (Neuroprobe, Cabin John, MD). Chemoattractants were loaded in a final volume of 31 μl at indicated concentrations in the lower compartment. The two compartments were separated by a polyvinylpyrollidone-free polycarbonate filter with 5-μm pores. The chemotaxis chamber was incubated at 37°C, 100% humidity, and 5% CO2 for 4 h. The filter was then removed, and the number of cells migrating into each bottom compartment was counted using a hemocytometer. All conditions were tested in triplicate.
Results and Discussion
Cloning of the Gene for CCR8.
Taking advantage of the observation that G protein–coupled receptor genes have conserved sequences and often lack introns in the coding region, we used degenerate PCR primers to amplify a novel human genomic sequence named CY6 related to CC chemokine receptors. A 1,953-bp fragment of a genomic clone containing the CY6 sequence was then isolated and sequenced. It extended to the 5′-end of the phage insert, and contained a 1,065-bp ORF, and 250 and 620 bp of 5′ and 3′ sequence, respectively. The deduced protein sequence is most closely related to CCR1–5 (39–43% identity) with lower identity (25–30%) to CXC chemokine receptors. To confirm the initiation codon, we first identified thymus as a rich natural source of CY6 mRNA and used it to amplify the 5′-UTR sequence by anchored PCR. The sequence revealed 120 bases 5′ of the putative ATG initiator (GenBank no. AF005210) with an in-frame terminator 15 bases 5′ of the ATG and residing at the 3′ end of an upstream exon, strongly supporting this codon as the initiator.
We have mapped the CY6 genomic fragment to human chromosome 3p22–p23 by FISH. 49 cells were examined, with 25 showing paired hybridization signal and 17 showing a single signal at 3p23–p22. Two-point linkage analysis of the radiation hybrid data by the Stanford Radiation Hybrid server gives a LOD of 11.5 for linkage to D3S3527 at a distance of 5.4 cR10000. This places the gene between the Genethon markers D3S1260 and D3S3522, and between the gene for CCR4 and a cluster of genes for CCR1, 2, 3, and 5 (22). Using FISH, Napolitano et al. (15) reported that TER1 maps to chromosome 3p21.
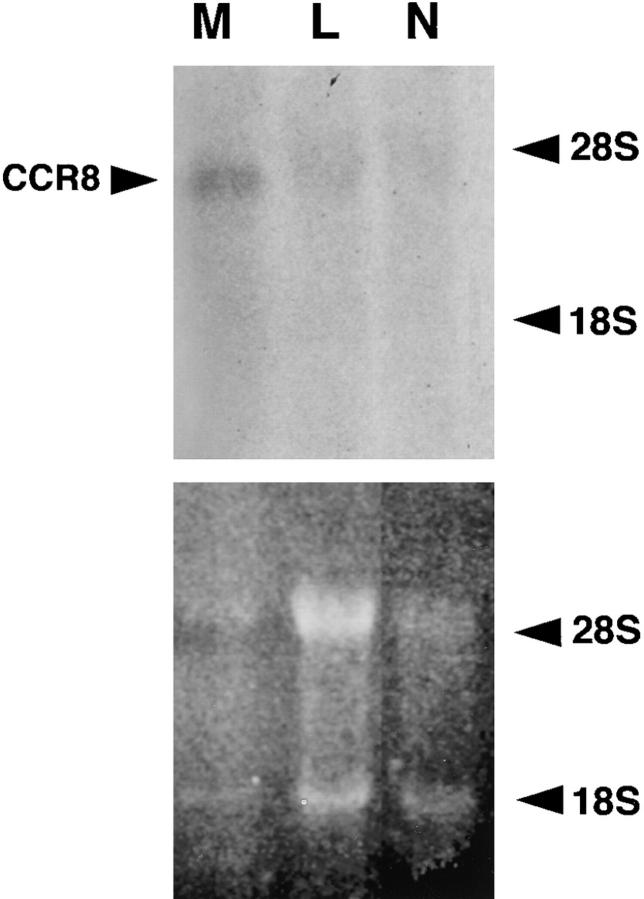
Using a multitissue Northern blot, mRNA was detected at high levels in thymus, and at lower levels in spleen, but not in 14 other tissues tested (data not shown). Using Northern blots from the same supplier, the same RNA distribution pattern has been reported by Napolitano et al. and Zaballos et al. (15, 16). Napolitano et al. (15) also detected transcripts in the MOLT-4 T cell line and the NK3.3 NK cell line, but not in primary NK cells, monocytes, neutrophils, or PHA/PMA-activated PBMCs. We were able to detect a 4.6-kb mRNA band in total RNA from adherent monocytes, consistent with the size in thymus, but not in neutrophil or lymphocyte samples (Fig. 1). Zaballos et al. (16) also detected mRNA in monocyte/macrophages, as well as in lymph node, and CD4+, CD8+, and CD19+ lymphocytes.
Figure 1.
RNA distribution of CCR8. Northern blots containing total RNA from the sources indicated above each lane were hybridized with a CCR8 ORF probe under high stringency conditions. M, monocyte/macrophages (PBMCs that remained adherent to plastic after 18-h overnight culture); L, lymphocytes (nonadherent PBMCs); N, neutrophils. The blots were exposed for 3 d to x-ray film using an intensifying screen.
Because of the functional specificity that we now describe, we have provisionally named the protein product of the CY6/TER1 open reading frame (ORF) CC chemokine receptor 8 or CCR8. This is in keeping with a nomenclature system accepted by consensus at the Second Gordon Conference on Chemotactic Cytokines (1997, Plymouth, NH).
Agonists for CCR8.
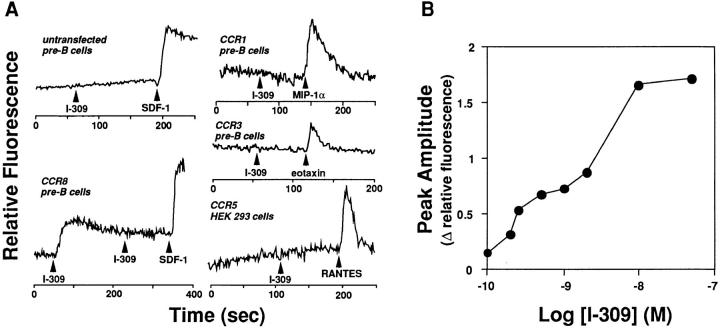
To identify a specific agonist, we screened a panel of chemoattractants for the ability to induce calcium flux in the mouse pre-B cell line 4DE4 before and after transfection with a plasmid encoding CCR8. Untransfected 4DE4 cells did not respond to any agonists tested except for the CXC chemokine SDF-1 (Fig. 2 A). 4DE4 cells transfected with the CCR8 plasmid exhibited [Ca2+]i transients in response to SDF-1 and I-309, but not in response to the following tested at 50 nM or greater: the CC chemokines HCC-1, MIP-1α, RANTES, MIP-1β, MCP-1, MCP-2, MCP-3, MCP-4, and eotaxin; the CXC chemokines IL-8, γIP-10, NAP-2, GRO-α, GRO-β, GRO-γ, and ENA-78; the C chemokine lymphotactin; and the nonchemokine leukocyte chemoattractants fMLP, C3a, and C5a (data not shown). I-309 did not induce calcium flux in 4DE4 cell lines expressing CCR1 or CCR3, or in HEK 293 cells stably expressing CCR5 (Fig. 2 A; data not shown). The CCR1, CCR3, and CCR5 cell lines all responded appropriately to previously described agonists (Fig. 2 A; data not shown).
Figure 2.
I-309 is an agonist for CCR8. (A) Receptor specificity and homologous densensitization. [Ca2+]i was monitored by ratio fluorescence of Fura-2–loaded pre–B cells or HEK 293 cells stably transfected with plasmids encoding CC chemokine receptors as indicated adjacent to each tracing. Cells were stimulated with chemokines 50 nM at the times indicated by arrowheads. Data are representative of at least three experiments with CCR8-expressing cells. (B) Potency. The amplitude of the peak of the calcium transient elicited by the indicated concentration of I-309 in CCR8 transfectants is shown. Data are representative of two separate experiments.
The threshold for the calcium flux response of CCR8-expressing cells to I-309 was 0.1 nM, and the EC50 was 2 nM (Fig. 2 B). These values are similar to those observed for other chemokine receptors (7–14). When the cells were pretreated with other ineffective chemokines or with SDF-1, there was no effect on the magnitude or kinetics of the I-309–induced calcium flux response, suggesting that other chemokines are not antagonists at CCR8 (data not shown). In contrast, when cells were sequentially stimulated with the same concentration of I-309, no response to the second application was observed (Fig. 2 A), suggesting homologous desensitization of the receptor.
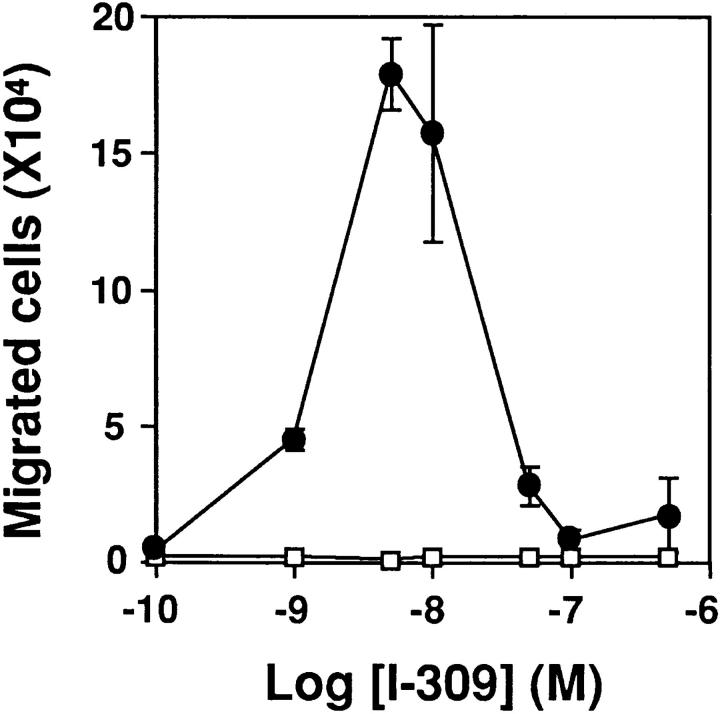
I-309 was able to induce transmigration of 4DE4 cells expressing CCR8, but not untransfected cells, across a filter in a modified Boyden chamber assay of chemotaxis (Fig. 3). The I-309 dose–response curve for chemotaxis was bell-shaped, which is typical for this response. I-309 was both highly potent (EC50 of 2 nM and an optimal concentration of 5 nM) and highly efficacious (∼40% of input cells migrated across the filter at the optimal concentration). Checkerboard analysis indicated that the response was chemotactic and not chemokinetic (data not shown).
Figure 3.
CCR8 is a chemotactic receptor. Untransfected pre-B cells (open squares) and cells stably expressing CCR8 (closed circles) were incubated in a microchemotaxis chamber and tested with the indicated concentrations of I-309. The number of input cells was 350,000/well. Data are the mean ± SEM of triplicate determinations, and are from a single experiment representative of two separate experiments. Checkerboard analysis indicated that the activity was chemotactic, not chemokinetic (data not shown).
Development of an HL-60 Cell Line Model for Endogenous CCR8.
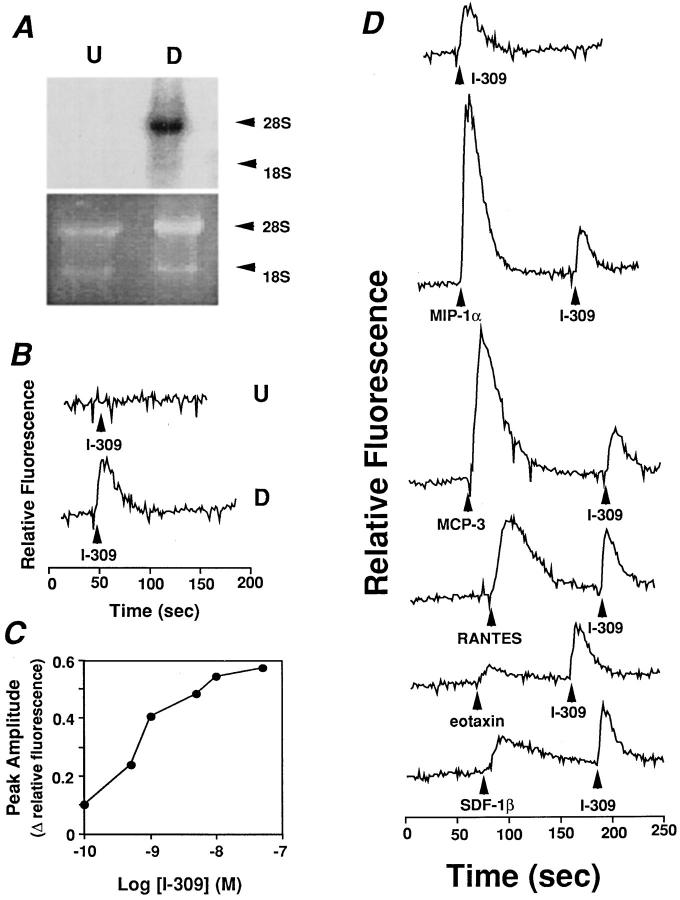
The clone 15 variant of HL-60 cells can be induced by butyric acid and IL-5 treatment to differentiate within 2 d to cells having many of the characteristics of peripheral blood eosinophils, including expression of eosinophil-specific granule proteins (20, 21). Using Northern blot analysis, we were unable to detect mRNA for CCR8 in the uninduced cells, and the cells did not respond to I-309 in either the calcium flux or chemotaxis assays (Fig. 4, A–C). However, when the cells were cultured in the presence of butyric acid and IL-5, a 4.6-kb band was detected by Northern blot using a CCR8 ORF probe (Fig. 4 A).
Figure 4.
An HL-60 clone 15 cell line model of endogenous CCR8 expression and function. HL-60 clone 15 cells were cultured for 6 d in the presence of butyric acid 0.5 μM and for the final 4 d in the presence of IL-5 10 ng/ml, which induces differentiation to an eosinophilic phenotype. (A) CCR8 mRNA expression. A Northern blot containing 10 μg total RNA from undifferentiated (U) and differentiated (D) cells was hybridized with a CCR8 ORF probe (top) and washed under high stringency conditions. The blot was then exposed to x-ray film using an intensifying screen for 20 h. The corresponding region of the ethidium bromide– stained gel is shown in the lower panel. (B) Calcium flux response to I-309. Fura-2–loaded undifferentiated (top tracing, U) and differentiated (lower tracing, D) cells were stimulated with I-309 50 nM. (C) Potency of I-309 for calcium flux. Data are from a single experiment representative of three separate experiments. (D) Distinct receptor usage by I-309 and other CC chemokines. Differentiated cells were stimulated with the indicated chemokines 50 nM and Fura-2 fluoresence was monitored.
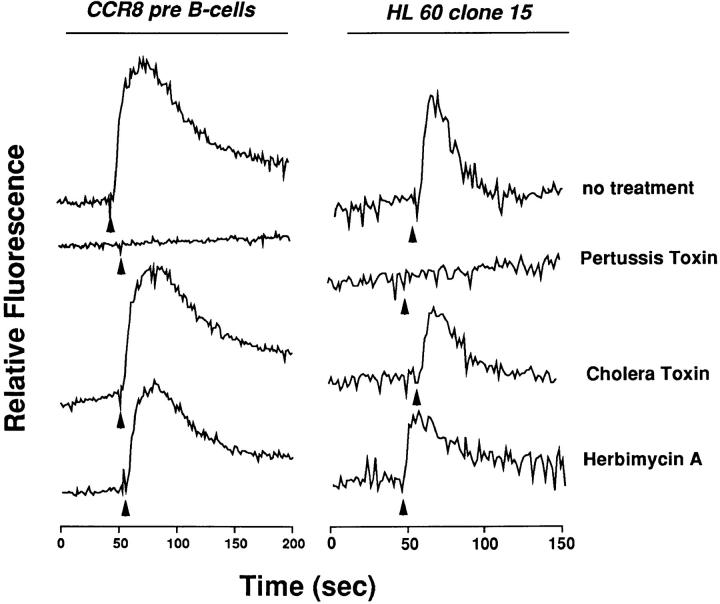
Induction of CCR8 mRNA correlated with acquisition of calcium flux responses to I-309 (Fig. 4 B). The EC50 was 1 nM (Fig. 4 C), similar to the value observed for I-309– induced calcium flux and chemotaxis in 4DE4 cells expressing recombinant CCR8. Induced cells also responded to MIP-1α, RANTES, MCP-3, and eotaxin, consistent with the induction of CCR1 and CCR3 mRNA (Tiffany, H.L., and P.M. Murphy, unpublished observations), but none of these chemokines was able to desensitize I-309– induced calcium flux, consistent with their lack of agonist and antagonist activity at CCR8 (Fig. 4 D). I-309–induced calcium flux in both differentiated HL-60 clone 15 cells and 4DE4 cells expressing CCR8 was abolished by treatment with pertussis toxin, but not by cholera toxin or herbimycin A, suggesting specific coupling of the receptor to G proteins of the Gi class in both cell types (Fig. 5). Although the HL-60 clone 15 cells are a useful model system for studying endogenous CCR8, it is important to point out that we have not been able to demonstrate CCR8 mRNA or I-309 responsiveness in primary human eosinophils, even when stimulated with IL-5.
Figure 5.
CCR8 couples to a Gi-type G protein. [Ca2+]i was measured as the relative fluorescence emitted by Fura-2–loaded pre–B cells stably transfected with CCR8 or HL-60 clone 15 cells differentiated with butyric acid and IL-5 after 4-h treatment with the inhibitors indicated to the right of each tracing. I-309 50 nM was added at the time indicated by the arrow. The results are from a single experiment representative of three separate experiments.
Identification of CCR8 is an essential first step in understanding the mechanism of action of I-309. The ability of recombinant CCR8 to support chemotaxis in transfected pre–B cells suggests that endogenous CCR8 may mediate the chemotactic activity of I-309 in monocytes. The pattern of constitutive CCR8 mRNA expression in tissues that we and others (15, 16) have observed is unique relative to known chemokine receptors, and suggests a role for CCR8 specifically in thymus. This is consistent with the ability of I-309 to inhibit dexamethasone-induced apoptosis in mouse thymic cell lines (6). Together, these observations suggest the importance of future studies to test the role of I-309 in thymocyte migration and development in vivo. This may be accomplished by targeted genetic disruption of mouse CCR8, which has not yet been identified, or of TCA3, which appears to be the mouse homologue of I-309. Like I-309, TCA3 induces monocyte chemotaxis. It has also been shown to induce degranulation, production of reactive nitrogen intermediates, and upregulation of adhesion molecules in monocytes, but, unlike I-309, has been reported to have parallel activities on neutrophils. TCA3 can also suppress the growth of certain tumors in both immunocompetent and immunodeficient mice (23–25).
Like other chemokines, I-309 is likely to provide directional information for orderly leukocyte trafficking in vivo (26). Like other chemokines, if it is dysregulated, I-309 has the potential to cause inappropriate inflammation and tissue injury. In this regard, our identification of an I-309 receptor may be useful in future research aimed at evaluating this pathway for development of potential anti-inflammatory therapies.
Footnotes
M. Locati is a recipient of a fellowship from the Italian Association for Cancer Research.
Address corespondence to Philip M. Murphy, M.D., Building 10, Room 11N113, National Institutes of Health, Bethesda, MD 20892. Phone: 301-496-2877; FAX: 301-402-4369; E-mail: pmm@nih.gov; or Tom I. Bonner, Ph.D. Building 36, Room 7A07, National Institutes of Health, Bethesda, MD 20892. Phone: 301-496-8906; FAX: 301-402-1748; E-mail: tibonner@helix.nih.gov
References
- 1.Baggiolini M, Dewald B, Moser B. Interleukin-8 and related chemotactic cytokines—CXC and CC chemokines. Adv Immunol. 1994;55:97–179. [PubMed] [Google Scholar]
- 2.Bazan JF, Bacon KB, Hardiman G, Wang W, Schall TJ. A new class of membrane-bound chemokine with a CX3C motif. Nature (Lond) 1997;385:640–644. doi: 10.1038/385640a0. [DOI] [PubMed] [Google Scholar]
- 3.Kelner GS, Kennedy J, Bacon KB, Kleyensteuber S, Largaespada DA, Jenkins NA, Copeland NG, Bazan JF, Moore KW, Schall TJ, Zlotnick A. Lymphotactin: a cytokine that represents a new class of chemokine. Science (Wash DC) 1994;266:1395–1398. doi: 10.1126/science.7973732. [DOI] [PubMed] [Google Scholar]
- 4.Miller MD, Wilson SD, Dorf ME, Seuanez HN, O'Brien SJ, Krangel MS. Sequence and chromosomal location of the I-309 gene. Relationship to genes encoding a family of inflammatory cytokines. J Immunol. 1990;145:2737–2744. [PubMed] [Google Scholar]
- 5.Miller MD, Krangel MS. The human cytokine I-309 is a monocyte chemoattractant. Proc Natl Acad Sci USA. 1992;89:2950–2954. doi: 10.1073/pnas.89.7.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Snick J, Houssiau F, Proost P, Van Damme J, Renauld J-C. I-309/T cell activation gene-3 chemokine protects murine T cell lymphomas against dexamethasone-induced apoptosis. J Immunol. 1996;157:2570–2576. [PubMed] [Google Scholar]
- 7.Neote K, DiGregorio D, Mak JY, Horuk R, Schall TJ. Molecular cloning, functional expression, and signaling characteristics of a C–C chemokine receptor. Cell. 1993;72:415–425. doi: 10.1016/0092-8674(93)90118-a. [DOI] [PubMed] [Google Scholar]
- 8.Gao J-L, Kuhns DB, Tiffany HL, McDermott D, Li X, Francke U, Murphy PM. Structure and functional expression of the human macrophage inflammatory protein-1α/RANTES receptor. J Exp Med. 1993;177:1421–1427. doi: 10.1084/jem.177.5.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charo I, Myers SJ, Herman A, Franci C, Connolly AJ, Coughlin SR. Molecular cloning and functional expression of two monocyte chemoattractant protein 1 receptors reveals alternative splicing of the carboxyl-terminal tails. Proc Natl Acad Sci USA. 1994;91:2752–2756. doi: 10.1073/pnas.91.7.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kitaura M, Nakajima T, Imai T, Harada S, Combadiere C, Tiffany HL, Murphy PM, Yoshie O. Molecular cloning of human eotaxin, an eosinophil-selective CC chemokine, and identification of a specific eosinophil eotaxin receptor, CC chemokine receptor 3. J Biol Chem. 1996;271:7725–7730. doi: 10.1074/jbc.271.13.7725. [DOI] [PubMed] [Google Scholar]
- 11.Ponath P, Qin S, Post TW, Wang J, Wu L, Gerard NP, Newman W, Gerard C, Mackay CR. Molecular cloning and characterization of a human eotaxin receptor expressed selectively on eosinophils. J Exp Med. 1996;183:2437–2448. doi: 10.1084/jem.183.6.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Power CA, Meyer A, Nemeth K, Bacon KB, Hoogewerf AJ, Proudfoot AEI, Wells TNC. Molecular cloning and functional expression of a novel CC chemokine receptor cDNA from a human basophilic cell line. J Biol Chem. 1995;270:19495–19500. doi: 10.1074/jbc.270.33.19495. [DOI] [PubMed] [Google Scholar]
- 13.Samson M, Labbe O, Mollereau C, Vassart G, Parmentier M. Molecular cloning and functional expression of a new human CC–chemokine receptor gene. Biochemistry. 1996;35:3362–3367. doi: 10.1021/bi952950g. [DOI] [PubMed] [Google Scholar]
- 14.Combadiere C, Ahuja SK, Tiffany HL, Murphy PM. Cloning and functional expression of CC CKR5, a human monocyte CC chemokine receptor selective for MIP-1α, MIP-1β, and RANTES. J Leukocyte Biol. 1996;60:147–152. doi: 10.1002/jlb.60.1.147. [DOI] [PubMed] [Google Scholar]
- 14a.Imai T, Baba M, Nishimura M, Kakizaki M, Takagi S, Yoshie O. The T cell-directed CC chemokine TARC is a highly specific biological ligand for CC chemokine receptor 4. J Biol Chem. 1997;272:15036–15042. doi: 10.1074/jbc.272.23.15036. [DOI] [PubMed] [Google Scholar]
- 14b.Baba M, Imai T, Nishimura M, Kakizaki M, Takagi S, Hieshima K, Nomiyama H, Yoshie O. Identification of CCR6, the specific receptor for a novel lymphocyte-directed CC chemokine LARC. J Biol Chem. 1997;272:14893–14898. doi: 10.1074/jbc.272.23.14893. [DOI] [PubMed] [Google Scholar]
- 14c.Yoshida R, Imai T, Hieshima K, Kusuda J, Baba M, Kitaura M, Nishimura M, Kakizaki M, Nomiyama H, Yoshie O. Molecular cloning of a novel human CC chemokine EBI1-ligand chemokine that is a specific functional ligand for EBI1, CCR7. J Biol Chem. 1997;272:13803–13809. doi: 10.1074/jbc.272.21.13803. [DOI] [PubMed] [Google Scholar]
- 15.Napolitano M, Zingoni A, Bernardini G, Spinetti G, Nista A, Storlazzi CT, Rocchi M, Santoni A. Molecular cloning of TER1, a chemokine receptor-like gene expressed by lymphoid tissues. J Immunol. 1996;157:2759–2763. [PubMed] [Google Scholar]
- 16.Zaballos A, Varona R, Gutierrez J, Lind P, Marquez G. Molecular cloning and RNA expression of two new human chemokine receptor-like genes. Biochem Biophys Res Commun. 1996;227:846–853. doi: 10.1006/bbrc.1996.1595. [DOI] [PubMed] [Google Scholar]
- 17.Song Z-H, Young WS, Brownstein MJ, Bonner TI. Molecular cloning of a novel candidate G protein– coupled receptor from rat brain. FEBS Lett. 1994;351:375–379. doi: 10.1016/0014-5793(94)00888-4. [DOI] [PubMed] [Google Scholar]
- 18.Lawn RM, Fritsch EF, Parker RC, Blake G, Maniatis T. The isolation and characterization of linked delta- and beta-globin genes from a cloned library of human DNA. Cell. 1978;15:1157–1174. doi: 10.1016/0092-8674(78)90043-0. [DOI] [PubMed] [Google Scholar]
- 19.Tory K, Latif F, Modi W, Schmidt L, Wei MH, Li H, Cobler P, Prcutt ML, Delisio J, Geil L, et al. A genetic linkage map of 96 loci on the short arm of human chromosome 3. Genomics. 1992;13:275–286. doi: 10.1016/0888-7543(92)90243-l. [DOI] [PubMed] [Google Scholar]
- 20.Fischkoff SA. Graded increase in probability of eosinophilic differentiation of HL-60 promyelocytic leukemia cells induced by culture under alkaline conditions. Leuk Res. 1988;12:679–686. doi: 10.1016/0145-2126(88)90103-8. [DOI] [PubMed] [Google Scholar]
- 21.Tiffany HL, Li F, Rosenberg HF. Hyperglycosylation of eosinophil ribonucleases in a promyelocytic leukemia cell line and in differentiated peripheral blood progenitor cells. J Leukocyte Biol. 1995;58:49–54. doi: 10.1002/jlb.58.1.49. [DOI] [PubMed] [Google Scholar]
- 22.Samson M, Soularue P, Vassart G, Parmentier M. The genes encoding the human CC-chemokine receptors CC-CKR1 to CC-CKR5 (CMKBR1–CMKBR5) are clustered in the p21.3–p24 region of chromosome 3. Genomics. 1996;36:522–526. doi: 10.1006/geno.1996.0498. [DOI] [PubMed] [Google Scholar]
- 23.Luo Y, Laning J, Devi S, Mak J, Dorf M. Biologic activities of the murine beta-chemokine TCA3. J Immunol. 1994;153:4616–4624. [PubMed] [Google Scholar]
- 24.Devi S, Laning J, Luo Y, Dorf ME. Biologic activities of the beta-chemokine TCA3 on neutrophils and macrophages. J Immunol. 1995;154:5376–5383. [PubMed] [Google Scholar]
- 25.Laning J, Kawasaki H, Tanaka E, Luo Y, Dorf ME. Inhibition of in vivo tumor growth by the beta chemokine, TCA3. J Immunol. 1994;153:4625–4635. [PubMed] [Google Scholar]
- 26.Springer TA. Traffic signals for lymphocyte recirculation and leukocyte emigration: the multistep paradigm. Cell. 1994;76:301–314. doi: 10.1016/0092-8674(94)90337-9. [DOI] [PubMed] [Google Scholar]