Abstract
The commitment, differentiation, and expansion of mainstream α/β T cells during ontogeny depend on the highly controlled interplay of signals relayed by cytokines through their receptors on progenitor cells. The role of cytokines in the development of natural killer (NK)1+ natural T cells is less clearly understood. In an approach to define the role of cytokines in the commitment, differentiation, and expansion of NK1+ T cells, their development was studied in common cytokine receptor γ chain (γc) and interleukin (IL)-7 receptor α (IL-7Rα)–deficient mice. These mutations block mainstream α/β T cell ontogeny at an early prethymocyte stage. Natural T cells do not develop in γc-deficient mice; they are absent in the thymus and peripheral lymphoid organs such as the liver and the spleen. In contrast, NK1+ T cells develop in IL-7Rα–deficient mice in the thymus, and they are present in the liver and in the spleen. However, the absolute number of NK1+ T cells in the thymus of IL-7Rα–deficient mice is reduced to ∼10%, compared to natural T cell number in the wild-type thymus. Additional data revealed that NK1+ T cell ontogeny is not impaired in IL-2– or IL-4–deficient mice, suggesting that neither IL-2, IL-4, nor IL-7 are required for their development. From these data, we conclude that commitment and/or differentiation to the NK1+ natural T cell lineage requires signal transduction through the γc, and once committed, their expansion requires signals relayed through the IL-7Rα.
Natural T (NT) cells are a distinct lineage of lymphocytes whose function is thought to be immunoregulatory in nature because they secrete a wide variety of cytokines, most notably IL-4, upon activation. They express both natural killer (NKR-P1) and T (α/β and γ/δ TCR) cell markers. In mice, they are present in the liver, bone marrow, spleen, lymph node, and postnatal thymus (for review see reference 1). The development of CD4+8− or CD4−8− NT cells depends on the expression of nonclassical antigen presenting molecules such as CD1d1 (2–4) and possibly H-2TL (5). They express highly conserved α/β TCR; a large majority of them express Vα14Jα281 (85%) paired with Vβ8.2 (up to 70%) (6, 7). A similar subset of T cells also exists among the human peripheral blood CD4−8− lymphocytes (8, 9). Hence, NT cells are predicted to serve an evolutionarily conserved function. Although this function of NT cells remains elusive, their selective absence in autoimmune prone mice, such as MRL-lpr/lpr, B6-lpr/lpr, C3H-gld/gld, and BWF1 (10, 11) as well as nonobese diabetic (NOD; 12), suggests a role in the control of the disease. Moreover, the ability to delay or prevent the onset of disease either by the adoptive transfer of NKR-P1+ splenocytes (10) that includes both natural killer and NT cells, or by the administration of recombinant IL-4 (13), underscore the importance of NT cells in the physiology of normal immune responses.
Here we report that: (a) commitment and/or differentiation to NT cell lineage requires signaling through the common cytokine receptor γ chain (γc) because γc0/0 mice do not develop NKR-P1+ T cells in the thymus or in the peripheral lymphoid organs, (b) the expansion of the committed NT cells requires signals relayed through the IL-7Rα chain because although IL-7Rα0/0 mice develop NKR-P1+ cells, they are dramatically reduced in numbers compared to the wild type, and (c), signal(s) transduced through γc is not mediated by IL-2, IL-4, or IL-7 because neither IL-20/0, IL-40/0, nor IL-7Rα0/0 mutations affect NT cell development.
Materials and Methods
Mice.
B6.IL-20/0 and IL-7Rα0/0 mice were purchased from the Jackson Laboratory (Bar Harbor, ME); B6.IL-20/0 were provided by D. Serreze (The Jackson Laboratory). R. Morawetz (National Institute of Allergy and Infectious Diseases, Bethesda, MD) provided B6.IL40/0 (14) and D. Roopenian (The Jackson Laboratory) provided B6.β2m0/0 and B6.IL-40/0 mice. B6.γc0/Y and B6.IL-7Rα0/0 mice were bred and genotyped at the National Heart, Lung and Blood Institute (Bethesda, MD; 15) and Immunex (Seattle, WA; 16), respectively.
Mononuclear Cell Preparation.
Mononuclear cells (MNC) from thymus and spleen were prepared by standard techniques. Intrahepatic MNC were prepared from minced liver lobes pressed through a wire mesh. The cells were suspended in 50 ml of RPMI-1640 (GIBCO BRL, Gaithersburg, MD) supplemented with 5% FCS (Hyclone Labs., Logan, UT) and allowed to settle for ∼10 min on ice. Liver MNC were separated from the hepatocytes by density centifugation over lympholyte M (Cederlane Labs. Ltd., Hornby, Canada). Cells at the interface were collected and washed with RPMI supplemented with 5% FCS.
Flow Cytometry.
Thymocytes (1–2 × 107) of individual animals 6 wk or older were stained for three-color flow cytometric analyses, as described previously (5), with anti–heat stable antigen (HSA)-PE (M1/69), anti–CD8α-PE (53-6.7), anti–CD44-FITC (IM7), and anti–NKR-P1 (NK1.1)-biotin (PK136), anti–TCR-β-biotin (H57-597), or anti–Vβ8.1,8.2-biotin (MR5-2). Thymocytes were also stained with anti–Ly6C-FITC (AL-21). When staining with anti–IL-2Rβ-PE (TM-β1), anti–HSA-FITC, and anti–CD8α-FITC were used to electronically gate in the HSAlow CD8low population. The biotinylated antibody was stained with streptavidin-RED670 (GIBCO BRL). HSAlowCD8low thymocytes were electronically gated and either CD44+NKR-P1+, CD44+ TCR-α/β+, and CD44+Vβ8.1,8.2+, Ly6ChighNKR-P1+, IL-2Rβ+ NKR-P1+, or IL-2Rβ+TCR-α/β+ NT cells were analyzed by flow cytometry using a FACScan® (Becton Dickinson, Mountain View, CA). Intrahepatic NT cells were stained with anti– NKR-P1-PE and anti–TCR-β-biotin or with anti–TCR-α/β-PE and anti–NKR-P1-biotin. Splenocytes were stained with anti–B220-FITC (RA3-6B2), anti–TCR-α/β-PE and anti–NKR- P1-biotin after blocking with anti-Fcγ III/II receptor antibody (2.4G2) for at least 15 min. NKR-P1+TCR-α/β+ cells were analyzed directly (liver) or after electronic gating within B220null MNC (spleen). All antibodies used in these experiments were purchased from PharMingen (San Diego, CA). Thymic NT cell number was calculated from the percentages of the double-positive CD44+ and NKR-P1+, TCR-α/β+ or Vβ8.1,8.2+ thymocytes within the HSAlowCD8low subset as described previously (5).
Results
NT Cell Ontogeny in the Thymus of γc, IL-7Rα–, IL-2– and IL-4–deficient Mice.
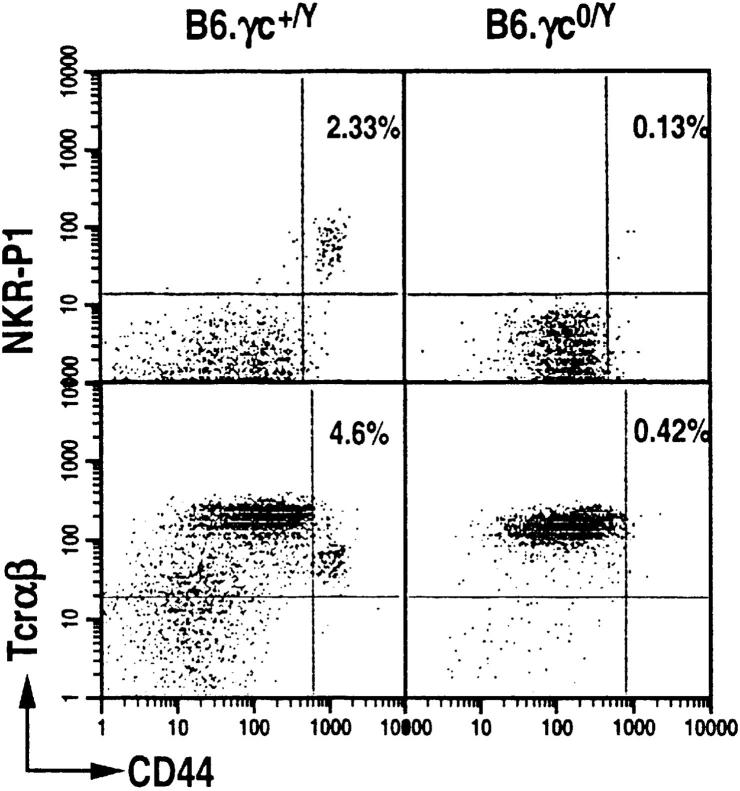
The loss of γc expression as found in X-linked severe combined immunodeficiency patients or in mice results in impaired development of mainstream α/β T cells (15, 17). To determine whether γc deficiency affects NT cell ontogeny, we examined their development in the thymus of γc0/Y mice backcrossed to C57BL/6 for 4–8 generations. γc0/Y mice, >6 wk old, do not develop HSAlowCD8low CD44highNKR-P1+ thymocytes, whereas they are detected in the γc+/Y wild-type littermates (Fig. 1). Thus, akin to mainstream T cells, signaling through γc is important for NT cell ontogeny.
Figure 1.
Common cytokine receptor γc-deficient mice do not develop thymic NTα/β cells. Dot plots displaying CD44+ NKR-P1+ and CD44+TCR-α/β+ T cells among HSAlow CD8low thymocytes of B6.γc0/Y (seventh generation backcross to C57BL/6) and B6.IL-2Rγc+/Y littermates. HSAlowCD8low thymocyte population was electronically gated and CD44highNKR-P1+ T cells were analyzed with a FACScan® flow cytometer.
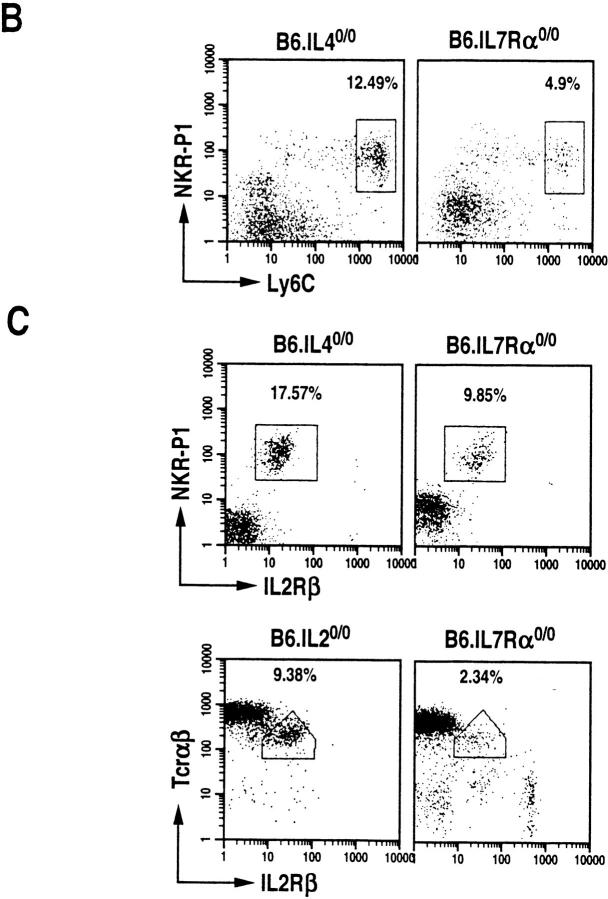
γc serves as an essential component of the multimeric receptors for IL-2, IL-4, IL-7, IL-9, and IL-15 (18). To determine which of the cytokines that use γc receptor affect NT cell development, the thymi of >6-wk-old B6.IL-20/0, B6.IL-40/0 and B6.IL-7Rα0/0 mice were analyzed. Mainstream T lymphocyte development is impaired in B6.IL-7 Rα0/0 mice, but proceeds normally in B6.IL-20/0 (19) and B6.IL-40/0 (20) mice. All three mutant mice develop CD44highNKR-P1+ T cells (Fig. 2 A). The repertoire of α/β-TCR is skewed towards Vβ8.1,8.2 usage in >40% (the percentage of TCR-α/β+ thymocytes that are Vβ8.1, 8.2+) of these NT cells (Fig. 2 A). Further data revealed that the NT cells that develop in B6.IL-40/0 and B6.IL-7 Rα0/0 mice also express Ly6C (Fig. 2 B) and that all the three mutant mice express IL-2Rβ (Fig. 2 C) similar to the NK1+ T cells that develop in the wild-type C57BL/6 mice. These data suggest that neither IL-2, IL-4, IL-7, nor thymic stromal-derived lymphopoietin (TSLP; IL-7Rα is a receptor component of IL-7 and TSLP; 16) are required for NT cell ontogeny.
Figure 2.
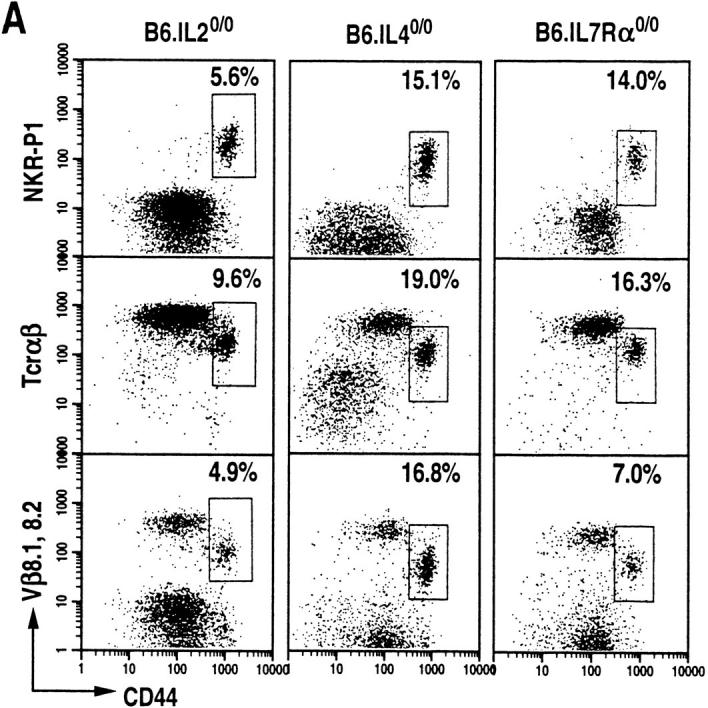
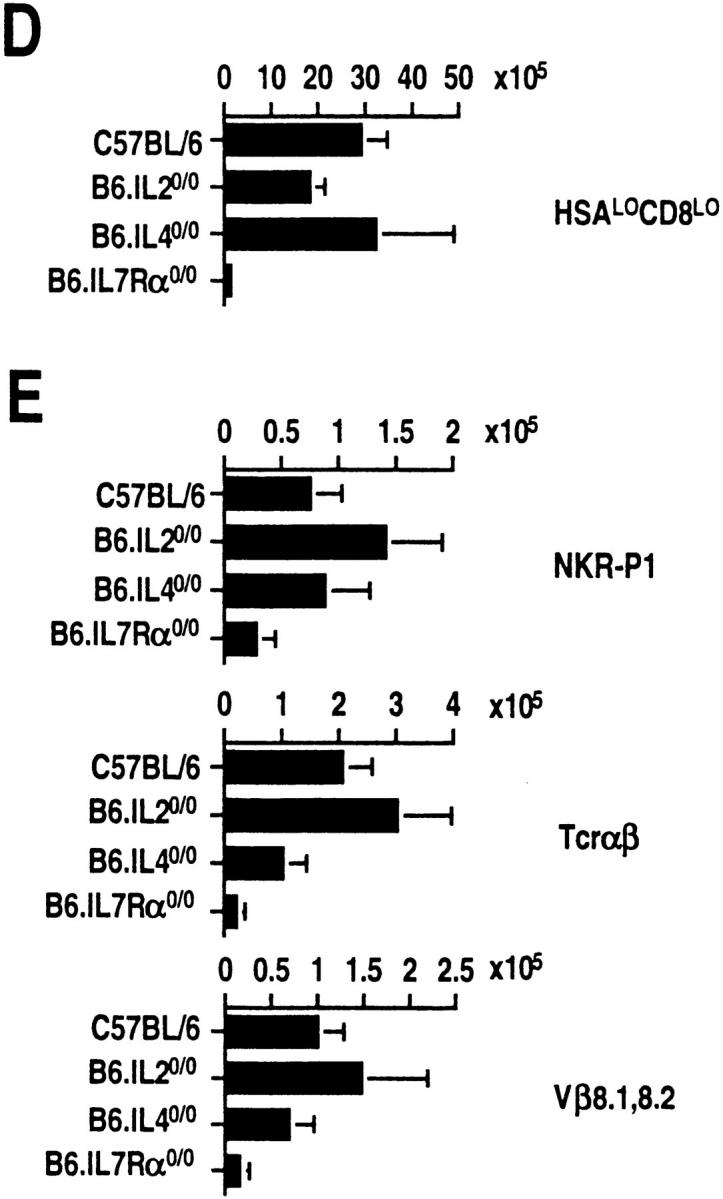
IL-2–, IL-4–, and IL-7Rα–deficient mice develop thymic NTα/β T cells. (A) Dot plots of CD44highNKR-P1+, CD44highTCR-α/βmed, and CD44highTCR-β8.1,8.2med T cells among HSAlowCD8low thymocytes of B6.IL-20/0 (n = 5), B6.IL-40/0 (n = 6) and B6.IL-7Rα0/0 mice (n = 6). (B) Dot plots of Ly6ChighNKR-P1+ T cells among HSAlowCD8low thymocytes. (C) Dot plots of IL-2 Rβ+NKR-P1+ (B6.IL-40/0 and B6.IL-7Rα0/0) and of IL-2Rβ+TCR-α/β+ (B6.IL-20/0 and B6.IL-7Rα0/0) T cells among HSAlowCD8low thymocytes. In B and C, the percentage of NT cells was almost equal in B6.IL-40/0, about half in B6.IL-7Rα0/0, or up to twofold greater in B6.IL-20/0 compared with those in the wild type. (D) Absolute numbers of HSAlowCD8low thymocytes calculated as the thymocyte number times the fraction of this subset. (E) Thymic NT cells in wild-type, IL-20/0, IL-40/0 and IL-7Rα0/0 mice. NT cell number was calculated from the percentages of the double-positive CD44+ and NKR-P1+, TCR-α/β+ or Vβ8.1,8.2+ thymocytes within the electronically gated HSAlowCD8low population in D as described previously (5).
To determine the absolute number of NT cells in the wild-type and the various mutant mice, thymocytes from them were electronically gated in the HSAlowCD8low population. The actual number of HSAlowCD8low cells was calculated from the observed percentages of this population, and the initial yield of thymocytes harvested from individual animals. Corresponding to the small size of the thymus (16), the number of HSAlowCD8low cells was dramatically reduced to ∼5% of the wild type in IL-7Rα0/0 mice (Fig. 2 D). The absolute number of CD44+NKR-P1+ T cells within the HSAlowCD8low population is reduced to ∼40% of wild type in IL-7Rα0/0 mice (Fig. 2 E, top). However, IL-7Rα0/0 thymus contains only up to 10% of NT cells that express TCR-α/β and Vβ8.1,8.2 compared to those of the wild type (Fig. 2 E, middle and bottom). The remaining CD44+NKR-P1+ thymocytes in the IL-7Rα0/0 mice are probably natural killer cells that also express these markers, but not the TCR-α/β (22). The reduced number of NT cells in the IL-7R0/0 mice then corresponds to the small size of its thymus (16). In contrast, the absolute number of NT cells in IL-20/0 mice is increased by 1.5–2-fold compared to the number of NKR-P1+ T cells in wild-type animals (Fig. 2 E). These data suggest that signals delivered through the IL-7Rα are not only important in maintaining thymic cellularity, but are also essential for the expansion of NT cells.
NT Cells in the Peripheral Lymphoid Organs of γc, IL-7Rα, and IL-2–deficient Mice.
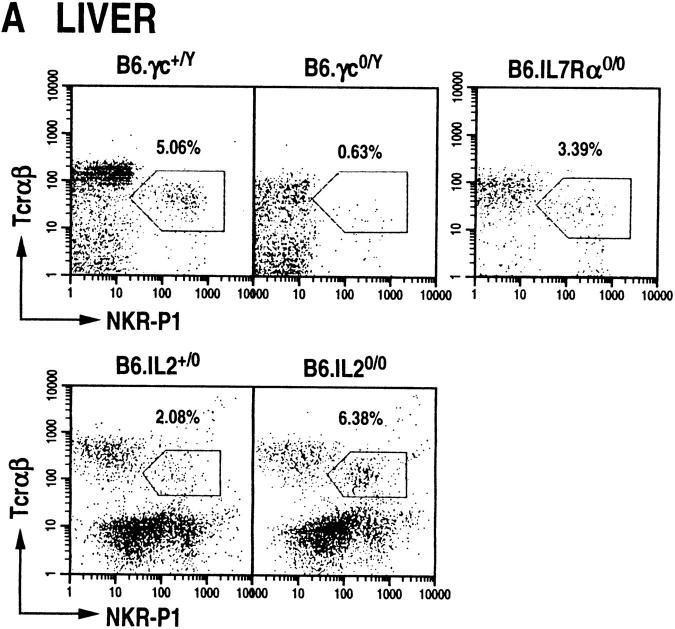
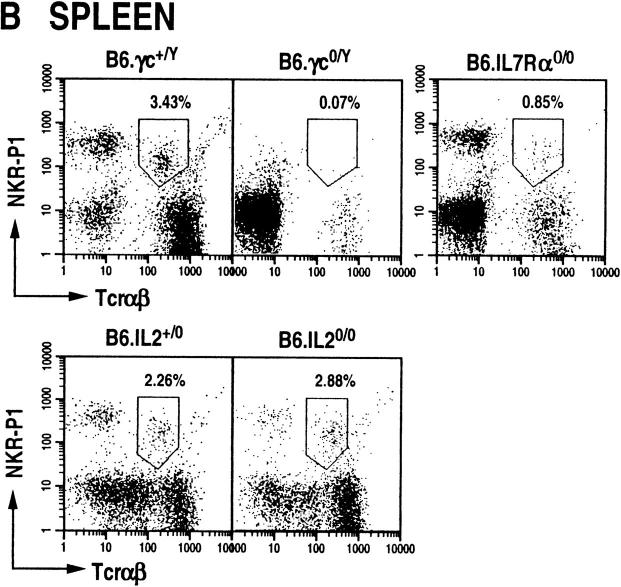
NT cells are also present in peripheral lymphoid organs such as the liver, spleen, bone marrow, and lymph nodes; they also form a subset of intraepithelial lymphocytes of the gut (23). It is at present not clear whether NT cells develop in situ in these organs or whether they home here after their genesis in the thymus (22, 24, 25). To determine whether γc, IL-7Rα, and IL-2 deficiency affect NT cell development and/or homing to the liver and spleen, the presence of NKR-P1+TCR-α/β+ cells was analyzed in these mutant animals. We find that γc null mice contain dramatically reduced levels of NT cells in the liver and spleen (Fig. 3). IL-7Rα0/0 mice contained reduced numbers of NT cells in the liver (∼60% of wild type; Fig. 3 A) and in the spleen (∼25% of wild type; Fig. 3 B). In contrast, IL-2 null mutations had increased levels of NKR-P1+ T cells in the liver, but remained about the same in the spleen (Fig. 3). Thus, although γc is required for the development and/or homing of this subset of T cells to the liver and spleen, the signaling for this process does not require IL-2. Additionally, it is unclear whether IL-7Rα null mutation affects the development and/or homing of NT cells to the periphery, or whether it affects the expansion of this T cell subset in the liver and spleen as it does in the thymus.
Figure 3.
NTα/β cells do not develop in peripheral lymphoid organs of γc-deficient mice, but develop in IL-7Rα0/0 and IL-20/0 animals. Dot plots displaying NKR-P1+TCR-α/β+ cells among mononuclear cells isolated from the liver (A) and spleen (B) of wild-type and mutant mice. Liver NTα/β cells were stained with anti–NKR-P1-PE and anti–TCR-β-biotin (B6.γc0/Y and B6.IL-7Rα0/0) or anti–TCR-β-PE and anti–NKR-P1-biotin (B6.IL-2+/0 and B6.IL-20/0). Splenic NTα/β cells were stained with anti–B220-FITC, anti-TCR-β-PE and anti-NKR-P1-biotin, and identified among B220null splenocytes. In all cases, the biotinylated antibodies were detected by staining with streptavidin-RED670. The staining pattern of intrahepatic NT cells was observed in over 20 different preparations (see also reference 4).
Discussion
The requirement for signal transduction through γc for NT cell development that does not involve IL-2 and IL-4 as well as, by extension, IL-7 and TSLP (evidenced by their development in IL-7Rα0/0 thymocytes) as the soluble mediator is noteworthy. Thus, the lack of NT cells in γc0/Y mice might be a reflection on the requirement for IL-9, IL-15, and/or an as yet unidentified cytokine as the signal transducer for their development. NKR-P1+ cells do not develop in mice with dysregulated expression of IL-2Rβ (26) or when the IL-2Rβ function is blocked with specific antibody (27). Whereas IL-9 uses only γc as part of its receptor complex (28, 29), IL-15 function depends on both the IL-2Rβ and γc (30). This suggests that IL-15 or a hithertofore undefined cytokine that uses both IL-2Rβ and γc specifies the soluble mediator function for NT cell ontogeny. If indeed an IL-2Rβ and γc-dependent cytokine(s) serves this function, how it selectively turns on NT cell development in fetal day 9 livers (25), and later in post-natal thymus (7), remains to be determined.
Human and mouse NK cells are predicted to develop from bipotential T/NK progenitor cells that commit to either T or NK cell lineage depending on the microenvironment of the developing precursor. Recent evidence suggests that the commitment to the NK lineage is strongly influenced by IL-15 (31–33). If NT and NK cells have a common precursor, then our data are consistent with the hypothesis that commitment to NKR-P1+ T cell lineage might be influenced by signals mediated by IL-15.
Two possible mechanisms can explain the lack of NT cell development in γc0/Y mice. Signaling through the γc may be critical for the commitment of the precursor cell to the NT cell lineage. An alternative mechanism would be that further differentiation of the already committed NT cells cannot proceed in the absence of signals from the γc. Current evidence supports the latter mechanism. It was recently demonstrated that γc null mice develop Vα14 and Vβ8 positive, the conserved TCR expressed by NT cells (6, 7), thymocytes within their CD4−8− subset (34). However, these thymocytes poorly respond to in vitro cross-linking of their TCR by secreting IL-4 (34), a characteristic of mature NT cells (35). This would suggest that commitment to NT cells has occurred in the thymus of γc null mice, but the committed precursor has not completely differentiated into this lineage.
Although IL-7Rα0/0 mice develop NT cells, their absolute numbers in the thymus are dramatically lower (∼10%) than those found in the wild-type mice. In vitro stimulation of unfractionated thymocytes with IL-7 results in the selective expansion of CD4+8− and CD4−8− thymocytes that express the Vβ8.2 TCR (36). In keeping with this and consistent with our results is the finding that IL-7 cytokine-deficient mice develop NT cells, but in reduced numbers (37). In this respect, it is also noteworthy that type I diabetes-prone NOD mice do not secrete IL-4 in response to in vivo cross-linking of TCR (12) suggesting that they do not develop NT cells. Type I diabetes is a Th1 disease that can be delayed or prevented by the administration of IL-4 to prediabetic NOD mice (13). The lack of IL-4 response of NOD mice to in vivo TCR cross-linking is reversible by IL-7 stimulation in vitro (12). Thus, in conclusion, akin to mainstream T cells, the commitment and/or differentiation to the NT cell lineage depends on signal transduction through the γc, and once committed, their expansion requires signals relayed through the IL-7Rα.
Acknowledgments
We thank M. Hunsinger and N. Schaffer for technical help; D. Roopenian for generous supply of B6.β2-m0/0 and B6.IL-40/0 mice; D. Serreze for generous supply of IL-20/0 and IL-2+/0 mice and R. Morawetz for B6.IL-40/0 mice for the initial experiments.
Footnotes
S. Joyce is the recipient of the American Cancer Society's Junior Faculty Research Award. This work is supported in part by grants to S. Joyce from the American Cancer Society (Institutional), the Juvenile Diabetes Foundation International and the National Institutes of Health (RO1 HL54977).
References
- 1.Vicari AP, Zlotnik A. Mouse NK1.1+T cells: a new family of T cells. Immunol Today. 1996;17:71–76. doi: 10.1016/0167-5699(96)80582-2. [DOI] [PubMed] [Google Scholar]
- 2.Smiley ST, Kaplan MH, Grusby MJ. Immunoglobulin E production in the absence of interleukin-4 secreting CD1 dependent cells. Science (Wash DC) 1997;275:977–979. doi: 10.1126/science.275.5302.977. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y-H, Chiu NM, Mandal M, Wang N, Wang C-R. Impaired NK1+T cell development and early IL-4 production in CD1 deficient mice. Immunity. 1997;6:459–467. doi: 10.1016/s1074-7613(00)80289-7. [DOI] [PubMed] [Google Scholar]
- 4.Mendiratta SK, Martin WD, Hong S, Boesteanu A, Joyce S, Van Kaer L. CD1d1mutant mice are deficient in natural T cells that promptly produce IL-4. Immunity. 1997;6:469–477. doi: 10.1016/s1074-7613(00)80290-3. [DOI] [PubMed] [Google Scholar]
- 5.Joyce S, Negishi I, Boesteanu A, DeSilva AD, Sharma P, Chorney MJ, Loh DY, Van Kaer L. Expansion of natural (NK1+) T cells that express αβ T cell receptors in transporters associated with antigen presentation-1 null and thymus leukemia antigen positive mice. J Exp Med. 1996;184:1579–1584. doi: 10.1084/jem.184.4.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lantz O, Bendelac A. An invariant T cell receptor α chain is used by a unique subset of major histocompatibility complex class I–specific CD4+ and CD8−T cells in mice and humans. J Exp Med. 1994;180:1097–1106. doi: 10.1084/jem.180.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bendelac A, Killeen N, Litman DR, Schwartz RH. A subset of CD4+thymocytes selected by MHC class I molecules. Science (Wash DC) 1994;263:1774–1778. doi: 10.1126/science.7907820. [DOI] [PubMed] [Google Scholar]
- 8.Porcelli SA, Yockey CE, Brenner MB, Balk SP. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4−8−α/β T cells demonstrates preferential use of several Vβ genes and an invariant TCR α chain. J Exp Med. 1993;178:1–16. doi: 10.1084/jem.178.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dellabona P, Padovan E, Casorati G, Brockhaus M, Lanzavecchia A. An invariant Vα24JαQ/Vβ11 T cell receptor is expressed in all individuals by clonally expanded CD4−8−T cells. J Exp Med. 1994;180:1171–1176. doi: 10.1084/jem.180.3.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takeda K, Dennert G. The development of autoimmunity in C57BL/6 lprmice correlates with the disappearance of the natural killer type 1 positive cells: evidence for their suppressive action on bone marrow stem cell proliferation, B cell immunoglobulin secretion, and autoimmune symptoms. J Exp Med. 1993;177:155–164. doi: 10.1084/jem.177.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mieza MA, Itoh T, Cui JQ, Makino Y, Kawano T, Tsuchida K, Koike T, Shirai T, Yagita H, Matsuzawa A, et al. Selective reduction of Vα14+NK T cells associated with disease development in autoimmune prone mice. J Immunol. 1996;156:4035–4040. [PubMed] [Google Scholar]
- 12.Gombert J-M, Tancrede-Bohin E, Hameg A, Leite-de-Moraes MDC, Vicari A, Bach J-F, Herbelin A. IL-7 reverses NK1.1+T cell defective IL-4 production in non-obese diabetic mouse. Int Immunol. 1996;8:1751–1758. doi: 10.1093/intimm/8.11.1751. [DOI] [PubMed] [Google Scholar]
- 13.Rapoport MJ, Jaramilo A, Zipris D, Lazarus AH, Serreze DV, Leiter EH, Cyopick P, Danska J, Delovitch TL. Interleukin-4 reverses T cell unresponsiveness and prevents the onset of diabetes in nonobese diabetic mice. J Exp Med. 1993;178:87–99. doi: 10.1084/jem.178.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morawetz RA, Gabriele L, Rizzo LV, Noben-Trauth N, Kuhn R, Rajewsky K, Muller W, Doherty TM, Finkelman F, Coffman RL, Morse HC., III Interleukin (IL)-4 independent immunoglobulin class switch to immunoglobulin (Ig)E in the mouse. J Exp Med. 1996;184:1654–1661. doi: 10.1084/jem.184.5.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao X, Shores EW, Hu-Li J, Anver MR, Kelsall BL, Russell SM, Drago J, Noguchi M, Grinberg A, Bloom ET, et al. Defective lymphoid development in mice lacking expression of the common cytokine receptor γ chain. Immunity. 1995;2:223–238. doi: 10.1016/1074-7613(95)90047-0. [DOI] [PubMed] [Google Scholar]
- 16.Peschon JJ, Morrissey PJ, Grabstein KH, Ramsdell FJ, Maraskovsky E, Gliniak BC, Park LS, Ziegler SF, Williams DE, Ware CB, et al. Early lymphocyte expansion is severely impaired in interleukin 7 receptor–deficient mice. J Exp Med. 1994;180:1955–1960. doi: 10.1084/jem.180.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noguchi M, Yi H, Rosenblatt HM, Filipovich AH, Adelstein S, Modi WS, McBride OW, Leonard WJ. Interleukin-2 receptor γ chain mutation results in X-linked severe combined immunodeficiency in humans. Cell. 1993;73:147–157. doi: 10.1016/0092-8674(93)90167-o. [DOI] [PubMed] [Google Scholar]
- 18.Leonard WJ, Shores EW, Love PE. Role of common cytokine receptor γ chain in cytokine signaling and lymphoid development. Immunol Rev. 1995;148:97–114. doi: 10.1111/j.1600-065x.1995.tb00095.x. [DOI] [PubMed] [Google Scholar]
- 19.Schorle H, Holtschke T, Hunig T, Schimpl A, Horak I. Development and function of T cells in mice rendered interleukin-2 deficient by gene targeting. Nature (Lond) 1991;352:621–624. doi: 10.1038/352621a0. [DOI] [PubMed] [Google Scholar]
- 20.Kuhn R, Rajewsky K, Muller W. Generation and analysis of interleukin-4 deficient mice. Science (Wash DC) 1991;254:707–710. doi: 10.1126/science.1948049. [DOI] [PubMed] [Google Scholar]
- 21.Yoshimoto T, Bendelac A, Hu-Li J, Paul WE. Defective IgE production by SJL mice is linked to the absence of CD4+, NK1.1+ T cells that prompty produce interleukin-4. Proc Natl Acad Sci USA. 1995;92:11931–11934. doi: 10.1073/pnas.92.25.11931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bendelac A. Mouse NK1+T cells. Curr Opin Immunol. 1995;7:367–374. doi: 10.1016/0952-7915(95)80112-x. [DOI] [PubMed] [Google Scholar]
- 23.Ohteki T, MacDonald HR. Major histocompatibility complex class I related molecules control the development of CD4+8− and CD4−8− subsets of natural killer 1.1+T cell receptor–α/β cells in the liver of mice. J Exp Med. 1994;180:699–704. doi: 10.1084/jem.180.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacDonald HR. NK1.1+T cell receptor–α/β cells: new clues to their origin, specificity and function. J Exp Med. 1995;182:633–638. doi: 10.1084/jem.182.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makino Y, Kanno R, Koseki H, Taniguchi M. Development of Vα14+NK T cells in the early stages of embryogenesis. Proc Natl Acad Sci USA. 1996;93:6516–6520. doi: 10.1073/pnas.93.13.6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suwa H, Tanaka T, Kitamura F, Shiohara T, Kuida K, Miyasaka M. Dysregulated expression of the IL-2 receptor β chain abrogates development of NK cells and Thy-1+dendritic epidermal cells in transgenic mice. Int Immunol. 1995;7:1441–1449. doi: 10.1093/intimm/7.9.1441. [DOI] [PubMed] [Google Scholar]
- 27.Takeuchi Y, Tanaka T, Hamamura K, Sugimoto T, Miyasaka M, Yagita H, Okumura K. Expression and role of interleukin-2 receptor β chain on CD4−CD8− T cell receptor αβ+cells. Eur J Immunol. 1992;22:2929–2935. doi: 10.1002/eji.1830221126. [DOI] [PubMed] [Google Scholar]
- 28.Russell SM, Johnson JA, Noguchi M, Kawamura M, Bacon CM, Friedmann M, Berg M, McVicar DW, Witthuhn BA, Silvennoinen O, et al. Interaction of IL-2Rβ and γc chains with Jak1 and Jak3: implications for XSCID and XCID. Science (Wash DC) 1994;266:1042–1045. doi: 10.1126/science.7973658. [DOI] [PubMed] [Google Scholar]
- 29.Kimura Y, Takeshita T, Kondo M, Ishii N, Nakamura M, Van Snick J, Sugamura K. Sharing of the IL-2 receptor gamma chain with the functional IL-9 receptor complex. Int Immunol. 1995;7:115–120. doi: 10.1093/intimm/7.1.115. [DOI] [PubMed] [Google Scholar]
- 30.Giri JG, Ahdieh M, Eisenman J, Shanenbeck K, Grabstein K, Kumaki S, Namen A, Park LS, Cosman D, Anderson D. Utilization of the β and γ chains of the IL-2 receptor by a novel cytokine IL-15. EMBO (Eur Mol Biol Organ) J. 1994;13:2822–2830. doi: 10.1002/j.1460-2075.1994.tb06576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cavazzana-Calvo M, Hacein-Bey S, de Saint G, Basile, De Coene C, Selz F, Le Deist F, Fischer A. Role of interleukin-2 (IL-2), IL-7, and IL-15 in natural killer cell differentiation from cord blood hematopoietic progenitor cells and from γc transduced severe combined immunodeficiency X1 bone marrow cells. Blood. 1996;88:3901–3909. [PubMed] [Google Scholar]
- 32.Leclercq G, Debacker V, De Smedt M, Plum J. Differential effects of interleukin-15 and interleukin-2 on the differentiation of bipotential T/natural killer progenitor cells. J Exp Med. 1996;184:325–336. doi: 10.1084/jem.184.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puzanav IJ, Bennett M, Kumar V. IL-15 can substitute for the marrow microenvironment in the differentiation of natural killer cells. J Immunol. 1996;157:4282–4285. [PubMed] [Google Scholar]
- 34.Lantz O, Sharara LI, Tilloy F, Anderson A, DiSanto JP. Lineage relationships and differentiation of natural killer (NK) T cells: intrathymic selection and interleukin (IL)-4 production in the absence of NKR-P1 and Ly49 molecules. J Exp Med. 1997;185:1395–1401. doi: 10.1084/jem.185.8.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshimoto T, Paul WE. CD4pos NK1posT cells promptly produced IL-4 in response to in vivo challenge with anti-CD3. J Exp Med. 1994;179:1285–1295. doi: 10.1084/jem.179.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vicari A, Liete-de-Moraes MDC, Gombert J-M, Dy M, Penit C, Papiernik M, Herbelin A. Interleukin 7 induces preferential expansion of Vβ8.2+CD4−8− and Vβ8.2+CD4+8−murine thymocytes positively selected by class I molecules. J Exp Med. 1994;180:653–661. doi: 10.1084/jem.180.2.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vicari A, Herbelin A, Leite-de-Moraes MDC, von Freedon-Jeffry U, Murray R, Zlotnik A. NK1.1+T cells from IL-7 deficient mice have normal distribution and selection but exhibit impaired cytokine production. Int Immunol. 1996;8:1759–1766. doi: 10.1093/intimm/8.11.1759. [DOI] [PubMed] [Google Scholar]