Abstract
We have previously shown that transgenic mice expressing the oncoprotein v-Rel under the control of a T cell–specific promoter develop T cell lymphomas. Tumor formation was correlated with the presence of p50/v-Rel and v-Rel/v-Rel nuclear κB-binding activity. Since experimental evidence has led to the suggestion of a potential tumor suppressor activity for IκBα, we have studied the role of IκBα in the transforming activity of v-Rel by overexpressing IκBα in v-rel transgenic mice. Overexpression of IκBα in v-rel transgenic mice resulted in an extended survival, and the development of cutaneous T cell lymphomas of CD8+CD4− phenotype. These phenotypic alterations were associated with a dramatic reduction of p50/v-Rel, but not v-Rel/v-Rel nuclear DNA binding activity and an increased expression of the intercellular adhesion molecule 1. Our results indicate that v-Rel homodimers are active in transformation and that the capacity of v-Rel–containing complexes to escape the inhibitory effect of IκBα may be a key element in its transforming capability.
The oncogene v-rel was originally identified as the transforming component of the avian retrovirus reticuloendotheliosis virus T. v-Rel induces neoplastic disease in birds, and compared to c-Rel, is missing 2 NH2-terminal and 118 COOH-terminal amino acids, and has several internal changes (1–5). v-Rel is a member of the Rel/NF-κB family of eukaryotic transcription factors, which includes c-Rel, RelA (p65), RelB, NF-κB1 (p50/p105), NF-κB2 (p52/p100), and the Drosophila proteins Dorsal, dorsal-related immunity factor, and Relish (6–12).
The finding that v-Rel and the corresponding protooncogene c-Rel are members of the Rel/NF-κB transcription factor family led to the suggestion that transformation resulted from v-Rel–induced changes in gene expression (1–5). Rel/NF-κB proteins are related through an ∼300– amino acid NH2-terminal region known as the Rel homology domain (RHD)1, which contains sequences important for dimerization, DNA binding, inhibitor binding, and nuclear localization. The activity of Rel/NF-κB complexes is modulated by their interaction with the IκB family of inhibitors, which contain ankyrin repeats. In unstimulated cells, Rel/NF-κB dimers remain in the cytoplasm as inactive complexes through association with IκB molecules that mask their nuclear localization signals. A wide variety of stimuli result in the rapid phosphorylation and degradation of IκB molecules and nuclear translocation of Rel/ NF-κB complexes (6–16).
The mechanism by which v-Rel induces oncogenic transformation is not clear, and it was originally believed that v-Rel could only transform avian cells (1–5). Recently, we demonstrated that v-Rel also has the capacity of transforming mammalian cells in vivo. Transgenic mice expressing v-Rel in thymocytes develop T cell lymphomas with very poor prognosis. In tumor cells from v-Rel transgenic mice, there are two major DNA-binding complexes containing v-Rel homodimers and p50/v-Rel heterodimers. However, when v-rel transgenic mice were crossed with p50-deficient animals, T cell leukemia appeared at an earlier stage, suggesting that the v-Rel homodimer is the essential transforming complex (17).
In this report we address the question of whether overexpression of the inhibitory IκBα protein, which has been suggested to have tumor suppresser activity (18), can reverse the transforming activity of v-Rel in v-rel transgenic mice. Overexpression of IκBα extended the survival of v-rel transgenic mice and reduced the severity of lymphomas. Surprisingly, IκBα overexpression resulted in a change in the clinical expression of the disease with an expansion of CD8+CD4− T cells in peripheral tissues. These T cell changes were associated with increased levels in the expression of the intercellular adhesion molecule 1 (ICAM-1), increased dermotropism and the development of cutaneous lymphoma. T cells from v-rel/ikba double transgenic mice presented a dramatic reduction of p50/v-Rel but not of v-Rel/v-Rel nuclear DNA-binding activity. Our results indicate that v-Rel homodimers are active in transformation and that v-Rel containing complexes have an intrinsic capability to escape the inhibitory effect of IκBα. We postulate that variations in the clinical expression of related lymphoid malignancies may reflect subtle changes in the nuclear composition and interplay among different Rel/ NF-κB and IκB molecules.
Materials and Methods
Plasmid Construction and Generation of Transgenic Mice.
The generation of v-rel transgenic mice has been previously described (17). A detailed description of the generation and characterization of ikba transgenic mice will be reported elsewhere (Perez, P., unpublished observations). Screening of IκBα transgenic mice was performed as described (19), and line 1 was selected for its high level of expression as determined by immunoblot analysis using a mouse monoclonal IκBα antibody. To generate double v-rel/ikba transgenic mice, ikba transgenic mice were bred to homozygocity and crossed with heterozygote v-rel transgenic animals. The mice obtained from these intercrosses were screened by PCR using a pair of specific v-rel oligonucleotides: 5′-TTTCTCACCAACCTCCGATTCACTG-3′ and 5′-ATCTCTGCAGCCTTTTCCAACTAGA-3′.
Histopathology and Immunofluorescence.
Mice were killed and tissues were immersion fixed in 10% buffered formalin. Tissues were embedded in paraffin blocks and processed by routine methods, sectioned at 5–10-μm thickness, stained with hematoxylin and eosin, and examined by light microscopy. Single cell suspensions from tumor-bearing spleens were prepared according to standard procedures and spun onto slides as previously described (20). For immunofluorescence, the cytospins were incubated at 4°C overnight with anti–v-Rel or anti-IκBα polyclonal antibodies. Anti–v-Rel and anti-IκBα antibodies were visualized with a donkey anti–rabbit immunoglobulin conjugated with Texas red (Amersham Corp. Arlington Heights, IL). For ICAM-1 immunofluorescence analysis, lymph node T cells were isolated on murine T cell enrichment columns (R&D Sys. Inc., Minneapolis, MN) according to manufacturer's recommendation from v-rel and v-rel/ikba transgenic mice. T cells were spun onto slides and incubated at 4°C overnight with anti–ICAM-1 FITC-labeled monoclonal antibody (PharMingen, San Diego, CA).
Flow Cytometric Analysis.
Single cell suspensions of spleens bearing tumors were placed in RPMI 1640 medium supplemented with 10% FCS, 4 mM glutamine, 50 mM β-mercaptoethanol, 50 U/ml penicillin, and 50 mg/ml streptomycin. Flow cytometry was performed with a flow cytometer and cell sorter (Epics Profile II; Coulter Corp., Hialeah, FL). Single cell suspensions from ikba and v-rel/ikba transgenic thymi and spleens were prepared and analyzed for surface expression of CD4 (clone H129.19), CD8 (clone 7D4), TCR-α/β (clone H57-597), B220 (clone RA3-6B2), HSA (clone J11d), Mac-1 (clone M17/70HL), and Gr-1 (clone RB6-8C5) as previously described (21). Monoclonal antibodies were obtained from GIBCO BRL (Gaithersburg, MD) and PharMingen.
Western Blot Assays.
Cytoplasmic and nuclear fractions from thymocytes were prepared as previously described (22). Aliquots of cytoplasmic and nuclear extracts (20 μg) were boiled in Laemmli buffer and run overnight on a 12.5% acrylamide-bisacrylamide (200:1) gel at 12 mA. Western blotting procedures and antibodies have been previously described (23). The proteins were transferred onto nitrocellulose membranes and transfer efficiency assessed by Ponceau S staining. Purity of the nuclear extracts was checked by incubating the membranes with an antiserum specific for the cytosolic enzyme lactate dehydrogenase.
Cell Labeling, Lysis, and Immunoprecipitation.
Isolated thymocytes were labeled for 2 h with 500 μCi/ml of [35S]methionine (1,000 Ci/mmol) in DMEM lacking methionine and containing 10% heat inactivated and dialyzed FCS. The labeling medium was removed and the cells were washed with cold PBS and lysed on ice by adding radio immunoprecipitation assay buffer (10 mM Tris-HCl, pH 7.5, 0.5% Nonidet P-40, 150 mM NaCl). Cell lysates were first cleared with a preimmune serum (3 μl) for 3 h and then immunoprecipitated with specific antisera as previously described (23).
Electrophoretic Mobility Shift Assay.
The palindromic κB site used for these assays has been previously described (23). Nuclear extracts (3 μg) were incubated with 20,000 cpm 32P-labeled probe, 3 μg poly (dI/dC) in buffer containing 20 mM Hepes, pH 7.9, 60 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol, 0.5 mM PMSF, and 17% glycerol in 25 ml final volume for 15 min on ice. Protein loading was checked by Oct 1 DNA-binding activity. Complexes were separated on 5.5% native polyacrylamide gels run in 0.25 × Tris base, boric acid, EDTA buffer, dried, and exposed to X-Omat AR film (Kodak, Rochester, NY) at −70°C.
Semiquantitative Reverse Transcription Analysis.
Semiquantitative PCR was performed as described previously (17). The PCR conditions were established such that amplification of the cDNAs was linearly dependent on the concentration of the corresponding messenger RNAs (mRNAs). This was achieved by performing the reactions in the presence of [32P]dCTP; due to the greater sensitivity of autoradiography as opposed to ethidium bromide staining, a relatively low number of PCR cycles was sufficient to detect the amplified product. In this way, the cDNA concentration remained the rate-limiting factor throughout the amplification procedure. Lymph node T cells derived from ikba, v-rel, and v-rel/ikba transgenic mice were isolated on murine T cell enrichment columns (R&D Sys. Inc.), and total RNA was prepared and quantified by absorbance at 260 nm. 10 μg of total RNA were used for cDNA synthesis using 1,000 U of reverse transcriptase (GIBCO BRL). After cDNA synthesis, each sample was diluted five times and 5 μl were taken for amplification with 25 cycles by using Taq polymerase in the presence of [32P]dCTP and sequence-specific primers. Specific oligonucleotides were obtained from Stratagene Corp. (La Jolla, CA). One-tenth of the PCR products were separated on 8% native polyacrylamide gels run in Tris base, boric acid, EDTA buffer, dried, and exposed to X-Omat AR film (Kodak) at −70°C.
Results
Generation of v-rel Transgenic Mice Overexpressing IκBα in the Thymus.
The transcriptional activity of Rel/NF-κB family of proteins is regulated, in part, by their association with inhibitory molecules (IκBs) that sequester them as inactive complexes in the cytoplasm. IκBα is the predominant inhibitory protein in most cell types (5, 13–16).
We recently generated transgenic mice specifically expressing v-Rel in thymocytes (17). These animals develop T cell multicentric aggressive lymphomas. To study the effect of IκBα on the transforming capability of v-Rel, we produced double transgenic v-rel/ikba mice by crossing transgenic mice expressing v-Rel with transgenic mice overexpressing IκBα. Both transgenes are under the control of the mouse T cell–specific lck proximal promoter.
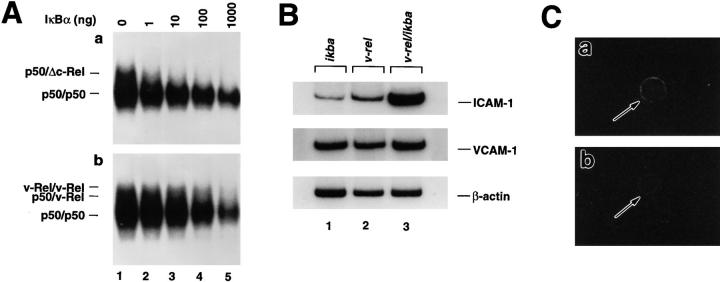
The expression of v-Rel and IκBα in thymocytes from the double transgenic mice was assessed by immunoprecipitation (Fig. 1 A). These analyses demonstrate the association of v-Rel complexes with IκBα (Fig. 1, lanes 1 and 3) and show that there is an excess of IκBα protein that is not associated with v-Rel (Fig. 1, lane 4). In addition, these data show that under these conditions, IκBβ, another IκB inhibitory protein, is not associated with v-Rel, suggesting that this molecule does not play a major role in the inactivation of v-Rel complexes (Fig. 1 A, lane 2).
Figure 1.
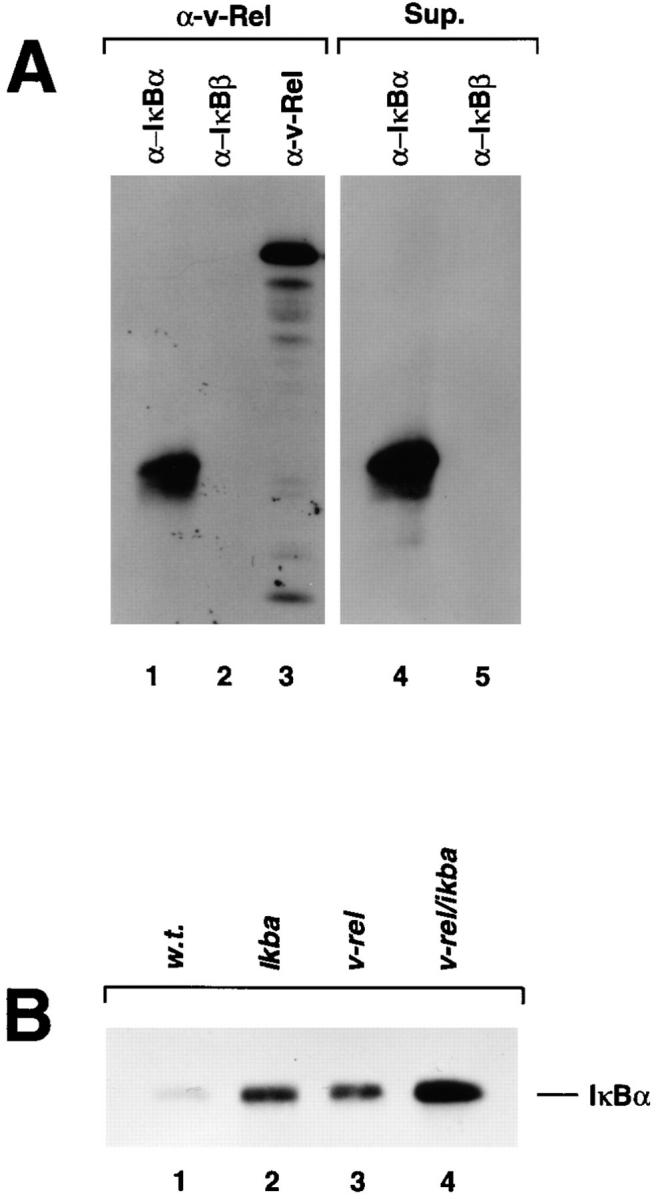
v-Rel/IκBα complexes (A) and IκBα protein expression (B) in v-rel/ikba double transgenic thymocytes. To generate v-rel/ikba double transgenic mice, ikba homozygous transgenic mice were crossed with v-rel heterozygous transgenic mice. Both transgenes were under the control of the lck promoter. Screening of double transgenic mice was done by PCR analysis using v-rel–specific oligonucleotides. (A) Immunoprecipitations were performed as previously described (31). [35S]methionine-labeled thymocytes were lysed under nondenaturing conditions, and total protein extract was immunoprecipitated with anti–v-Rel antibodies. The immunoprecipitate was denatured to dissociate the v-Rel–containing immune complex and then reprecipitated, first with IκBα and subsequently with IκBβ and v-Rel antibodies (left). The supernatant of the total protein extract treated with anti–v-Rel antibody was subsequently reprecipitated with anti-IκBα and anti-IκBβ antibodies (right). (B) The levels of IκBα protein expression were analyzed by Western blot in total protein extracts from control (w.t.), ikba transgenic, v-rel transgenic and v-rel/ikba double transgenic thymocytes.
The levels of IκBα protein expression in v-rel/ikba double transgenic mice was determined by Western blot with whole protein extract prepared from transgenic thymocytes and compared with the levels of IκBα protein expression in wild-type, ikba transgenic, and v-rel transgenic thymocytes (Fig. 1 B). The highest levels of IκBα protein expression were detected in v-rel/ikba double transgenic thymocytes (lane 4). This represents almost two times the amount of IκBα present in ikba transgenic (lane 2) and v-rel transgenic thymocytes (lane 3). As described previously, there is an increase in the levels of IκBα in v-rel transgenic thymocytes when compared with control thymocytes (lane 1) due to IκBα protein stabilization by v-Rel–containing complexes (17).
IκBα Overexpression Changes v-Rel Nucleus/cytoplasm Distribution.
To examine whether the overexpression of IκBα in v-rel transgenic mice alters the nuclear–cytoplasmic distribution of v-Rel, we performed Western blot analyses using thymocyte extracts from wild-type, v-rel transgenic, and v-rel/ikba double transgenic mice (Fig. 2 A). In v-rel transgenic thymocytes, a significant amount of v-Rel, ∼30% of the total protein, was localized in the nucleus (lanes 3 and 4). In contrast, in v-rel/ikba double transgenic thymocytes, >90% of the v-Rel protein is retained in the cytoplasm, and only a small amount translocates to the nucleus (lanes 5 and 6). The blot containing lanes 5 and 6 was overexposed to detect the min amounts of v-Rel protein in the nucleus. Incubation of the same membrane with antiserum specific for the cytosolic enzyme lactic dehydrogenase demonstrated no cross-contamination of nuclear and cytoplasmic extracts (data not shown).
Figure 2.
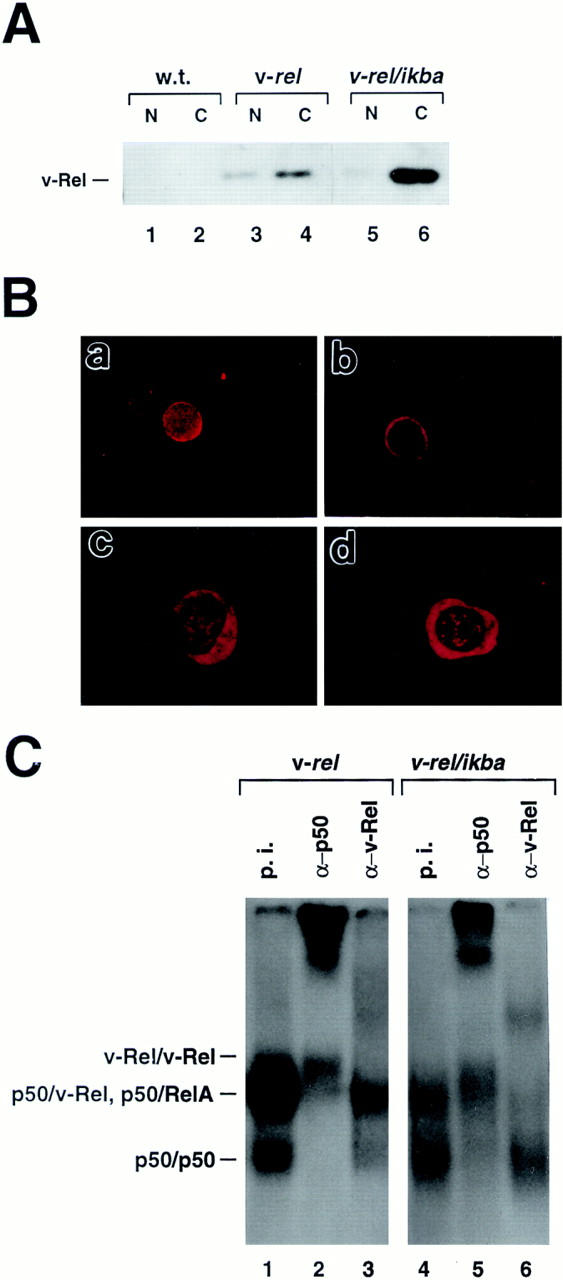
Change of nucleus/cytoplasm distribution of v-Rel in v-rel/ ikba double transgenic thymocytes. (A) Western blot analysis. Cytoplasmic (C) and nuclear (N) fractions from thymocytes and Western blot assays were performed as previously described (31). (B) Immunofluoresence. Thymocytes from 6-wk-old v-rel (a) and v-rel/ikba (b) transgenic mice were spun onto slides, incubated with anti–v-Rel antibodies, and visualized with donkey anti–rabbit Texas red–labeled antibodies. T cells from spleen-bearing tumors of v-rel/ikba double transgenic mice were spun onto slides, incubated with anti–v-Rel (c) or anti-IκBα (d) antibodies, and visualized with donkey anti–rabbit Texas red–labeled antibodies. (C) Decreased p50/v-Rel κB-binding activity in v-rel/ikba double transgenic thymocytes. Electrophoretic mobility shift assays were performed as previously described (23) using nuclear protein extracts from v-rel and v-rel/ikba double transgenic thymocytes. Lanes 1 and 4 were treated with preimmune sera (p.i.), lanes 2 and 5 were treated with anti-p50 antibody (α-p50), and lanes 3 and 6 were treated with anti–v-Rel antibody (α-v-Rel).
The changes in the nuclear/cytoplasmic distribution of v-Rel protein in the double transgenics were further confirmed by immunofluoresence (Fig. 2 B). v-Rel protein in v-rel transgenic thymocytes is detected in both nucleus and cytoplasm; in contrast, in v-rel/ikba double transgenic thymocytes, v-Rel is mainly detected in the cytoplasm (a and b, respectively). In addition, the distribution of v-Rel and IκBα proteins in transformed T cells from v-rel/ikba double transgenic mice is also mainly in the cytoplasm (c and d, respectively).
IκBα Overexpression Decreases p50/v-Rel Heterodimer, but Not v-Rel Homodimer DNA-binding Activity.
To investigate whether the κB-binding activity was altered in thymocytes of v-rel/ikba double transgenic mice compared to v-rel transgenic mice, thymocyte protein extracts were analyzed by electrophoretic mobility shift assays (EMSA) using a palindromic κB site (Fig. 2 C).
The p50/v-Rel DNA binding activity in nuclear extracts from v-rel/ikba double transgenic thymocytes presents a dramatic reduction compared to nuclear protein extracts from v-rel transgenic thymocytes (compare lanes 1 and 4). However, the v-Rel/v-Rel DNA-binding activity remains almost unchanged (compare lanes 2 and 5). The amount of nuclear protein extract added to the DNA binding reaction was controlled by measuring Oct 1 DNA-binding activity. Oct 1 DNA-binding activity was comparable between the different protein samples (data not shown).
These results indicate that v-Rel homodimers are significantly less sensitive than p50/v-Rel heterodimers to the inhibitory effect of IκBα. The reduced affinity of v-Rel for IκBα may be one of the key molecular basis for v-Rel transformation.
IκBα Overexpression Extends Survival and Changes the Clinical Expression of v-rel Transgenic Mice.
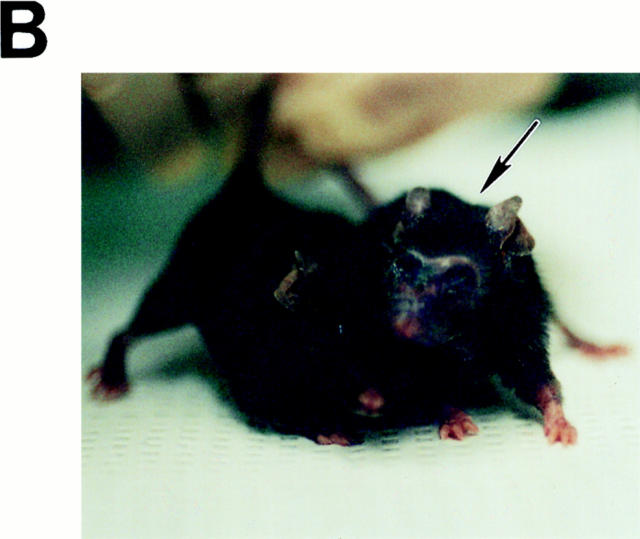
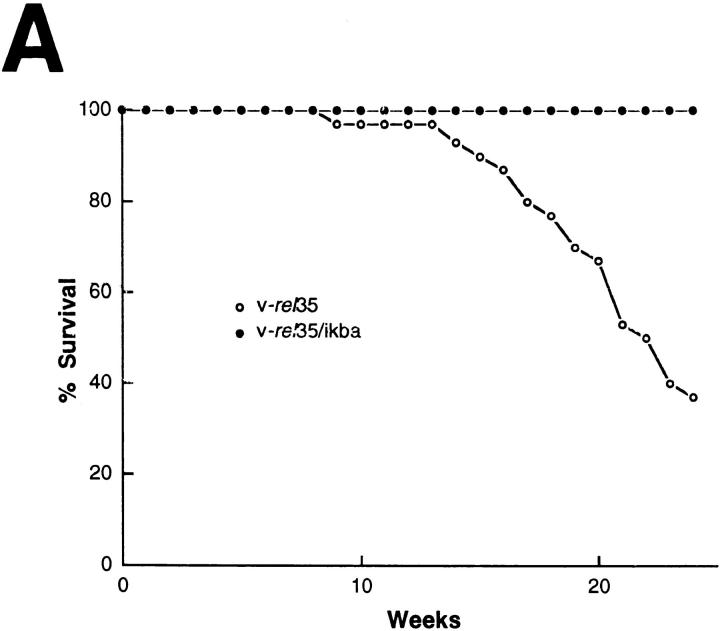
To investigate the consequences of the changes in κB binding activity in v-rel/ikba double transgenic mice, a colony of these animals was bred and their health status monitored over time in a nongerm-free environment (Fig. 3). The v-rel/ikba double transgenic mice presented a longer survival and appeared completely healthy for a longer period of time when compared with v-rel transgenic mice. The mortality curve demonstrates that high levels of IκBα extended the survival of v-rel transgenic mice (75% of v-rel transgenic mice succumbed before 25 wk of age, whereas 100% of the v-rel/ ikba double transgenic mice were unaffected at that time). Young v-rel/ikba double transgenic mice, between 3 and 10 wk old, appeared normal as assessed by habits, weight, posture, and histopathology. However, all v-rel/ikba double transgenic mice developed skin lesion in face, ears, tail, and feet after 15 wk of age. These lesions, characterized by loss of hair, thickening of the skin, and exfoliative plaques (Fig. 3 B) progressed slowly with no major compromise in the health status of the mice. Evaluation for a longer period of time revealed that v-rel/ikba double transgenic were susceptible to bacterial infections and that they succumbed due to secondary infections by pathogens that colonize the skin; antibiotic prophylactic treatment extended their survival for an additional 2–4 wk (data not shown). Autopsies of v-rel/ikba double transgenic mice did not reveal massive tumoral cell burden in lung, liver, lymph nodes, and spleen, indicating that in contrast to v-rel transgenic mice, massive tumor cellular infiltration was not the primary reason for organ failure in the double trangenic animals (17).
Figure 3.
Overexpression of IκBα increases survival of v-rel transgenic mice and changes the clinical expression. (A) Mortality curve. Deaths in v-rel/ikba transgenic animals occurred at later times and were the result of secondary opportunistic infection according to pathologic analysis and microbiology. Autopsy, and histologic examination of v-rel/ikba transgenic mice revealed a less severe lymphomatous infiltrate than in v-rel transgenic mice. n = 29 for v-rel 35, and n = 13 for v-rel/ikba. (B) T-cell cutaneous lymphoma in v-rel/ikba double transgenic mice. All v-rel/ikba transgenic mice developed skin lesions characterized by thickening and exfoliative plaques. Arrow indicates a v-rel/ikba transgenic mouse; the control mouse is an ikba transgenic littermate. (The lesions were never observed in v-rel transgenic mice.)
T Cell Cutaneous Lymphoid Infiltrates in v-rel/ikba Double Transgenic Mice.
The earliest manifestation of disease in most of the v-rel/ikba double transgenic mice was the appearance of facial skin lesions that progressed slowly (Fig. 3 B). Light microscopy revealed cellular infiltration of the dermis by large lymphocytes of irregular nuclei (Fig. 4, a–d). Isolated foci of epidermal infiltration by tumor cells were occasionally seen, but typical Pautrier's microabcesses and massive epidermal infiltration were not observed (Fig. 4 e). The histologic appearance of this lymphoid infiltration resembles one seen in cutaneous T cell lymphomas (24–27). In contrast to v-rel/ikba transgenic mice, dermal compromise was rarely seen in v-rel transgenic mice (17). Histopathology of v-rel/ikba transgenic animals revealed much less severe tumoral cellular infiltration in other tissues besides skin (17). For instance, in the lung of v-rel transgenic mice, tumoral cellular infiltration around bronchi and alveoli was severe enough to cause organ failure. In v-rel/ikba transgenic mice, lung compromise was much less severe with only tumoral cell infiltration around the bronchi, but not in the parenchyma (17; Fig. 4 e). The liver in v-rel transgenic mice was always enlarged and massively infiltrated by tumor cells that frequently plugged the vessels and produced extensive areas of ischemia and tissue necrosis. In contrast, in v-rel/ikba double transgenic mice, the liver was not enlarged, and lymphoid infiltration was seen in hepatic sinusoids and around the portal triad, but areas of necrosis were not observed (Fig. 4 f ). The spleen in v-rel transgenic mice was dramatically enlarged with complete distortion of the normal architecture and massive areas of tissue necrosis. On the other hand, in v-rel/ikba double transgenic mice, the spleen was moderately enlarged and remnants of the normal splenic architecture with demarcation of white and red pulp areas were always present (Fig. 4 g). Lymph nodes in v-rel/ikba double transgenic mice were not severely compromised, even at very late stages of disease, as was the case in v-rel transgenic mice (Fig. 4 h, and data not shown). Thymic atrophy was a constant observation in both v-rel and v-rel/ikba transgenic mice (data not shown). These histopathological studies demonstrate that v-rel/ikba double transgenic mice developed lymphoma, but the magnitude of the visceral compromise was much less than in v-rel transgenic mice of the same age. These results indicate that IκBα overexpression does not prevent tumor formation, but does delay the appearance of tumors, the progression of the disease, and decreases the severity of the lymphomatous state observed in v-rel transgenic mice. In addition, these results indicate that IκBα overexpression induces transcriptional changes in v-Rel–transformed T cells that resulted in their increased tropism for the skin.
Figure 4.
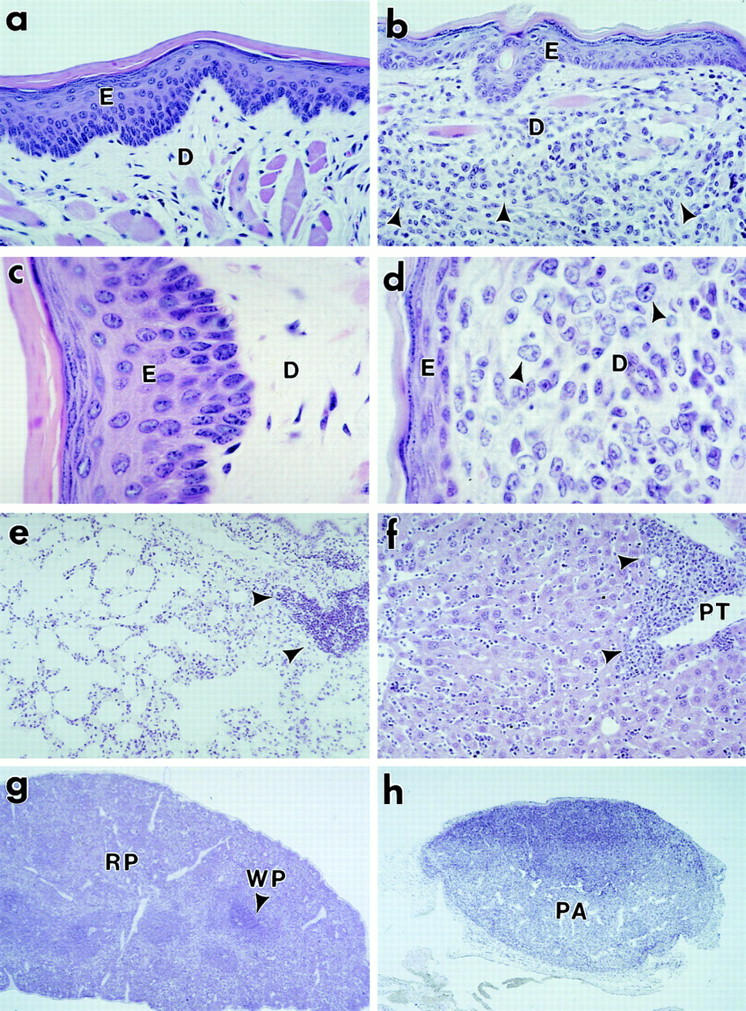
Tumor pathology in v-rel/ikba transgenic mice. Tissue blocks were fixed in 10% neutral buffered formalin and processed for paraffin embedding. 5-μm sections were stained with hematoxylin and eosin. Sections from skin (a, b, c, and d), lung (e), liver ( f ), spleen (g), and lymph node (h) are shown. Arrow heads indicate sites of lymphoid infiltration. Photomicrographs were taken at ×100 (a, b, e, and f ), ×250 (c and d), ×12.5 (g and h). E, epidermis; D, dermis; PA, paracortical area; PT, portal triad; RP, red pulp; WP, white pulp.
Flow Cytometric Analysis of Lymphoid Cells from v-rel/ikba Double Transgenic Mice.
To understand the changes in the clinical expression of v-rel/ikba double transgenics compared to v-rel transgenic mice, we analyzed lymphoid cell populations by flow cytometry (Fig. 5). Flow cytometric analysis of lymphoid cells in v-rel transgenic mice have been previously described (17). Analyses of thymuses and spleens from young v-rel/ikba double transgenic animals (3–8 wk) did not reveal any alterations when compared to ikba transgenic littermates (Fig. 5 A, a–d, and data not shown). ikba transgenic littermates were used as a control, since no differences in lymphoid populations have been detected in ikba transgenic mice when compared to wild-type animals (Perez, P., unpublished results and data not shown). Analysis of spleens from moribund and euthanized v-rel/ ikba double transgenic animals showed an expanded T cell population characterized by intermediate and low levels of the TCR α and β chains (Fig. 5 B, a and b). In addition, a striking difference between v-rel transgenic and v-rel/ikba double transgenic mice was a significant increase in CD8+ single positive T cells in v-rel/ikba transgenic mice, a population of cells that was not always increased in v-rel transgenic mice. These results indicate that IκBα overexpression in v-rel transgenic mice alters the developmental profile of the v-rel transformed T cells, allowing the T cells to reach a more mature phenotype. Only a moderate increase of T cells coexpressing markers associated with an immature phenotype (HSA) was detected in the double transgenic (Fig. 5 B, e and f ), in contrast to the dramatic changes that we previously observed in v-rel transgenic mice. An increased population of granulocytes as a result of infection was detected in v-rel/ikba double transgenic mice (Fig. 5 B, g and h).
Figure 5.
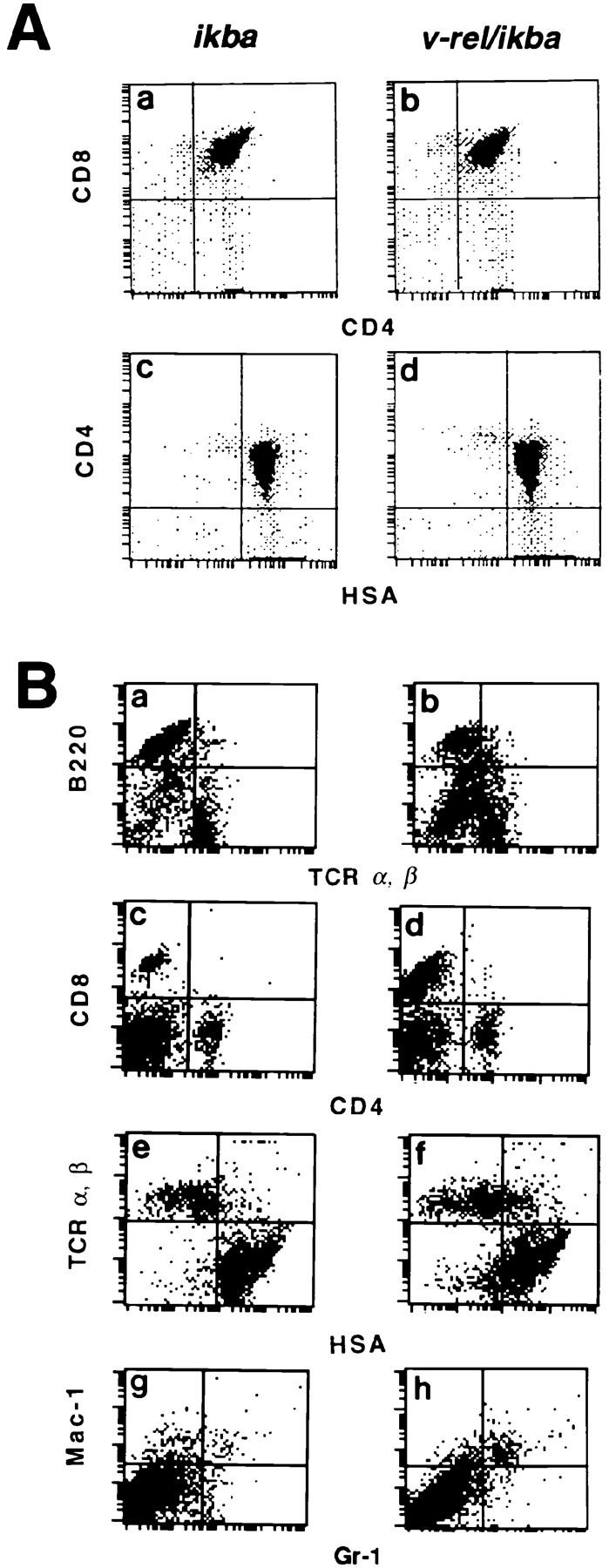
Flow cytometric analysis in v-rel/ikba transgenic mice. (A) Flow cytometric analysis of CD4 and CD8 surface markers in thymocytes from 6-wk-old ikba (a and c) and v-rel/ikba (b and d) transgenic mice. (B) Flow cytometric analysis of surface markers in splenocytes from 6-mo-old ikba (a, c, e, and g) and v-rel/ikba (b, d, f, and h) transgenic mice. Flow cytometry was performed using a flow cytometer (Epics Profile II) and cell sorter (Coulter Corp.). Single cell suspensions from thymocytes and splenocytes were prepared and analyzed for surface markers expression of CD4, CD8, HSA, TCR-α/β, B220, Mac-1, and Gr-1 as previously described (8).
v-Rel–containing Complexes Have Lower Affinity for IκBα than Δc-Rel–containing Complexes.
To understand why higher levels of IκBα are required to reduce nuclear translocation of v-Rel and p50/v-Rel κB-binding activity in transgenic thymocytes, we compared the inhibitory effect of IκBα on κB binding activity in nuclear protein extract derived from v-rel transgenic (Fig. 6 A) and Δc-rel transgenic thymocytes. Δc-rel transgenic animals express a truncated version of the mouse c-Rel protein (Δc-Rel) under the control of the lck promoter, and, like the v-Rel oncogenic protein, lacks part of the COOH-terminal transcriptional activation domain keeping the RHD intact. The generation of Δc-rel transgenic mice has been previously described (17). Increasing levels of purified baculovirus expressed IκBα protein were added to identical amounts of nuclear protein extracts from Δc-rel transgenic thymocytes (Fig. 6 A a) and v-rel transgenic thymocytes (Fig. 6 A b) before starting the κB-binding reaction. The total amount of protein extract used in the reaction was tested by Oct 1 DNA-binding activity and the identification of the complexes by antibody supershifts (data not shown). In the absence of added IκBα, similar amounts of total κB binding activities were detected in Δc-rel transgenic (Fig. 6 A a, lane 1) and in v-rel transgenic thymocytes (Fig. 6 A b, lane 1). However, when increased amounts of IκBα protein were added to the binding reaction, the binding of p50/Δc-Rel was dramatically reduced (Fig. 6 A a, lanes 2 and 3), whereas the binding of v-Rel containing complexes was slightly affected (Fig. 6 A b, lanes 2 and 3). At 100 ng of IκBα, the binding of v-Rel–containing complexes is significantly reduced (Fig. 6 A b, lane 4), and only when the highest amounts of IκBα were used (1 μg) and when endogenous p50/p50 κB binding also began to be affected (Fig. 6 A a, lane 5), was the binding of v-Rel–containing complexes completely reduced (Fig. 6 A b, lane 5). These results demonstrate that the binding of v-Rel–containing complexes is more resistant than the binding of complexes containing the cellular homologue Δc-Rel to the inhibitory effect of IκBα, and offer an attractive explanation for the molecular mechanisms involved in the transforming activity of v-Rel. This is in agreement with previous in vitro studies that demonstrated a reduced IκBα inhibition of DNA binding by the oncogenic v-Rel protein when compared to the nononcogenic c-Rel protein (28).
Figure 6.
Reduced IκBα inhibition of v-Rel DNA binding activity (A) and increased levels of ICAM-1 in v-rel/ikba double transgenic T cells (B and C). (A) Equal amounts of nuclear protein extracts from Δc-rel (a) or v-rel (b) transgenic thymocytes were incubated in the presence of increased amounts of IκBα protein and analyzed by EMSA using a palindromic κB-binding site. (B) Total RNA was prepared from ikba transgenic, v-rel transgenic or v-rel/ikba double transgenic lymph node–derived cells. After cDNA synthesis, one-tenth of the reaction mixture was amplified in the presence of 0.1 μCi of [32P]dCTP using Taq polymerase and sequence-specific primers. One-fifth of the amplified products were separated on 6% nondenaturing polyacrilamide gels. Gels were dried and exposed to x-ray film. (C) Purified T cells from lymph nodes of v-rel transgenic (a) and v-rel/ikba (b) double transgenic mice were spun onto slides and incubated with anti–ICAM-1 FITC-labeled antibody.
Increased Levels in the Expression of a Cell Adhesion Molecule ICAM-1 in v-rel/ikba Double Transgenic Thymocytes.
To correlate the changes in the clinical expression and the cutaneous infiltration observed in v-rel/ikba double transgenic mice, we examined the level of expression of κB responsive genes encoding adhesion molecules that may be involved in the increased tropism of v-rel/ikba double transgenic T cells for the skin (22, 24–27, 29, 30). We analyzed mRNA expression levels of ICAM-1 by reverse transcription PCR in ikba, v-rel, and v-rel/ikba transgenic T cells (Fig. 6 B). cDNA was prepared from total mRNA extracted from single cell suspension from ikba normal lymph nodes and from v-rel and v-rel/ikba tumor-bearing lymph nodes. The cDNA was amplified with 25 PCR cycles in the presence of [32P]dCTP and the appropriate pairs of specific oligonucleotides. The same amount of total mRNA was present in all the samples, as demonstrated by equal amounts of β-actin PCR products (Fig. 6 B). By comparing the amount of PCR products detected in ikba (lane 1), v-rel (lane 2), and v-rel/ikba lymph node–derived T cells (lane 3), it is evident that there is a dramatic increase in the expression of ICAM-1 in v-rel/ikba transgenic T cells. No major changes in the expression of vascular cell adhesion molecule 1 and dorsal midline–globulin-restricted axonal surface protein were detected between v-rel and v-rel/ikba– transformed T cells (Fig. 6 B and data not shown). Increased levels of ICAM-1 expression in v-rel/ikba transgenic T cells were also detected on cytospin preparation by indirect immunofluorescence using anti–ICAM-1 FITC-labeled antibody (Fig. 6 C). The difference in the level of ICAM-1 expression between v-rel and v-rel/ikba may explain the increased tropism for the skin of v-rel/ikba–transformed T cells.
Discussion
In this work we have demonstrated that overexpression of the Rel/NF-κB inhibitory protein, IκBα, delays the development of T cell lymphomas in a transgenic mouse model that expresses the oncogenic v-Rel protein under the control of the lck T cell–specific promoter (17). Previous studies indicate that v-Rel transformation requires its translocation to the nucleus and binding to a set of specific Rel/NF-κB responsive genes (1–5).
Role of IκBα in v-Rel Transformation.
Previous studies have suggested a potential tumor supresser activity for IκBα (4). IκBα-deficient mice displayed elevated levels of nuclear Rel/NF-κB activity in hemopoietic tissues. Unfortunately, these animals do not survive long enough for evaluation of appearance of lymphoid malignancies (31, 32).
In this work, we have demonstrated that although v-rel/ ikba double transgenic thymocytes expressed IκBα protein at very high levels, there was still some nuclear v-Rel protein in the double transgenic cells, suggesting an intrinsic ability of v-Rel to escape the inhibitory effect of IκBα. This observation was correlated with an increased resistance of v-Rel DNA-binding activity to the inhibitory effect of IκBα when compared to a truncated version of c-Rel that lacks the transcriptional activation domain. The reduced affinity of v-Rel for IκBα may be associated with some of the internal mutations present in the Rel homology domain of v-Rel (1–5). It is noteworthy to mention that transgenic mice overexpressing either RelA or RelB in T cells under the control of the lck did not develop lymphomas. In the case of rela transgenic mice, the absence of tumor formation was correlated with the fact that there was no increased DNA-binding activity in T cells, because RelA protein was efficiently retained in the cytoplasm by IκBα (19). However, relb transgenic thymocytes presented a dramatic increase in DNA-binding activity due to the fact that RelB complexes were not efficiently retained in the cytoplasm because of their low affinity for IκBα (33), indicating that increased nuclear levels of Rel/NF-κB activity appear to be insufficient for transformation. Therefore, together with the low affinity for IκBα, it appears that v-Rel possesses additional and unique properties that render a potent transforming potential in mammalian cells.
The fact that IκBα overexpression extended the survival but did not prevent tumor formation in v-rel transgenic mice indicates that it would be impossible to achieve in vivo the levels of IκBα required to compensate for the reduced affinity of v-Rel for IκBα to prevent v-Rel transformation.
ICAM-1 Overexpression in Lymphoproliferative Disorders.
When nuclear κB-binding activity was assessed by EMSA in v-rel/ikba double transgenic thymocytes, the most striking observation was the dramatic reduction in the p50/v-Rel DNA-binding activity with almost no change in v-Rel/ v-Rel DNA binding activity. The reduction in the p50/ v-Rel DNA binding correlated with the changes observed in the clinical expression of v-rel/ikba double transgenic mice, and supports our previous indications that v-Rel may be active as a p50/v-Rel heterodimer and as a homodimer (17), with the p50/v-Rel heterodimeric form being more susceptible to IκBα inhibition. Because of the changes in the clinical expression and the differences observed in p50/ v-Rel and v-Rel/v-Rel DNA-binding activity in v-rel/ ikba double transgenic T cells, it is possible to speculate that v-Rel either as a heterodimeric complex with p50 or as a homodimeric complex may target different sets of κB-regulated genes. An example of this differential regulation is the observation of increased levels of ICAM-1 in T cells derived from v-rel/ikba double transgenic mice, but not in T cells derived from v-rel transgenic mice. Interestingly, strong induction of ICAM-1 has been observed in human lymphoproliferative disorders (29, 34), and ICAM-1 overexpression in v-rel/ikba double transgenic mice may be responsible for the increased tropism of T cells for the skin observed in these animals (25, 22).
Rel/NF-κB Activity in Cutaneous T Cell Lymphomas.
The lymphomatous infiltration observed in v-rel/ikba transgenic mice resembles that seen in T cell cutaneous lymphomas: the Sezary syndrome, mycosis fungoides, or other related disorders (35). Our observation that v-rel/ikba double transgenic mice developed cutaneous T cell lymphomas is particularly interesting in light of the observation that chromosomal translocations associated with structural alterations of the Rel/NF-κB family of proteins have been documented in several cases of T cell cutaneous lymphomas in human patients (26, 35–40). In particular, rearrangement and altered expression of the NFKB-2 gene have been identified in the HUT 78 human cutaneous T cell leukemia line (39, 40). In addition, structural alterations of the NFKB-2 gene have also been identified very commonly (14%) in neoplasms derived from mature T cells such as mycosis fungoides and Sezary syndrome (24, 38). These structural alterations may contribute to lymphomagenesis by determining a constitutive activation of the NF-κB system and, in particular, of NFKB-2 target genes (36, 37). This data, in addition to our observations, strongly supports the notion that structural alterations of other Rel/NF-κB family members may play a role in lymphomagenesis. Furthermore, our results indicate that variations in the clinical expression of lymphoid malignancies such us mycosis fungoides and Sezary syndrome may be based on subtle changes in the nuclear composition and interplay among the different Rel/NF-κB and IκB molecules. In this sense, v-rel and v-rel/ikba transgenic mice constitute a potential model to study the molecular mechanisms involved in the generation of lymphoid malignancies.
Acknowledgments
We would like to thank C. Raventos-Suarez and K. Class for FACS® analysis; K. Dorfman and L. Chen for technical assistance; S. Lira, M. Swerdel, and P. Zalamea for the generation of the transgenic mice, and all the staff in Veterinary Sciences of Bristol-Myers Squibb for their excellent support; C. Gelinas for anti– v-Rel antibodies, Roger M. Perlmutter for the mouse lck promoter, and Dimitris Kioussis for the human CD2 LCR sequences; and Janet Cheng for her valuable comments on this manuscript.
Footnotes
1 Abbreviations used in this paper: EMSA, electrophoretic mobility shift assay; ICAM-1, intercellular adhesion molecule 1; mRNA, messenger RNA; RHD, Rel homology domain.
These studies were performed according to guidelines established by the Bristol-Myers Squibb Animal Care and Use Committee.
References
- 1.Bose HR. The Rel family: models for transcriptional regulation and oncogenic transformation. Biochim Biophys Acta. 1992;1114:1–17. doi: 10.1016/0304-419x(92)90002-g. [DOI] [PubMed] [Google Scholar]
- 2.Enrietto PJ. The relgene and its role in oncogenesis. Semin Cancer Biol. 1990;1:399–405. [PubMed] [Google Scholar]
- 3.Gilmore TD. Malignant transformation by mutant Rel proteins. TIG (Trends Genet) 1991;7:318–322. doi: 10.1016/0168-9525(91)90421-l. [DOI] [PubMed] [Google Scholar]
- 4.Gilmore TD. Role of relfamily genes in normal and malignant lymphoid cell growth. Cancer Surv. 1992;15:69–87. [PubMed] [Google Scholar]
- 5.Gilmore TD, Doedood M K, Piffat KA, White DW. Rel/NF-κB/IκB proteins and cancer. Oncogene. 1996;13:1367–1378. [PubMed] [Google Scholar]
- 6.Baeuerle PA, Baltimore D. NF-κB: ten years after. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 7.Baeuerle PA, Henkel T. Function and activation of NF-κB in the immune system. Annu Rev Immunol. 1994;12:141–179. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 8.Finco TS, Baldwin AS. Mechanistic aspects of NF-κB regulation: the emerging role of phosphorylation and proteolysis. Immunity. 1995;3:263–272. doi: 10.1016/1074-7613(95)90112-4. [DOI] [PubMed] [Google Scholar]
- 9.Grilli M, Chiu J-S, Lenardo MJ. NF-κB and Rel-participants in a multiform transcriptional regulatory system. Int Rev Cytol. 1993;143:1–62. doi: 10.1016/s0074-7696(08)61873-2. [DOI] [PubMed] [Google Scholar]
- 10.Kopp EB, Ghosh S. NF-κB and Rel proteins in innate immunity. Adv Immunol. 1995;58:1–27. doi: 10.1016/s0065-2776(08)60618-5. [DOI] [PubMed] [Google Scholar]
- 11.Miyamoto S, Verma IM. Rel/NF-κB story. Adv Cancer Res. 1995;66:255–292. [PubMed] [Google Scholar]
- 12.Siebenlist U, Franzoso G, Brown K. Structure, regulation and function of NF-κB. Annu Rev Cell Biol. 1994;10:405–455. doi: 10.1146/annurev.cb.10.110194.002201. [DOI] [PubMed] [Google Scholar]
- 13.Baldwin AS. The NFκB and Iκβ proteins, new discoveries and insights. Annu Rev Immunol. 1996;14:649–681. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 14.Beg AA, Baldwin AS., Jr The IκB proteins: multifunctional regulators of Rel/NF-κB transcription factors. Genes Dev. 1993;7:2064–2070. doi: 10.1101/gad.7.11.2064. [DOI] [PubMed] [Google Scholar]
- 15.Gilmore TD, Morin PJ. The IκB proteins: members of a multifunctional family. TIG (Trends Genet) 1993;9:427–433. doi: 10.1016/0168-9525(93)90106-r. [DOI] [PubMed] [Google Scholar]
- 16.Verma IM, Stevenson JK, Schwarz EM, Van Antwerp D, Miyamoto S. Rel/NF-κB/IκB family: intimate tales of association and dissociation. Genes Dev. 1995;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- 17.Carrasco D, Rizzo CA, Dorfman K, Bravo R. The v-reloncogene promotes malignant T-cell leukemia/ lymphoma in transgenic mice. EMBO (Eur Mol Biol Organ) J. 1996;15:3640–3650. [PMC free article] [PubMed] [Google Scholar]
- 18.Beauparlant P, Kwan I, Bitar R, Chou P, Koromilas AE, Sonenberg N, Hiscott J. Disruption of IκBα regulation by antisense RNA expression leads to malignant transformation. Oncogene. 1994;9:3189–3197. [PubMed] [Google Scholar]
- 19.Perez P, Lira SA, Bravo R. Overexpression of RelA in transgenic mouse thymocytes: specific increase on levels of the inhibitor IκBα. Mol Cell Biol. 1995;15:3523–3530. doi: 10.1128/mcb.15.7.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carrasco D, Weih F, Bravo R. Developmental expression of the mouse c-relprotooncogene in hematopoietic organs. Development (Camb) 1994;120:2991–3004. doi: 10.1242/dev.120.10.2991. [DOI] [PubMed] [Google Scholar]
- 21.Weih F, Carrasco D, Durham SK, Barton DS, Rizzo CA, Ryseck R-P, Lira SA, Bravo R. Multiorgan inflammation and hematopoietic abnormalities in mice with a targeted disruption of RelB, a member of the NF-κB/ Rel family. Cell. 1995;80:331–340. doi: 10.1016/0092-8674(95)90416-6. [DOI] [PubMed] [Google Scholar]
- 22.Weih F, Carrasco D, Bravo R. Constitutive and inducible Rel/NF-κB activities in mouse thymus and spleen. Oncogene. 1994;9:3289–3297. [PubMed] [Google Scholar]
- 23.Diamandidou E, Cohen PR, Kurzrock R. Mycosis fungoides and Sezary syndrome. Blood. 1996;88:2385–2409. [PubMed] [Google Scholar]
- 24.Fivenson DP, Hanson CA, Nickoloff BJ. Localization of clonal T cells to the epidermis in cutaneous T-cell lymphoma. J Am Acad Dermatol. 1994;31:717–723. doi: 10.1016/s0190-9622(94)70231-4. [DOI] [PubMed] [Google Scholar]
- 25.Lutzner M, Edelson R, Schein P, Green I, Kirkpatrick C, Ahmed A. Cutaneous T-cell lymphomas: the sezary syndrome, mycosis fungoides, and related disorders. Ann Intern Med. 1975;83:534–552. doi: 10.7326/0003-4819-83-4-534. [DOI] [PubMed] [Google Scholar]
- 26.Schein PS, Macdonald JS, Edelson R. Cutaneous T-cell lymphoma. Cancer (Phila) 1976;38:1859–1861. doi: 10.1002/1097-0142(197610)38:4<1859::aid-cncr2820380466>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 27.Diehl JA, McKinsey T, Hannink M. Differential pp40IκB-β inhibition of DNA binding by relproteins. Mol Cell Biol. 1993;13:1769–1778. doi: 10.1128/mcb.13.3.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fukudome K, Furuse M, Fukuhara N, Orita S, Imai T, Takagi S, Nagira M, Hinuma Y, Yoshie O. Strong induction of ICAM-1 in human T cells transformed by human T-cell-leukemia virus type 1 and depression of ICAM-1 or LFA-1 in adult T-cell-leukemia–derived cell lines. Int J Cancer. 1992;52:418–427. doi: 10.1002/ijc.2910520316. [DOI] [PubMed] [Google Scholar]
- 29.Singer KH, Le PT, Denning SM, Whichard LP, Haynes BF. The role of adhesion molecules in epithelial–T-cell interactions in thymus and skin. J Invest Dermatol. 1990;94:85S–90S. doi: 10.1111/1523-1747.ep12876038. [DOI] [PubMed] [Google Scholar]
- 30.van de Stolpe A, van der Saag PT. Intercellular adhesion molecule–1. J Mol Med. 1996;74:13–33. doi: 10.1007/BF00202069. [DOI] [PubMed] [Google Scholar]
- 31.Beg AA, Sha WC, Bronson RT, Baltimore D. Constitutive NF-κB activation, enhanced granulopoiesis, and neonatal lethality in IκBα-deficient mice. Genes Dev. 1995;9:2736–2746. doi: 10.1101/gad.9.22.2736. [DOI] [PubMed] [Google Scholar]
- 32.Klement JF, Rice NR, Car BD, Abbondanzo SJ, Powers GD, Bhatt H, Chen C-H, Rosen CA, Stewart CL. IκBα deficiency results in a sustained NF-κB response and severe widespread dermatitis in mice. Mol Cell Biol. 1996;16:2341–2349. doi: 10.1128/mcb.16.5.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weih F, Lira SA, Bravo R. Overexpression of RelB in transgenic mice does not affect IκBα levels: differential regulation of RelA and RelB by the inhibitor protein. Oncogene. 1996;12:445–450. [PubMed] [Google Scholar]
- 34.Maio M, Pinto A, Carbone A, Zagonel V, Gloghini A, Marotta G, Cirillo D, Colombatti A, Ferrara F, Del Vecchio L. Differential expression of CD54/intercellular adhesion molecule–1 in myeloid leukemias and in lymphoproliferative disorders. Blood. 1990;76:783–790. [PubMed] [Google Scholar]
- 35.Neri A, Fracchiolla NS, Rosecetti E, Garatti S, Trecca D, Boletini A, Perletti B, Bardini L, Maiolo AT, Berti E. Molecular analysis of cutaneous B- and T-cell lymphomas. Blood. 1995;86:3160–3172. [PubMed] [Google Scholar]
- 36.Chang C-C, Zhang J, Lombardi L, Neri A, Dalla-Favera R. Mechanism of expression and role in transcriptional control of the proto-oncogene NFKB-2/LYT-10. Oncogene. 1994;9:923–933. [PubMed] [Google Scholar]
- 37.Chang C-C, Zhang J, Lombardi L, Neri A, Dalla-Favera R. Rearranged NFKB-2genes in lymphoid neoplasms code for constitutively active nuclear transactivators. Mol Cell Biol. 1995;15:5180–5187. doi: 10.1128/mcb.15.9.5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fracchiolla NS, Lombardi L, Salina M, Migliazza A, Baldini L, Berti E, Cro L, Polli E, Maiolo AT, Neri A. Structural alterations of the NF-κB transcription factor lyt-10 in lymphoid malignancies. Oncogene. 1993;8:2839–2845. [PubMed] [Google Scholar]
- 39.Thakur S, Lin H-C, Tseng W-T, Kumar S, Bravo R, Foss F, Gélinas C, Rabson AB. Rearrangement and altered expression of the NFKB-2gene in human cutaneous T-lymphoma cells. Oncogene. 1994;9:2335–2344. [PubMed] [Google Scholar]
- 40.Zhang J, Chang C-C, Lombardi L, Dalla-Favera R. Rearranged NFKB2 gene in the HUT78 T-lymphoma cell line codes for a constitutively nuclear factor lacking transcriptional repressor functions. Oncogene. 1994;9:1931–1937. [PubMed] [Google Scholar]