Abstract
The T cell receptor for antigen (TCR) is a multisubunit complex that consists of at least seven polypeptides: the clonotypic, disulfide-linked α/β heterodimer that is noncovalently associated with the invariant polypeptides of the CD3 complex (CD3-γ, -δ, -ε) and ζ, a disulfide-linked homodimer. We achieved the complete assembly of the human TCR in an in vitro transcription/translation system supplemented with dog pancreas microsomes by simultaneous translation of the messenger RNAs encoding the TCR-α, -β and CD3-γ, -δ, -ε, and -ζ subunits. CD3-ε, one of the subunits that initiates the assembly of the TCR in living cells, forms misfolded, disulfide-linked homooligomers when translated alone. However, co-translation of one of its first binding partners in the course of assembly, CD3-γ or -δ, led to the expression of mainly monomeric and correctly folded ε subunits, the only form we could detect as part of a properly assembled TCR complex. In the absence of these subunits, the ER-resident chaperone calnexin interacted with oligomeric, i.e. misfolded, structures of CD3-ε in a glycan-independent manner. A glycan-dependent interaction between CD3-ε and calnexin was mediated by CD3-γ and concerned only monomeric CD3-ε complexed with CD3-γ, but was dispensable for proper folding of CD3-ε. We suggest that in addition to its signaling function, CD3-ε serves as a monitor for proper subunit assembly of the TCR.
Most T lymphocytes express on their plasma membrane the TCR–CD3 complex. This multisubunit receptor has served as a paradigm for the analysis of the biogenesis of multimembrane proteins, and shown how, in the absence of a properly assembled complex, the remaining subunits are purged from the endoplasmic reticulum (ER)1 (1).
At the cell surface, the clonotypic TCR-α and -β subunits appear as a disulfide-linked heterodimer that constitutes the true ligand binding unit and determines the specificity of the receptor. To transduce extracellular signals into the cytoplasm, the TCR associates noncovalently with a number of accessory polypeptides, jointly referred to as the CD3 complex. This complex consists of the evolutionarily related CD3-γ, -δ, and -ε subunits, all of which belong to the Ig gene family (2–4), and a disulfide-linked homodimer of the TCR-ζ subunit. The ζ chain, a member of the gene family that also includes the γ chain of the high affinity IgE receptor, lies largely on the cytoplasmic side of the plasma membrane and has an extracellular domain of only nine residues (5). Although the occurrence of γε, δε, δεα, γεβ, αβγδε2, and αβγδε2ζ2 structures has been reported in vivo (6, 7), the exact stoichiometry of a completely assembled TCR–CD3 complex remains to be determined accurately (8).
Besides fulfilling signaling functions, the CD3 subunits and the ζ chain are also required for cell surface expression of the TCR-α/β heterodimer (9, 10). The efficiency of TCR assembly in the ER determines receptor density at the cell surface of T cells; single subunits that fail to join a complex are retained in the ER and subsequently degraded (11), whereas partial complexes are targeted to lysosomal compartments for destruction (10).
The molecular determinants underlying the subunit-specific interactions that promote assembly and the degradation of single TCR–CD3 subunits in the ER have been the subject of intensive research, but are still not fully understood. Experimental evidence supports an assembly model based on salt bridges formed in the lipid bilayer between charged residues within the transmembrane of the individual TCR subunits (12). The presence of these charged residues in the transmembrane domains of single TCR-α, -β, and CD3-δ subunits has been shown to play a key role in rapid ER degradation (13). In addition, the role of extracellular domains in the assembly of the TCR subunits has been well documented in several studies (14, 15). Furthermore, the ζ homodimer seems to monitor the quaternary structure of the partial TCR–CD3 complex to ensure that only functionally active complexes are displayed at the cell surface. It is seen in association only with completely assembled receptor complexes and, in the absence of ζ2 homodimers, surface expression of TCR is compromised (10). The ζ2 homodimer may be associated with the TCR complex only peripherally, as it can apparently be exchanged for ζ subunits that reside at the cell surface (16).
The molecular chaperone calnexin (IP90, p88) is also involved in the assembly of TCR. Originally discovered in association with partially assembled TCR complexes devoid of ζ subunits and in association with MHC class I molecules (17, 18), calnexin and its close relative calreticulin facilitate protein folding in the ER (19). Although calnexin can act exclusively as a lectin, binding to monoglucosylated trimming intermediates of N-linked glycans attached to the target polypeptide (20), the initially glycan-dependent calnexin–MHC class I interaction was maintained after the removal of the N-linked glycans (21). Furthermore, glycan-independent binding between calnexin and aggregates of the vesicular stomatitis virus G protein (VSV–G protein) has been reported (22). Because calnexin associates with all TCR subunits except TCR-ζ (17, 18, 23, 24), it has been suggested that it promotes the assembly of αβγδε2 complexes. The half-life of newly synthesized TCR-α proteins that fail to join a TCR complex and become a target of ER degradation is significantly prolonged due to their interaction with calnexin (23). However, the extent to which calnexin is involved in the folding of the individual TCR subunits and the assembly of the TCR–CD3 complex is not known.
The role of the CD3 subunits in TCR assembly has been explored mostly in transfection-based expression systems. The requirement for the simultaneous presence of all CD3 subunits for proper complex formation has been convincingly demonstrated. However, the contribution of the individual TCR subunits to folding, assembly, and quality control, such as the monitoring of proper quaternary structure, has received far less attention.
We have studied the assembly of the human TCR–CD3 complex with a particular focus on early events of this process, using a cell-free translation system supplemented with dog pancreas microsomes. Optimization of the translation conditions allow the simultaneous translation of nine different messenger RNAs (mRNAs) with high efficiency. If proper redox conditions are imposed on the ER-derived microsomes, we observe correct folding of translated polypeptides, which are assembled in a subunit-specific manner into TCR–CD3 complexes indistinguishable from those reported for living cells. We have used this in vitro system to monitor the folding of CD3-ε in an early stage of TCR assembly, a process influenced dramatically by the presence of related CD3 subunits. Translation of CD3-ε alone gives rise mainly to misfolded, disulfide-linked homooligomeric structures, the formation of which was substantially suppressed when CD3-γ or -δ were translated in the same reaction. The molecular chaperone calnexin was found to bind oligomeric forms of CD3-ε in a glycan-independent manner. When CD3-γ, which carries two N-linked glycans, was translated simultaneously, monomeric CD3-ε interacted with calnexin exclusively in a glycan-dependent manner. We propose a role for CD3-ε in sensing the quarternary structure of the TCR–CD3 complex.
Materials and Methods
DNA Constructs.
The cDNAs encoding the HA1.7 TCR-α and -β chains were provided by Dr. M.J. Owen (Imperial Cancer Research Fund, London, UK; reference 25). We obtained the cDNAs encoding CD3-γ, -δ and -ε (2–4) and TCR-ζ (5, 26) from Dr. C. Terhorst (Beth Israel Hospital, Boston, MA). The cDNAs encoding HLA-DR1-α (27) and -β chains (28) were a gift from Dr. Trowsdale (Human Immunogenetics Group, Imperial Cancer Research Fund, London, UK). The sequences which encode the cleavable signal peptide of the TCR-α, -β, CD3-γ, -δ, -ε, TCR-ζ, and HLA-DR1–β were exchanged for the signal peptide encoding sequence of the mouse class I heavy chain H2-Kb. The CD4 encoding cDNA (29) was cloned into pSP64 (Promega Corp., Madison, WI) under the control of the SP6 promotor. All other cDNAs were cloned into pSP72 (Promega Corp.) under the control of the T7 promotor.
Antibodies and Reagents.
The following mouse mAbs were used in this study: Tü36 (30) recognizes properly conformed HLA- DR1 heterodimers, OKT4 (31, 32), which is reactive against properly conformed human CD4 and OKT3 (31), recognizes CD3 (both obtained from American Type Culture Collection, Rockville, MD). JOVI.1 and JOVI.3 were raised against human TCR-β (Vβ3Cβ1; 33) and TIA-2 recognizes preferentially human TCR-ζ homodimers (34; gift from Dr. P. Anderson, Dana-Farber Cancer Institute, Boston, MA).
The following polyclonal rabbit antisera were used: antisera raised against the cytoplasmic tails of CD3-γ, -δ and -ε were provided by Dr. J. Coligan (National Institutes of Health, Bethesda, MD) and the antisera raised against the human TCR-α and -β chain (35) were a gift from Dr. O. Acuto (Institut Pasteur, Paris, France). The rabbit polyclonal antiserum against the COOH terminus of canine calnexin was purchased from StressGen Biotechnologies Corp. (Victoria, Canada).
The HLA-DR1–presentable peptide from influenza virus HA306–318 (36) has been synthesized in the lab by 9-fluorenylmethoxycarbonyl chemistry. Cycloheximide and iodoacetamide were purchased from Sigma Chemical Co. (St. Louis, MO).
Gel Electrophoresis.
SDS-glycine electrophoresis (SDS-PAGE) was performed as described (37). SDS-glycine gels were fluorographed using DMSO–2,5-diphenyl oxazole (Sigma Chemical Co.) and exposed to XAR-5 films (Kodak, Rochester, NY).
In Vitro Transcription and In Vitro Translation.
In vitro transcriptions were performed using T7 or SP6 RNA polymerase (Promega Corp.). RNA was equipped with a G5′ppp5′ N-cap structure either co- or posttranscriptionally according to the vendor's instructions (Promega Corp.). RNA was stored in water at −80°C. The optimal amount of RNA for translation was determined empirically for each stock of RNA prepared. Before its use in translations, the RNA was heated (80°C) for 2 min and subsequently chilled on ice.
In vitro translations were previously described (38). Reaction times were indicated in the results section. Reticulocyte lysate without supplemented dithiothreitol (DTT) (Flexi™) was purchased from Promega. Microsomes were prepared from dog pancreas as described (39).
Immunoprecipitation and Endoglycosidase H Digestion.
After translation, microsomes were pelleted by centrifugation (5415C centrifuge; Eppendorf North America, Inc., Madison, WI; 14,000 rpm, 10 min) and subsequently lysed for 30 min on ice in 1 ml NP-40 lysis buffer (0.5% NP-40, 50 mM Tris-HCl, pH 7.4, 5 mM MgCl2) or digitonin (Wako, Kyoto, Japan) lysis mix (1% digitonin, 150 mM NaCl, 50 mM Tris-HCl, pH 7.4) containing 1 mM phenylmethylsulfonyl fluoride (Sigma Chemical Co.). Insoluble debris was removed by centrifugation (14,000 rpm, 10 min). Supernatants were precleared twice with either normal rabbit or mouse serum and formalin fixed Staphylococcus aureus and then immunoprecipitated with the indicated antibodies as described elsewhere (38). Digestions with endoglycosidase H (EndoH)f (New England Biolabs, Beverly, MA) were performed on immune complexes bound to S. aureus according to the vendor's instruction.
Results
Exchange of the 5′ Noncoding and Signal Peptide–encoding Sequence Leads to Improved Translation Efficiency.
The translational performance of the mRNAs encoding the TCR subunits was optimized to ensure the likelihood of subsequent assembly of subunits translated simultaneously. We substituted the 5′ untranslated sequence as well as the signal peptide encoding sequence of all cDNAs with the corresponding sequence of the murine MHC class I molecule Kb, the in vitro transcript of which had proven to be translated with high efficiency (40). A comparison of the translation products of both sets of mRNAs bearing either the endogenous or the Kb signal sequence shows that in all cases this substitution led to an enhanced translation efficiency (Fig. 1). This holds true as well for the translation of CD3-δ, the nonmodified in vitro transcript that failed to be translated (data not shown). Since sequence substitution as described above did not affect the mobility of the translation products in SDS-PAGE, we performed all subsequent experiments with modified mRNAs.
Figure 1.
Exchange of the 5′ noncoding and signal peptide–encoding sequence leads to improved translation efficiency. In vitro transcripts of indicated TCR subunits, equipped either with the endogenous 5′ untranslated region and the sequence or with the corresponding sequence of the murine MHC class I molecule Kb (end. Kb) were translated in the presence of microsomes for 1 h at 30°C. Total microsome-associated material was analyzed by SDS-PAGE (12.5%) performed under reducing conditions.
Proper N-linked Glycosylation of TCR Subunits In Vitro.
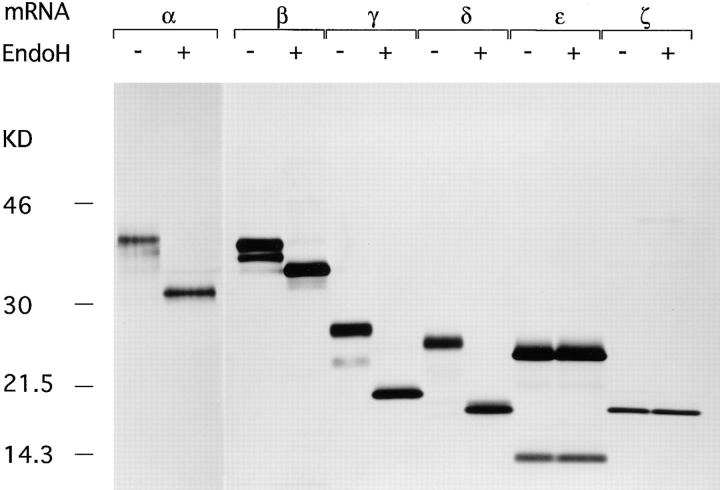
In living cells, TCR-α, -β, CD3-γ and δ, but not CD3-ε and the ζ subunit are co-translationally N-glycosylated in the lumen of the ER. To confirm proper glycosylation in vitro, we translated the mRNAs of all TCR subunits individually in the presence of microsomes, followed by recovery of the translation products by immunoprecipitation using the appropriate antibodies. Immunoprecipitates were digested with EndoH or mock digested to remove N-linked glycans (Fig. 2). The number of glycans attached could be estimated for in vitro translated TCR-α (four glycans), -β, CD3-γ and -δ (two glycans), and CD3-ε and -ζ (no glycans), all in agreement with in vivo data (2–5, 25).
Figure 2.
Proper N glycosylation of in vitro translated TCR subunits. In vitro transcripts of the indicated TCR subunits were translated for 1 h at 30°C. Immunoprecipitates obtained with the appropriate antibodies were either mock digested or digested with EndoH, and subsequently analyzed by SDS-PAGE (12.5%) performed under reducing conditions. The lower band in the anti–CD3-ε immunoprecipitates may constitute a degradation product of CD3-ε.
Proper Formation of Intra- and Intermolecular Disulfide Bridges Depends on Redox Conditions and Is Essential for Correct Folding.
The formation of disulfide bridges occurs in the lumen of the ER both co- and posttranslationally, and requires an oxidizing environment. Disulfide bonds constrain the tertiary structure and help maintain the conformation of a polypeptide chain (41–43). In this study, we used conformation-specific mAbs to monitor disulfide bond–dependent folding. TCR-β and CD4 translated in the presence of the reducing agent DTT do not acquire their native conformation, demonstrated by their failure to bind to JOVI.1, JOVI.3 (anti–TCRβ mAbs) and OKT4 (anti-CD4 mAb), respectively. However, the epitopes recognized by all three antibodies were recovered upon inclusion of oxidized glutathion (GSSG) (Fig. 3 A) in a dose-dependent manner.
Figure 3.
Redox conditions determine the proper formation of intra- and intermolecular disulfide bonds in vitro. (A) Formation of intrachain disulfide bridges and proper folding of TCR-β and CD4 is dependent on an oxidizing environment; TCR-β and CD4 were translated for 1 h at 30°C in the presence of DTT and GSSG at the indicated concentrations. Immunoprecipitations were performed with the indicated conformation-specific antibodies after lysis of the microsomes in NP-40 lysis buffer in the presence of 50 mM iodoacetamide. Total microsome-associated material as well as immunoprecipitates were analyzed by SDS-PAGE (12.5%), performed under nonreducing conditions. (B) The formation of disulfide-linked ζ dimers occurs under proper redox conditions: TCR-ζ was translated for 1 h at 30°C in the presence of DTT and GSSG at indicated concentrations. Lysates were prepared in NP-40 lysis buffer in the presence of 50 mM iodoacetamide and subjected to immunoprecipitation using the mAb TIA-2. The ζ homodimer migrates in SDS-PAGE (12.5%) performed under nonreducing as a complex with a molecular weight of 30 kD but is converted to monomeric TCR-ζ if analyzed by reducing SDS-PAGE. Total microsome-associated material was analyzed by reducing SDS-PAGE and serves as a control for the quantity of TCR-ζ expressed.
Similarly, disulfide bond–mediated homodimerization of the ζ subunit is dependent on proper redox conditions. In the presence of 4 mM DTT, ζ homodimers could not be detected with TIA-2 (anti-ζ mAb, reacting preferentially with dimeric ζ) but were recovered upon addition of GSSG (Fig. 3 B). We conclude that the proper formation of intra- and intermolecular disulfide bonds in vitro requires an oxidizing environment and is essential for correct folding.
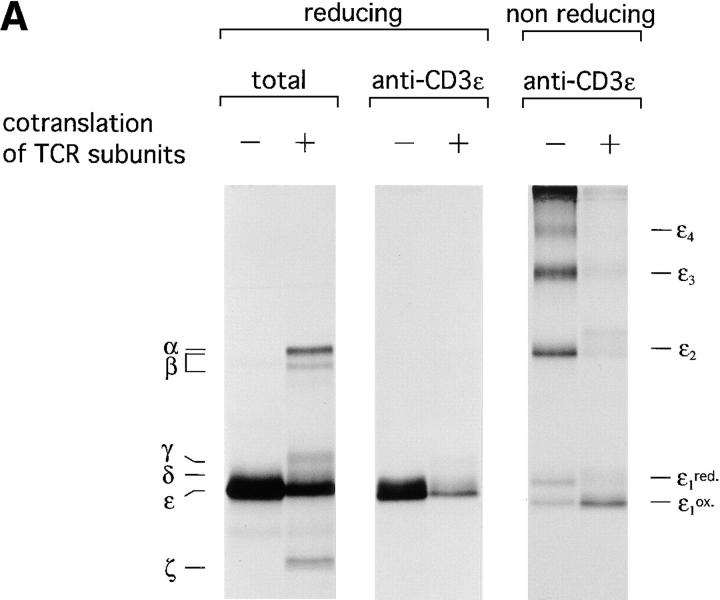
Assembly of the Human TCR Complex Translated In Vitro Is a Subunit-specific Process and Requires the Simultaneous Translation of All Polypeptides Involved.
Because the complexity of polypeptides resulting from in vitro translation is limited and their specific radioactivity is high, specificity of immunoprecipitation must be rigorously established. All TCR subunits were therefore translated separately (Fig. 4 A, left), and the microsomes lysed in digitonin lysis buffer. Lysates of the different translations were pooled and subjected to immunoprecipitation using the antibodies as indicated (Fig. 4 A), or lysates of the individual translations were immunoprecipitated with a mixture of antibodies lacking only the antibody specific for the translated polypeptide (Fig. 4 A, middle).
Figure 4.
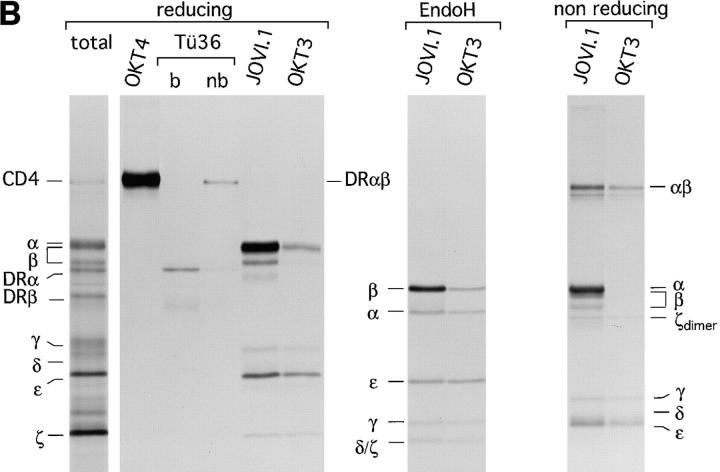
Assembly of the TCR–CD3 complex is a subunit-specific process and does not occur after lysis. (A) Test for antibody specificity confirms that in vitro assembly of the TCR–CD3 complex does not occur after lysis. (Left) Indicated mRNAs were translated individually for 1 h at 30°C, and an aliquot of the total microsome-associated material of each reaction was directly analyzed by reducing SDS-PAGE (12.5%). (Middle) Microsomes of individual translation reactions (left) were lysed in digitonin lysis buffer. Lysates were pooled to generate a mixture of labeled subunits, and subjected to immunoprecipitation using the antibodies indicated. (Right) Digitonin lysates of the individual translation reactions (left) were subjected to immunoprecipitation performed with a mixture of all antibodies used (middle), but lacking the indicated antibodies specific for the translated subunit. The lack of recovery of labeled subunits in this experiment indicates that none of the antibodies used react with the inappropriate subunits. (B) Subunit specific assembly of the TCR–CD3 complex in vitro. Nine mRNAs encoding all subunits of the TCR–CD3 complex as well as CD4 and HLA-DRα and HLA-DRβ were translated simultaneously. Microsomes were either analyzed directly by SDS-PAGE (12.5%) or subjected to immunoprecipitation using OKT4 (anti-CD4), Tü36 (anti–HLA-DR), JOVI.1 (anti–TCR-β) or OKT3 (anti-CD3). Precipitates obtained with OKT4 and Tü36 were analyzed by SDS-PAGE (12.5%) performed under reducing conditions. b, boiled; nb, non-boiled. SDS-stable DR dimers are indicated as DRαβ. Immunoprecipitates obtained with JOVI.1 and OKT3 were analyzed by SDS-PAGE (12.5%) performed under reducing and nonreducing conditions. To resolve TCR-α from TCR-β, these samples were also first digested with EndoH and then separated by SDS-PAGE (12.5%).
All of the antibodies used precipitated exclusively the appropriate chain from a polypeptide mix (Fig. 4 A, middle) but failed to recover inappropriate polypeptides (Fig. 4 A, right). This result also demonstrates that assembly of the TCR does not occur artefactually after lysis, but requires the presence of all subunits in a shared membrane compartment (see below).
To examine subunit-specific assembly of the TCR in vitro, we translated simultaneously all chains of the TCR as well as three unrelated polypeptides; the human CD4 coreceptor and the α and β chain of the human MHC class II molecule HLA-DR1 (DR). The peptide of the influenza hemagglutinin (HA306-318) was added to the reaction as a ligand of DR to promote the formation of peptide-bound SDS-stable MHC class II molecules, which migrate as SDS-stable heterodimer in SDS-PAGE (44). This control serves to show the ability of the in vitro system to support the proper assembly of this heterodimer, in accord with our earlier studies (40). After translation, microsomes were solubilized in digitonin lysis buffer, subjected to immunoprecipitation using OKT4 (anti-CD4 mAb), Tü36 (anti-DR mAb), JOVI.1 (anti–TCR-β mAb), or OKT3 (anti-CD3 mAb). The resulting immunoprecipitates were analyzed by SDS-PAGE under reducing conditions (Fig. 4 B, left). CD4 and DR were recovered only by their appropriate antibodies. A sizable proportion of DR displayed the SDS-stable phenotype, indicative of proper peptide loading and specific assembly. The immunoprecipitates obtained with JOVI.1 (anti–TCR-β) and OKT3 (anti-CD3) contained subunits of the TCR exclusively and were devoid of co-translated CD4 or DR.
Disulfide Bonding of TCR Subunits Translated In Vitro.
To assess the formation of disulfide-linked subunits, complexes recovered with JOVI.1 and OKT3 were analyzed by SDS-PAGE under nonreducing conditions (Fig. 4 B, middle). We observed the presence of α/β heterodimers and ζ homodimers, as well as the absence of monomeric α, β, and ζ subunits in complexes immunoprecipitated with OKT3. To resolve TCR-α from TCR-β, immunoprecipitates from JOVI.1 and OKT3 were digested with EndoH before separation by reducing SDS-PAGE (Fig. 4 B, right). The methionine content (TCR-α, 4 methionines; TCR-β, 6 methionines) and relative intensity of both subunits recovered with OKT3 strongly suggest a 1:1 stoichiometry for both chains, in accordance with in vivo data.
We conclude that the formation of TCR complexes as monitored by coimmunoprecipitation of the relevant subunits occurs in a specific manner. The recovery of TCR-ζ with both JOVI.1 and OKT3 indicate the formation of complete TCR complexes, since this subunit joins the TCR during the last stage of assembly (10). Moreover, specific assembly of multimolecular complexes (TCR versus DR) can occur simultaneously in an in vitro translation system, as complexes containing inappropriate subunits were not observed.
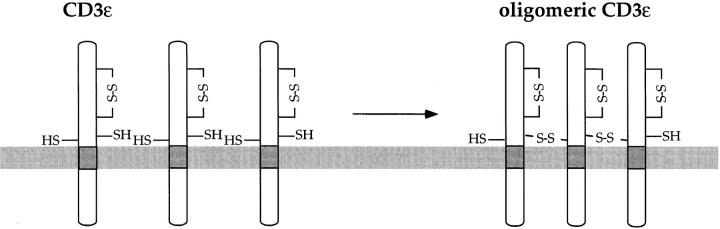
CD3-ε Translated Oligomerizes in the Absence of Related CD3 Subunits.
The CD3-ε chain contains two cysteine residues presumably involved in stabilization of the Ig fold, and two more cysteines in the lumenal domain (2), one of which is located within a few residues of the lipid bilayer (Fig. 5). The occurrence of disulfide-linked ε homodimers has been reported in vivo (45, 46), but the monomer is the prevalent form found in living cells. We investigated the biochemical properties of CD3-ε translated in vitro under both reducing and oxidizing conditions. Immunoprecipitations were performed with the anti–CD3-ε antibody in the presence of 50 mM iodoacetamide to prevent the formation of disulfide bridges after lysis. As visualized by SDS-PAGE under nonreducing conditions, translation of CD3-ε performed under oxidizing conditions resulted in oligomeric structures that collapsed to the monomer when analyzed by SDS-PAGE under reducing conditions (Fig. 6 A). Moreover, the difference in mobility for the oxidized and reduced monomeric form of CD3-ε is clearly visible (Fig. 6 A, left). We noted that it is only the oxidized form of CD3-ε that coimmunoprecipitated with fully assembled TCR complexes. We do not know whether the more rapidly migrating, oxidized form of CD3-ε contains any remaining free sulfhydryl groups, but we consider this unlikely. Inclusion of DTT in the translation reaction abrogated oligomerization but led to a monomeric polypeptide with a slightly decreased mobility, as a consequence of alkylation by iodoacetamide upon lysis. We confirmed the homooligomeric nature of CD3-ε by two-dimensional gel electrophoresis (first dimension: isoelectric focusing under nonreducing conditions; second dimension: reducing SDS-PAGE; data not shown). We conclude that CD3-ε translated in vitro in the absence of related subunits tends to form mostly disulfide-linked oligomers, a process that appears to be redox dependent.
Figure 5.
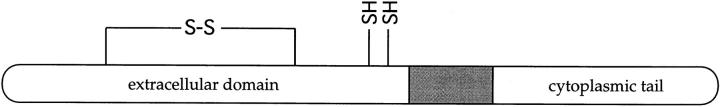
Primary structure of human CD3-ε and its proposed intra-chain disulfide bonding pattern. *Cysteine residues in the extracellular domain. The transmembrane region is underlined in the sequence and shaded in the schematic figure. A disulfide bond that stabilizes the Ig fold involves cysteine residues 27 and 76 (57). Two remaining cysteines are located close to the membrane and may be available for the formation of interchain disulfide bonds
Figure 6.
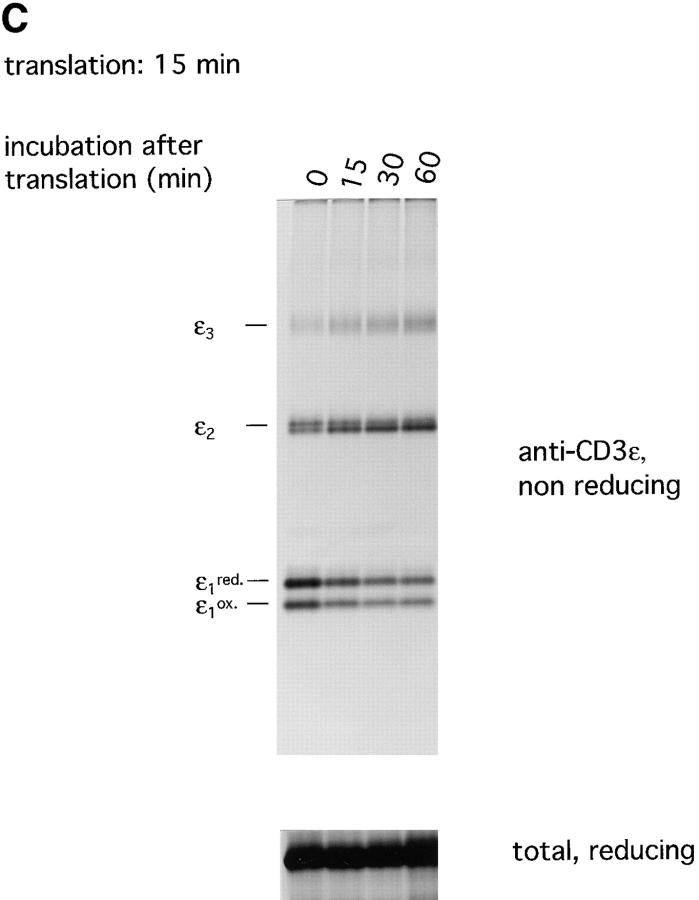
CD3-ε tends to form disulfide linked homo-oligomers. (A) CD3-ε was translated for 1 h at 30°C in the presence of DTT and GSSG at indicated concentrations. Microsomes were lysed in NP-40 lysis buffer in the presence of 50 mM iodoacetamide and immunoprecipitations were performed using an antiserum raised against the cytoplasmic tail of CD3-ε. Samples were analyzed by SDS-PAGE (12.5%) performed under both nonreducing and reducing conditions. The number of covalently linked ε subunits are indicated. red, reduced; ox, oxidized. (B) mRNAs of CD3-ε and HLA-DRβ were translated simultaneously at indicated ratios for 1 h at 30°C. Microsome-associated material as well as immunoprecipitates obtained in the presence of 50 mM iodoacetamide with the anti–CD3-ε antiserum were analyzed directly by reducing and nonreducing SDS-PAGE (12.5%). (C ) Translation of CD3-ε was stopped after 15 min by the addition of cycloheximide (20 μM) and aliquots of the translation reaction were further incubated at 30°C for the times indicated. Immunoprecipitations were performed in the presence of 50 mM iodoacetamide using the anti–CD3-ε antiserum. Microsomes were analyzed directly by SDS-PAGE (12.5%) performed under reducing conditions and immunoprecipitates were subjected to SDS-PAGE (12.5%) performed under nonreducing conditions.
To investigate whether the quantity of CD3-ε expressed in microsomes exerts any influence on the formation of disulfide-linked ε oligomers, we translated CD3-ε together with increasing amounts of an unrelated chain, the DR-β chain. Total microsome-associated material as well as immunoprecipitates of CD3-ε were analyzed by SDS-PAGE performed under both reducing and nonreducing conditions (Fig. 6 B). As the ratio between monomeric and disulfide-linked oligomeric CD3-ε does not depend on the amount of translated CD3-ε (and DR-β), the formation of homooligomers must be an inherent property of the CD3-ε subunit.
We addressed the kinetics of this reaction (Fig. 6 C) by terminating the translation of CD3-ε after 15 min by the addition of cycloheximide. Aliquots of microsomes were lysed in the presence of iodoacetamide at indicated times after translation and subjected to immunoprecipitation with anti–CD3-ε antibodies. Analysis of immune complexes by SDS-PAGE performed under nonreducing conditions reveals that dimeric CD3-ε was already prevalent at the end of the translation period, but higher order forms gradually increased with time.
The Presence of Related TCR Subunits Prevents the Formation of Disulfide Linked ε Oligomers.
The propensity of CD3-ε to form disulfide-linked oligomers was examined by translating CD3-ε either alone or together with the other subunits of the TCR. Total microsome-associated material as well as immunoprecipitates obtained from lysed microsomes were analyzed by SDS-PAGE performed under both reducing and nonreducing conditions (Fig. 7 A). Lysis in 0.5% NP-40 facilitates the visualization of CD3-ε as it leads to the disruption of all noncovalent interactions between the subunits of the TCR, but not those between CD3-ε, -γ, and -δ (6).
Figure 7.
Co-translation of related TCR subunits prevents the formation of disulfide-linked ε homooligomers. (A) CD3-ε was either translated alone or along with the remaining subunits of the TCR–CD3 complex for 1 h at 30°C. Microsomes were lysed in NP-40 lysis buffer in the presence of 50 mM iodoacetamide. Total microsome-associated material as well as immunoprecipitates obtained with the anti–CD3-ε antiserum were analyzed by SDS-PAGE (12.5%) performed as indicated under reducing and nonreducing conditions. (B) CD3-ε was translated alone and pairwise with one subunit of the TCR or with HLA-DRβ as indicated, for 1 h at 30°C. Microsomes were lysed in NP-40 lysis buffer in the presence of 50 mM iodoacetamide. Total microsome-associated material as well as immunoprecipitates obtained with the anti–CD3-ε antiserum were analyzed by SDS-PAGE (12.5%) performed under reducing and nonreducing conditions as indicated.
As shown above, CD3-ε translated on its own is found primarily in oligomeric structures. However, its behavior changes dramatically in the presence of TCR subunits, where CD3-ε is now predominantly monomeric. The role of each individual subunit in “chaperoning” CD3-ε was studied by translating ε alone and pairwise with other subunits of the TCR or with DR-β. Analysis of anti–CD3-ε immunoprecipitates by nonreducing SDS-PAGE (Fig. 7 B) shows that in the presence of TCR-α, -β, -ζ and DR-β the formation of ε oligomers is not affected. However, co-translation of CD3-γ and CD3-δ drastically shifts the equilibrium towards the formation of ε monomers. The strong interaction between CD3-ε and CD3-γ or -δ (which survives solubilization in 0.5% NP-40) may account for this effect. As binding of both γ and δ to CD3-ε appears to compete with the formation of ε homooligomers, the high degree of homology between CD3-γ, -δ, and, to some degree, CD3-ε may be particularly noteworthy and suggests the presence of a common binding motif for complex formation. Since Ig-like domains are present not only in the CD3 subunits, but also in the clonotypic α/β heterodimer and in DR-β, the mere presence of Ig-like domains is clearly not sufficient to confer the ability of blocking homooligomerization of CD3-ε.
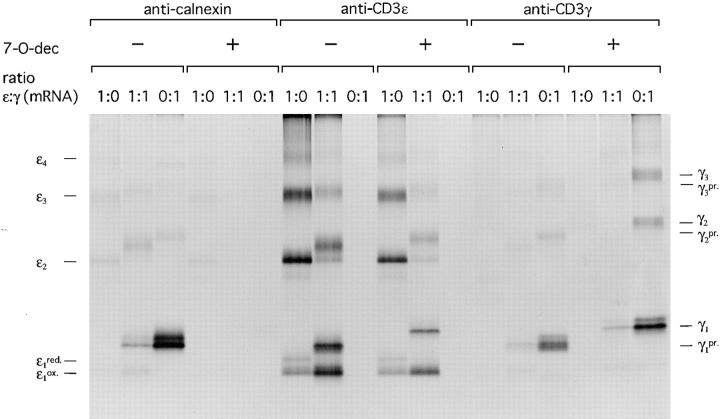
Complex Formation of Calnexin and Monomeric CD3-ε Can Be Mediated by CD3-γ, but Is Dispensable for Proper Folding of CD3-ε.
Calnexin, a molecular chaperone, binds preferentially to monoglucosylated trimming intermediates in the ER (21). Interestingly it also interacts with free CD3-ε and CD3-ε complexed with CD3-γ or -δ, although CD3-ε lacks N-linked glycans (24, 47). How calnexin binds to CD3-ε is not known. To study calnexin–CD3-ε interactions and their importance for proper folding of CD3-ε, we designed the following experiment: CD3-ε alone, ε and γ, or γ alone were translated in the absence and presence of the glucosidase inhibitor N-7-oxadecyl-dNM (7-O-dec), which blocks glucose trimming of N-linked glycans (48) and consequently prevents a glycan-dependent interaction between calnexin and glycoproteins. The change in mobility for the glycosylated γ subunit upon translation in the presence of 7-O-dec is due to glucose retention and is clearly visible on SDS-PAGE. After translation (1-h duration), microsomes were lysed in digitonin lysis buffer. Immunoprecipitates obtained with antibodies directed against calnexin, CD3-ε, and -γ were analyzed by SDS-PAGE under nonreducing conditions (Fig. 8).
Figure 8.
Calnexin interacts with monomeric CD3-ε in a glycan-dependent manner and with oligomeric forms of CD3-ε in a glycan-independent manner. CD3-ε and -γ were translated at indicated ratios for 1 h at 30°C in the absence and presence of the glucosidase inhibitor 7-O-dec. After lysis in NP-40 lysis buffer containing 50 mM iodoacetamide, immunoprecipitates were obtained with the indicated antibodies and analyzed by SDS-PAGE (12.5%) performed under nonreducing conditions. The number of covalently linked ε or γ subunits are indicated. pr., processed N-linked glycans.
Translated alone, oligomeric ε subunits could be recovered with the anticalnexin antibody regardless of the absence or presence of 7-O-dec in the translation reaction. However, the γ subunit strongly interacted with calnexin in a glycan-dependent manner, as this interaction appeared to be 7-O-dec sensitive. Monomeric CD3-ε was co-immunoprecipitated with calnexin in the presence of CD3-γ. Complete sensitivity of this interaction to 7-O-dec implies a role of CD3-ε as a bridging intermediate for CD3-γ in this binding.
Notably, 7-O-dec did not exert any effect on the oligomerization of CD3-ε translated alone or together with CD3-γ. Therefore, for both glycan-dependent and -independent modes of binding, the interaction of calnexin with the CD3-ε does itself not influence the oligomerization of CD3-ε. Hence, we conclude that the folding of ε is guided by the formation of a complex with CD3-γ and most likely also with CD3-δ, two subunits competing for binding to CD3-ε at an early stage of TCR assembly.
Discussion
Our understanding of the requirements for the assembly of the TCR stems largely from studies in which the individual subunits were transfected into suitable recipient cell lines, e.g., mutant T cell lines deficient in one of the six subunits or by transfections into nonlymphoid cells. The expression of all subunits of the TCR–CD3 complex is necessary and sufficient to accomplish its assembly in the ER and its subsequent expression at the cell surface (49, 50), without obvious involvement of lymphoid-specific factors.
The analysis of heterooligomerization of the TCR-CD3 complex in living cells is complicated by the presence of widely different pool sizes of the participating subunits, and by the egress of incompletely assembled receptors out of the ER, a complication avoided in the present study. We sought to reproduce the early stages of the TCR assembly in a complete in vitro system, with particular emphasis on the role of the CD3-ε subunit. We optimized the translation efficiency of all mRNAs involved to allow the simultaneous translation of multiple mRNAs, without having to consider the confounding effects of the 5′ untranslated sequence and signal peptides on the efficiency of translation and membrane insertion. In this fashion, we successfully translated nine different mRNAs simultaneously, and, provided proper redox conditions were imposed on the ER-derived microsomes, observed specific complex formation amongst the appropriate subunits produced in vitro. Specifically, the co-translation of class II HLA-DR–α and –β subunits in the presence of an appropriate peptide ligand, along with CD4 and all subunits of the TCR–CD3 complex, resulted in the formation of properly assembled TCR–CD3 complexes, correctly folded CD4 molecules, as well as peptide-loaded class II molecules, without cross-contamination of any of these complexes by inappropriate subunits. To our knowledge, no oligomeric membrane proteins of greater complexity have been generated by in vitro translation. The ability to obtain such complexes testifies to the robustness of these microsome-supplemented in vitro systems. The actual concentrations of the TCR subunits obtained by in vitro translation are difficult to estimate, but likely to be quite low, compared to the concentrations of the proteins in the lumen of the microsomes used. The remarkably efficient assembly and folding of the polypeptides analyzed thus suggests the possibility that these processes might be limited to small numbers of “insertion sites”, where conditions for folding and assembly are favorable. Through association of the translocon with accessory proteins (N-oligosaccharyl transferase, peptidyl disulfide isomerase, calnexin), some specialization through accomplishing this goal within the ER could occur.
The sequence of subunit interactions that leads to a complete TCR–CD3 complex has been addressed for living cells (7, 9). Given the complexity of this receptor, it is perhaps not surprising that assembly occurs in a well ordered and stepwise fashion; the formation of CD3-γ/ε and δ/ε pairs takes place early and depends on the specific interaction between their ectodomains (15). Both CD3-γ and CD3-δ compete for binding to CD3-ε (51). The crucial sites in the extracellular portion of CD3-γ for its interaction with CD3-ε are also conserved in the primary structure of CD3δ, which suggests that both CD3-γ and -δ share a common binding motif in CD3-ε (15). Next, pairs of CD3-δ/ε and γ/ε recruit TCR-α and -β, respectively. Upon formation of an intrachain disulfide bridge between TCR-α and -β, trimeric complexes merge to a hexameric εγβ–αδε complex, into which the ζ homodimer is integrated as the last remaining subunit. It is noteworthy that the stoichiometry of the CD3–TCR complex is still considered tentative (8).
When CD3-ε is translated alone, it tends to form disulfide-linked homooligomers; moreover, a substantial amount of monomeric CD3-ε is found in a reduced, i.e., incompletely, folded state. The quantity of CD3-ε translated and inserted into microsomes does not influence the ratio between monomeric (reduced and oxidized) and oligomeric structures. In all likelihood, homooligomerization of CD3-ε is an inherent property of the ε subunit; it is fast and occurs under physiological redox conditions. However, co-translation of TCR subunits under conditions that support complex formation leads to a dramatic change in the balance of newly synthesized CD3-ε subunits towards oxidized and properly folded monomers, the only form of CD3-ε recovered with the conformation-dependent mAb OKT3. The mere co-translation of CD3-γ or -δ was sufficient to keep CD3-ε primarily in an oxidized and monomeric state, indicating that CD3-γ and -δ guide the folding of CD3-ε at the initial stage of TCR assembly.
Although the disulfide bonding status for individual cysteine residues in CD3-ε has not been established, we consider it likely that the Ig fold may facilitate formation of the disulfides between the cysteines at position 27 and position 76, which stabilize this structure. The extent to which the remaining extracellular cysteine residues (at position 97 and 100) are involved in disulfide bonding in the mature TCR–CD3 complex is not known. Our data suggest that when no DTT is added, some CD3-ε may not acquire a proper disulfide bonding status. We therefore cannot indicate which cysteine residues are involved in the formation of these ε oligomers.
The presence of disulfide-linked ε homodimers in a completely assembled and functional TCR complex has been reported for several human and murine T cell lines (45, 46). However, the presence of these ε homodimers was established either using conformation-independent anti–CD3-ε reagents (46) or by examining immunoprecipitates obtained with the conformation-specific antibody OKT3 after cell surface labeling under oxidative conditions (45). Although we observe oligomerizaton of CD3-ε as a major pathway in the course of its biosynthesis in the absence of CD3-γ and -δ, we could not detect these structures as part of a complete TCR–CD3 complex by immunoprecipitation with the mAb OKT3. This antibody, the epitope of which has not been mapped accurately, only recovered CD3 subunits in immunoprecipitations when these were translated simultaneously with TCR-α and -β under redox conditions that allow proper folding (data not shown). Intracellular immunostaining of CD3-ε transiently expressed in COS cells could only be achieved with OKT3 when either CD3-γ or CD3-δ were coexpressed (52).
Furthermore, using OKT3 we could only co-immunoprecipitate TCR-α and -β as disulfide-linked heterodimers, despite the presence of free TCR-α, -β, and CD3 pairs in the lysate. Hence, OKT3 most likely recognizes a conformation-dependent epitope on CD3-ε that becomes available when CD3-γ or -δ is present, and remains stable after solubilization in detergent when the hexameric εγβ–αδε structure has been formed. A conformational change of murine CD3-ε, dependent on the presence of murine CD3-γ or -δ, has been reported (53). Taken together, our data strongly support the notion that CD3-ε requires the presence of CD3-γ or -δ to fold correctly and to allow the assembly of the TCR–CD3 complex to proceed.
CD3-ω, a nonglycosylated polypeptide of 28 kD, is expressed in human T cells and transiently interacts with pairs of CD3-δ/ε at an early stage of TCR assembly (54). Upon maturation and egress of the nascent TCR from the ER, it is released from the complex (54). CD3-ω is commonly regarded as a T cell–specific molecular chaperone and hence, is likely to be absent from the canine microsomes used in this study. The function of CD3-ω is unknown since the expression of CD3-ω is dispensable for the assembly and cell surface expression of the TCR–CD3 complex.
We have also investigated the role of the molecular chaperone calnexin in the folding of CD3-ε. Calnexin binds in a prolonged fashion to misfolded proteins or single subunits that fail to join a complex, suggesting that it may participate in the ER quality control machinery that prevents egress of incompletely folded or unassembled polypeptides from the ER (24, 55). A characteristic of calnexin is its preference for monoglucosylated trimming intermediates of N-linked glycoproteins (21). Calnexin can bind exclusively in a lectin-like fashion, i.e., independent of the folding state of the substrate (20), an observation that ties the cycle of deglucosylation and reglucosylation to glycoprotein folding (56). Although the interaction between heavy chains of MHC class I molecules and calnexin is only initiated by partially trimmed glycans, removal of glycans of class I–calnexin complexes by treatment with EndoH does not lead to the dissociation of this complex (21). Calnexin-dependent ER retention has been observed for the nonglycosylated CD3-ε subunit transfected into COS cells, based on colocalization of CD3-ε with mutant forms of calnexin, engineered with respect to their retention in the ER (24). The mode of binding between calnexin and CD3-ε therefore appears to deviate from that between calnexin and other molecules.
When we translated CD3-ε in the absence of CD3-γ and -δ, we could only detect oligomeric CD3-ε subunits in a complex with calnexin. Theoretically, this interaction could involve an unlabeled ER-resident protein that resides in the microsomes used in the translation experiments and would escape detection. Calnexin can interact in a glycan-independent manner with aggregates of the glycosylated VSV–G protein (22). Possibly some structural attribute of such VSV–G protein aggregates might be mimicked by the CD3-ε oligomer. It will be important to define more accurately and in molecular terms, how calnexin can engage in glycan-independent interactions, and whether the outcome of these interactions could, under certain circumstances, lead to the resolution of the aggregates.
In conclusion, we have demonstrated that the oligomerization of CD3-ε is a pathway essentially not used when CD3-γ and/or CD3-δ subunits are available. In this sense, the CD3-ε could serve as a subunit that monitors proper oligomerization of the CD3 complex.
Figure 9.
Homooligomerization of CD3-ε. The formation of CD3-ε homooligomers in all likelihood involves the membrane-proximal cysteine residues not involved in stabilization of the Ig fold.
Acknowledgments
We would like to thank Dr. Armin Rehm for continuous discussions and all present lab members for helpful suggestions. We thank Dr. O. Acuto, Dr. P. Anderson, Dr. J. Coligan, Dr. M. Owen, Dr. C. Terhorst, and Dr. J. Trowsdale for providing essential reagents.
Footnotes
Johannes B. Huppa was supported by fellowship of the Daimler-Benz-Stiftung (Ladenburg, Germany) and is currently a fellow of the Boehringer-Ingelheim-Fonds (Stuttgart, Germany). This work received support from the National Institutes of Health (1 PO1 AI 37833-01).
Abbreviations used in this paper: 7-O-dec, N-7-oxadecyl-dNM; DTT, dithiothreitol; EndoH, endoglycosidase H; ER, endoplasmic reticulum; GSSG, oxidized glutathion; HA, hemagglutinin; mRNA, messenger RNA; VSV–G protein, vesicular stomatitis virus G protein.
References
- 1.Klausner RD, Lippincott-Schwartz J, Bonifacino JS. The T cell antigen receptor: insights into organelle biology. Annu Rev Cell Biol. 1990;6:403–431. doi: 10.1146/annurev.cb.06.110190.002155. [DOI] [PubMed] [Google Scholar]
- 2.Gold DP, Puck JM, Pettey CL, Cho M, Coligan J, Woody JN, Terhorst C. Isolation of cDNA clones encoding the 20K non-glycosylated polypeptide chain of the human T-cell receptor/T3 complex. Nature (Lond) 1986;324:431–434. doi: 10.1038/321431a0. [DOI] [PubMed] [Google Scholar]
- 3.Krissansen GW, Owen MJ, Verbi W, Crumpton MJ. Primary structure of the T3 gamma subunit of the T3/T cell antigen receptor complex deduced from cDNA sequences: evolution of the T3γ and δ subunits. EMBO (Eur Mol Biol Organ) J. 1986;5:1799–1808. doi: 10.1002/j.1460-2075.1986.tb04429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van den Elsen P, Shepley B-A, Borst J, Coligan JE, Markham AF, Orkin S, Terhorst T. Isolation of cDNA clones encoding the 20K T3 glycoprotein of human T-cell receptor complex. Nature (Lond) 1984;312:413–418. doi: 10.1038/312413a0. [DOI] [PubMed] [Google Scholar]
- 5.Weissman AM, Hou D, Orloff DG, Modi WS, Seuanez H, O'Brian SJ, Klausner RD. Molecular cloning and chromosomal localization of the human T-cell receptor ζ chain: distinction from the molecular CD3 complex. Proc Natl Acad Sci USA. 1988;85:9709–9713. doi: 10.1073/pnas.85.24.9709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koning F, Maloy W, Coligan J. The implication of subunit interactions for the structure of the T cell receptor-CD3 complex. Eur J Immunol. 1990;20:299–305. doi: 10.1002/eji.1830200211. [DOI] [PubMed] [Google Scholar]
- 7.Kearse KP, Roberts JL, Singer A. TCRα-CD3δε association is the initial step in α/β dimer formation in murine T cells and is limiting in immature CD4+CD8+thymocytes. Immunity. 1995;2:391–399. doi: 10.1016/1074-7613(95)90147-7. [DOI] [PubMed] [Google Scholar]
- 8.Punt JA, Roberts JL, Kearse KP, Singer A. Stoichiometry of the T cell antigen (TCR) receptor: each TCR/CD3 complex contains one TCR α, one TCR β, and two CD3ε chains. J Exp Med. 1994;180:587–593. doi: 10.1084/jem.180.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall C, Berkhout B, Alarco J, Wileman T, Terhorst C. Requirements for cell surface expression of the human TCR/CD3 complex in non-T cells. Int Immunol. 1991;3:359–368. doi: 10.1093/intimm/3.4.359. [DOI] [PubMed] [Google Scholar]
- 10.Sussman JJ, Bonifacino JS, Lippincott-Schwartz J, Weissman AM, Saito T, Klausner RD, Ashwell JD. Failure to synthesize the T cell ζ chain: structure and function of a partial T cell receptor complex. Cell. 1988;52:85–95. doi: 10.1016/0092-8674(88)90533-8. [DOI] [PubMed] [Google Scholar]
- 11.Chen C, Bonifacino JS, Yuan L, Klausner RD. Selective degradation of T cell antigen receptor chains retained in a pre-Golgi compartment. J Cell Biol. 1988;107:2149–2161. doi: 10.1083/jcb.107.6.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cosson P, Lankford SP, Bonifacino JS, Klausner RD. Membrane protein association by potential intermembrane charge pairs. Nature (Lond) 1991;351:414–416. doi: 10.1038/351414a0. [DOI] [PubMed] [Google Scholar]
- 13.Bonifacino JS, Cosson P, Shah N, Klausner RD. Role of charged transmembrane residues in targeting proteins for retention and degradation within the endoplasmic reticulum. EMBO (Eur Mol Biol Organ) J. 1991;10:2783–2793. doi: 10.1002/j.1460-2075.1991.tb07827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wileman T, Kane LP, Young J, Carson GR, Terhorst C. Associations between subunit ectodomains promote T cell antigen receptor assembly and protect against degradation in the ER. J Cell Biol. 1993;122:67–78. doi: 10.1083/jcb.122.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dietrich J, Neisig A, Hou X, Wegener A-MK, Gajhede M, Geisler C. Role of CD3-γ in T cell receptor assembly. J Cell Biol. 1996;132:299–310. doi: 10.1083/jcb.132.3.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kishimoto H, Kubo RT, Yorifuji H, Nakayama T, Asano Y, Tada T. Physical dissociation of the TCR– CD3 complex accompanies receptor ligation. J Exp Med. 1995;182:1997–2006. doi: 10.1084/jem.182.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.David V, Hochstenbach F, Rajagopalan S, Brenner MB. Interaction with newly synthesized and retained proteins in the endoplasmic reticulum suggests a chaperone function for human integral membrane protein IP90 (calnexin) J Biol Chem. 1993;268:9585–9592. [PubMed] [Google Scholar]
- 18.Hochstenbach F, David V, Watkins C, Brenner MB. Endoplasmic reticulum resident protein of 90 kilodaltons associates with the T-and B-cell antigen receptors and major histocompatibility complex antigens during their assembly. Proc Natl Acad Sci USA. 1992;89:4734–4738. doi: 10.1073/pnas.89.10.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vassilakos A, Cohen-Doyle MF, Peterson PA, Jackson MR, Williams DB. The molecular chaperone calnexin facilitates folding and assembly of class I histocompatibility molecules. EMBO (Eur Mol Biol Organ) J. 1996;15:1495–1506. [PMC free article] [PubMed] [Google Scholar]
- 20.Zapun A, Petrescu SM, Rudd PM, Dwek RA, Thomas DY, Bergeron JM. Conformation-independent binding of monoglycosylated ribonuclease B to calnexin. Cell. 1997;88:29–38. doi: 10.1016/s0092-8674(00)81855-3. [DOI] [PubMed] [Google Scholar]
- 21.Ware FE, Vassilakos A, Peterson PA, Lehrman MA, Williams DB. The molecular chaperone calnexin binds Glc1Man9GlcNAc2 oligosaccharides as an initial step in recognizing unfolded glycoproteins. J Biol Chem. 1995;270:4697–4704. doi: 10.1074/jbc.270.9.4697. [DOI] [PubMed] [Google Scholar]
- 22.Cannon K, Hebert DN, Helenius A. Glycan-dependent and -independent association of vesicular stomatitis virus G protein with calnexin. J Biol Chem. 1996;271:14280–14284. doi: 10.1074/jbc.271.24.14280. [DOI] [PubMed] [Google Scholar]
- 23.Kearse KP, Williams DB, Singer A. Persistence of glucose residues on core oligosaccharides prevents association of TCR α and TCR β proteins with calnexin and results specifically in accelerated degradation of nascent TCR α proteins within the endoplasmic reticulum. EMBO (Eur Mol Biol Organ) J. 1994;13:3678–3686. doi: 10.1002/j.1460-2075.1994.tb06677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajagopalan S, Xu Y, Brenner MB. Retention of unassembled components of integral membrane proteins by calnexin. Science (Wash DC) 1994;263:387–390. doi: 10.1126/science.8278814. [DOI] [PubMed] [Google Scholar]
- 25.Hewitt CR, Lamb JR, Hayball J, Hill M, Owen MJ, O'Hehir RE. Major histocompatibility complex independent clonal T cell anergy by direct interaction of Staphylococcus aureusenterotoxin B with the T cell antigen receptor. JExp Med. 1992;175:1493–1499. doi: 10.1084/jem.175.6.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moingeon P, Stebbins CC, D'Adamio L, Lucich J, Reinherz EL. Human natural killer cells and mature T lymphocytes express identical CD3 ζ subunits as defined by cDNA cloning and sequence analysis. Eur J Immunol. 1990;20:1741–1745. doi: 10.1002/eji.1830200818. [DOI] [PubMed] [Google Scholar]
- 27.Larhammer D, Gustafsson K, Claesson L, Bill P, Wiman K, Schenning L, Sundelin J, Widmark E, Peterson PA, Rask L. Alpha chain of HLA-DR transplantation antigens is a member of the same protein superfamily as the immunoglobulins. Cell. 1982;30:153–161. doi: 10.1016/0092-8674(82)90021-6. [DOI] [PubMed] [Google Scholar]
- 28.Bell JI, Estess P, John TS, Saiki R, Watling DL, Erlich HA, McDevitt HO. DNA sequence and characterization of human class II major histocompatibility complex β chains from the DR1 haplotype. Proc Natl Acad Sci USA. 1985;82:3405–3409. doi: 10.1073/pnas.82.10.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maddon PJ, Littman DR, Godfrey M, Maddon DE, Chess L, Axel R. The isolation and nucleotide sequence of a cDNA encoding the T cell surface protein T4: a new member of the immunoglobuline gene family. Cell. 1985;42:93–104. doi: 10.1016/s0092-8674(85)80105-7. [DOI] [PubMed] [Google Scholar]
- 30.Shaw S, Ziegler A, DeMars R. Specificity of monoclonal antibodies directed against human and murine class II histocompatibility antigens as analyzed by binding to HLA-deletion mutant cell lines. Hum Immunol. 1985;12:191–211. doi: 10.1016/0198-8859(85)90336-2. [DOI] [PubMed] [Google Scholar]
- 31.Kung P, Goldstein G, Reinherz EL, Schlossman SF. Monoclonal antibodies defining distinct human T cell surface antigens. Science (Wash DC) 1979;206:347–349. doi: 10.1126/science.314668. [DOI] [PubMed] [Google Scholar]
- 32.Reinherz EL, Kung PC, Goldstein G, Schlossman SF. Separation of functional subsets of human T cells by a monoclonal antibody. Proc Natl Acad Sci USA. 1979;76:4061–4065. doi: 10.1073/pnas.76.8.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viney JL, Prosser H, Hewitt CRA, Lamb JR, Owen MJ. Generation of monoclonal antibodies against a human T cell receptor β chain expressed in transgenic mice. Hybridoma. 1992;11:701–713. doi: 10.1089/hyb.1992.11.701. [DOI] [PubMed] [Google Scholar]
- 34.Anderson P, Blue ML, O'Brian C, Schlossman SF. Monoclonal antibodies reactive with the T cell receptor ζ chain: production and characterization using a new method. J Immunol. 1989;143:1899–1904. [PubMed] [Google Scholar]
- 35.Fabbi M, Acuto O, Bensussan A, Poole CB, Reinherz E. Production and characterization of antibody probes directed at constant regions of the alpha and beta subunit of the human T cell receptor. Eur J Immunol. 1985;15:821–827. doi: 10.1002/eji.1830150815. [DOI] [PubMed] [Google Scholar]
- 36.Rothbard JB, Lechler RI, Howland K, Bal V, Eckels DD, Sekaly R, Long EO, Taylor WR, Lamb JR. Structural model of HLA-DR1 restricted T cell antigen recognition. Cell. 1988;52:515–523. doi: 10.1016/0092-8674(88)90464-3. [DOI] [PubMed] [Google Scholar]
- 37.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (Lond) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 38.Bijlmakers MJ, Benaroch P, Ploegh H. Mapping functional regions in the lumenal domain of the class II associated invariant chain. J Exp Med. 1994;180:623–629. doi: 10.1084/jem.180.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walter P, Blobel G. Preparation of microsomal membranes for cotranslational protein translocation. Methods Enzymol. 1983;96:84–93. doi: 10.1016/s0076-6879(83)96010-x. [DOI] [PubMed] [Google Scholar]
- 40.Bijlmakers MJE, Neefjes JJ, Wojcik-Jacobs EHM, Ploegh HL. The assembly of H2-Kb class I molecules translated in vitrorequires oxidized glutathione and peptide. Eur J Immunol. 1993;23:1305–1313. doi: 10.1002/eji.1830230618. [DOI] [PubMed] [Google Scholar]
- 41.Braakman I, Helenius J, Helenius A. Role of ATP and disulphide bonds during protein folding in the endoplasmic reticulum. Nature (Lond) 1992;356:260–262. doi: 10.1038/356260a0. [DOI] [PubMed] [Google Scholar]
- 42.Braakman I, Helenius J, Helenius A. Manipulating disulfide formation and protein folding in the endoplasmic reticulum. EMBO (Eur Mol Biol Organ) J. 1992;11:1717–1722. doi: 10.1002/j.1460-2075.1992.tb05223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yilla M, Doyle D, Sawyer JT. Early disulfide bond formation prevents heterotypic aggregation of membrane proteins in a cell-free translation system. J Cell Biol. 1992;118:245–252. doi: 10.1083/jcb.118.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Springer TA, Kaufman JF, Siddoway LA, Mann DL, Strominger JL. Purification of HLA-linked B lymphocyte alloantigens in immunologically active form by preparative sodium dodecyl sulfate-gel electrophoresis and studies on their subunit association. J Biol Chem. 1977;252:6201–6207. [PubMed] [Google Scholar]
- 45.Jin Y-J, Koyasu S, Moingeon P, Steinbrich R, Tarr GE, Reinherz EL. A fraction of CD3-ε subunits exists as disulfide–linked dimers in both human and murine T lymphocytes. J Biol Chem. 1990;265:15850–15853. [PubMed] [Google Scholar]
- 46.Sancho J, Chatila T, Wong RCK, Hall C, Blumberg R, Alarcon B, Geha RS, Terhorst C. T-cell antigen receptor (TCR)-α/β heterodimer formation is a prerequisite for association of CD3-ζ2into functionally competent TCR– CD3 complexes. J Biol Chem. 1989;264:20760–20769. [PubMed] [Google Scholar]
- 47.Wiest DL, Burgess WH, McKean D, Kearse KP, Singer A. The molecular chaperone calnexin is expressed on the surface of immature thymocytes in association with clonotype-independent CD3 complexes. EMBO (Eur Mol Biol Organ) J. 1995;14:3425–3433. doi: 10.1002/j.1460-2075.1995.tb07348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan A, van den Broek L, Bolscher J, Vermaas DJ, Pastoors L, van Boeckel C, Ploegh H. Introduction of oxygen into the alkyl chain of N-decyl-dNM decreases lipophilicity and results in increased retention of glucose residues on N-linked oligosaccharides. Glycobiology. 1994;4:141–149. doi: 10.1093/glycob/4.2.141. [DOI] [PubMed] [Google Scholar]
- 49.Berkhout B, Alarcon B, Terhorst C. Transfection of genes encoding the T cell receptor–associated CD3 complex into COS cells results in assembly of the macromolecular structure. J Biol Chem. 1988;263:8528–8536. [PubMed] [Google Scholar]
- 50.Carson G, Kuestner R, Ahmed A, Pettey C, Concino M. Six chains of the human T cell antigen receptor– CD3 complex are necessary and sufficient for processing the receptor heterodimer to the cell surface. J Biol Chem. 1991;266:7883–7887. [PubMed] [Google Scholar]
- 51.Geisler C. Failure to synthesize the CD3-γ chain. Consequences for the T cell antigen receptor assembly, processing and expression. J Immunol. 1992;145:2437–2445. [PubMed] [Google Scholar]
- 52.Salmeron A, Sanchez-Madrid F, Ursa MA, Fresno M, Alarcon B. A conformational epitope expressed upon association of CD3 ε with either CD3 δ or CD3 γ is the main target for recognition by anti-CD3 monoclonal antibodies. J Immunol. 1991;147:3047–3052. [PubMed] [Google Scholar]
- 53.Bonifacino JS, Suzuki CK, Lippincott-Schwartz J, Weissman AM, Klausner RD. Pre-Golgi degradation of newly synthesized T-cell antigen receptor chains: intrinsic sensitivity and the role of subunit assembly. J Cell Biol. 1989;109:73–83. doi: 10.1083/jcb.109.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neisig A, Vangsted A, Zeuthen J, Geisler C. Assembly of the T-cell antigen receptor. Participation of the CD3 ω chain. J Immunol. 1993;151:870–879. [PubMed] [Google Scholar]
- 55.Jackson MR, Cohen-Doyle MF, Peterson PA, Williams DB. Regulation of MHC class I transport by the molecular chaperone calnexin (p88, IP90) Science (Wash DC) 1994;263:384–387. doi: 10.1126/science.8278813. [DOI] [PubMed] [Google Scholar]
- 56.Hebert DN, Foellmer B, Helenius A. Glucose trimming and reglucosylation determine glycoprotein association with calnexin in the endoplasmic reticulum. Cell. 1995;81:425–433. doi: 10.1016/0092-8674(95)90395-x. [DOI] [PubMed] [Google Scholar]
- 57.Gold DP, Clevers H, Alarcon B, Dunlap S, Novotny J, Williams A, Terhorst C. Evolutionary relationship between the T3 chain of the T-cell receptor complex and the immunoglobulin gene superfamily. Proc Natl Acad Sci USA. 1987;84:7649–7653. doi: 10.1073/pnas.84.21.7649. [DOI] [PMC free article] [PubMed] [Google Scholar]