Abstract
Recent experiments have strongly suggested that the process of somatic mutation is linked to transcription initiation. It was postulated that a mutator factor loads onto the RNA polymerase and, during elongation, causes transcriptional arrest that activates DNA repair, thus occasionally causing errors in the DNA sequence. We report the analysis of the role of one of the known DNA repair systems, nucleotide excision repair (NER), in somatic mutation. Epstein–Barrvirus-transformed B cells from patients with defects in NER (XP-B, XP-D, XP-V, and CS-A) were studied. Their heavy and light chain genes show a high frequency of point mutations in the variable (V), but not in the constant (C) regions. This suggests that these B cells can undergo somatic hypermutation despite significant defects in NER. Thus, it is doubtful that NER is an essential part of the mechanism of somatic hypermutation of Ig genes. As an aside, NER seems also not involved in Ig gene switch recombination.
Ahigh level of Ig variable (V)1 region diversity arises in mature B cells during somatic hypermutation of the expressed Ig genes (1). The mutations are restricted to the V region and its immediate 5′ and 3′ flanks, extending to about 1.5 kb from the start of the V gene, whereas the promoter-upstream region and the constant (C) region generally are not mutated (2–4).
The molecular basis of somatic hypermutation is not known. Several studies have suggested that transcriptional control elements may be required for somatic mutation (3, 5, 6). To determine whether transcription was directly invovled, we created a light chain transgene whose transcriptional promoter was duplicated and placed upstream of the C region (7). B cells of these mice showed similar levels of somatic point mutations in both the V-joining (J) region and the C region, but not in the intron between these. The data strongly suggested that somatic mutation is linked to transcription initiation. To explain the link to transcription, we have proposed a model of transcription-coupled DNA repair (7). In this model, a mutator factor is bound to the RNA polymerase during initiation and remains associated with the polymerase during elongation. It is postulated that the mutator factor would cause prolonged stalling of the polymerase at some point along the first 1.5 kb of transcribed DNA. This would call into action a DNA repair complex that would cause the excision of the DNA sequence in which the stalled complex is located. Replication of the excised segment of DNA would result in occasional errors, which would eventually be preserved as point mutations.
There are a number of known repair systems, as well as possibly novel ones, that may be involved. We report the analysis of the role of certain proteins that are known to be required for nucleotide excision repair (NER) (8). The Ig genes expressed in EBV transformed B cell lines from four patients were the subjects of the study.
Materials and Methods
EBV-transformed Lymphocytes.
All cell cultures were from National Institute of General Medical Sciences Human Genetic Mutant Cell Repository, Coriell Institute for Medical Research, (Camden, NJ). Three EBV-transformed lymphocyte cultures under analysis were from xeroderma pigmentosum (XP) patients: XP-B, XP-D, XP-V, and one from a Cockayne syndrome (CS-A) patient (Table 1). The cells were grown in RPMI-1640 medium with 15% FCS. From each sample, genomic DNA and total RNA were isolated.
Table 1.
NER-defective EBV-transformed B Cell Lines Analyzed for Somatic Mutation
| XP-B | −/−: 31-yr-old female; | Cockayne symptoms: UV sensitivity; mutation in RNA splice site of XP-B gene | ||
| * (GM02252A) | ‡(3′ → 5′ DNA helicase; component of transcription factor TFIIH) | |||
| XP-D | −/−: 5-yr-old male; | Cockayne symptoms; neurological symptoms; UV sensitivity | ||
| (GM03249) | (5′ → 3′ DNA helicase; component of transcription factor TFIIH) | |||
| XP-V | −/−: 9-yr-old female; | photosensitivity; Cockayne symptoms: microcephaly; growth retardation; ataxia | ||
| (GM10902 | (function unknown; involved in NER and postreplication DNA repair) | |||
| CS-A | −/−: 13-yr-old male; | Cockayne syndrome; low UV recovery | ||
| (GM01857A) | (repair of transcriptionally active DNA; interacts with TFIIH) |
Cell repository catalog number;
Putative function of the gene product.
Primers.
(For ease of reading, the primers are written in triplets that however, are unrelated to coding triplets). CgRT, 5′-TCT TGT CCA CCT TGG TGT TGC T-3′; Cmb (reference 9; 5′-GGG GAA TTC TCA CAG GAG ACG AGG GGG AA-3′; Cgb (9) 5′-ATC AGT CGA CAA GAC CGA TGG GCC CTT GGT GGA-3′; CmRT 5′-GCC AGC TGT GTC GGA CAT GAC-3′; VH4 (10), 5′-ACT AGT CGA CCC TGT CCC TCA CCT GC(A/G)CTG TC-3′; CkRT (11), 5′-CAT CAG ATG GCG GGA AGA T-3′; HUVK1FOR (12), 5′-GGT GCA GCC ACA GTA CGT TAG ATC TCC A-3′; VH6 (13), 5′-GTA AAG CTT CAG CAG TCA GGT-3′; V26-FW1 (14), 5′-CGC CGG ATC CTG TGA GGT GCA GCT GT-3′; hc3pall (15), 5′-GAG AGA CCC CTC CCC TGG GA-3′; hclam reverse (15), 5′-AGT GTG GCC TTG TTG GCT TG-3′.
Cloning of Heavy Chain cDNAs.
Expressed IgH chain genes were first cloned by using the VH family–specific primers listed above. First-strand cDNA synthesis was carried out with either the CgRT primer, which anneals to the 3′ end of human Cγ1, Cγ2, Cγ3, and Cγ4, or the CmRT primer, which anneals to the 3′ end of human Cμ. Then, a portion of the cDNA was used as template in the PCR step carried out with one of three family-specific primers, VH4, VH6, or V26-fw1, as the 5′ primer and one of the two constant region primers, CgRT or CmRT, as 3′ primers. High fidelity DNA polymerase PFU (Stratagene, La Jolla, CA) was used. The PCR products were cloned into the BluescriptII KS+ vector (Stratagene). Automated sequencing was carried out using T3 and T7 vector primers and a dye terminator method.
Additional heavy chain clones were obtained by a rapid amplification of cDNA ends (RACE) kit (GibcoBRL, Gaithersburg, MD). Amplification of cDNA fragments was done as indicated in the manual of the RACE kit. Taq DNA polymerase was used for PCR because inosine incorporated in the anchor primer will not allow the use of PFU DNA polymerase, which has exonuclease activity. The PCR product was cloned into the EcoRV site of BluescriptII KS+ by the TA cloning method (16). Automated and manual (Sequenase kit from GIBCO BRL; T3 and T7 vector primers and anchor primer) DNA sequencing was done.
Cloning of κ and λ Chain cDNAs.
Expressed Vκ and Vλ sequences were cloned by RACE. Primers used in the reverse transcription (RT) step were hC3pall, specific for the 3′ end of human Cλ, and CkRT, specific for human Cκ. Hclam reverse primer used in amplification of Vλ is complementary to a region highly homologous in all human λ C regions, about 60 to 80 bp downstream of the J–C splice site. The HUVK1FOR primer used to amplify Vk sequences is complementary to the 3′ end of five human κ light chain J segments and to the first 14 nucleotides of the human Cκ region.
Alignment of cDNA Sequences to Genomic Sequences.
All cDNA sequences were compared with germline reference sequences of V region genes to determine base changes due to somatic hypermutation. The Vλ sequences (17) were retrieved from IMGT, the ImMunoGeneTics database (http://imgt.cnusc.fr:8104) and were aligned using the multiple sequence alignment tool Clustal W (1.4) (18) with default parameters (gap open penalty = 10), on unix station. The germline VH sequences are unpublished (F. Matsuda, unpublished data). The germline Vκ sequences are from the EMBL/GenBank.
Results
Clonality of EBV-transformed Lymphocyte Cultures.
Southern blot analysis was carried out to determine the clonality of the lymphocyte cultures (data not shown). All four cell cultures under analysis are oligoclonal. Specifically, there were two major rearranged JH-containing bands for CS-A, four major bands for XP-V, two major bands for XP-B, and four major bands for XP-D. This information led us to employ the RACE method in order to clone out all, or most of the expressed Ig genes in addition to using a conventional PCR method with V gene–family specific primers. Also, RT-PCR showed that the XP-B lymphocyte culture contains no detectable Igμ or Igκ transcript. The other three, XP-V, XP-D, and CS-A, contained Igμ and Igγ, as well as Igλ and Igκ transcripts (data not shown).
Cloning and Sequencing IgV Region Genes.
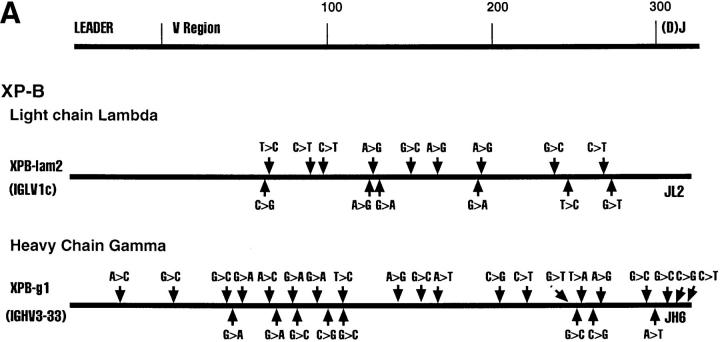
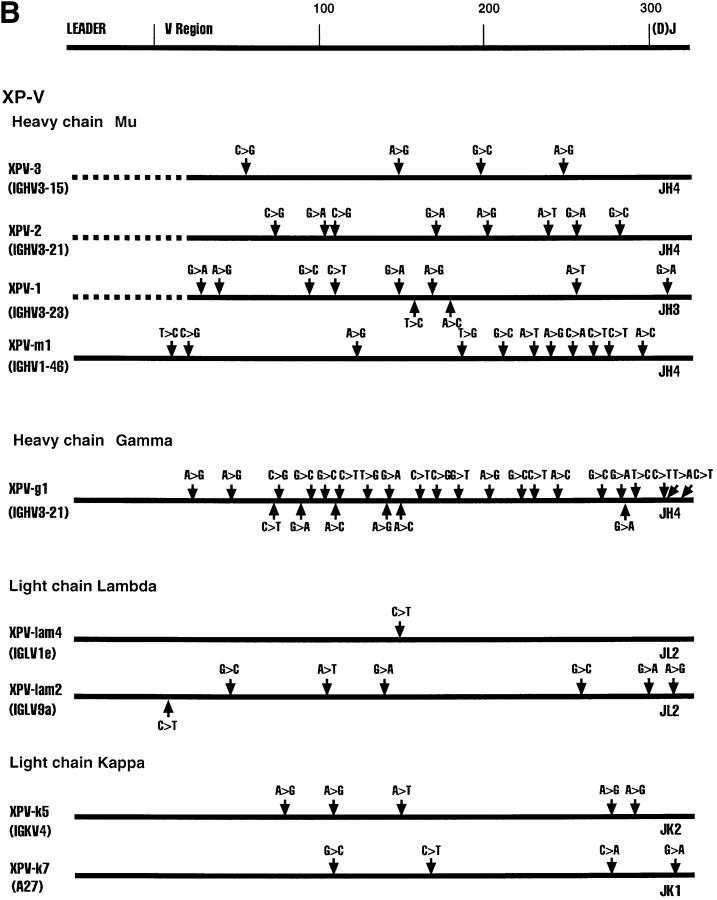
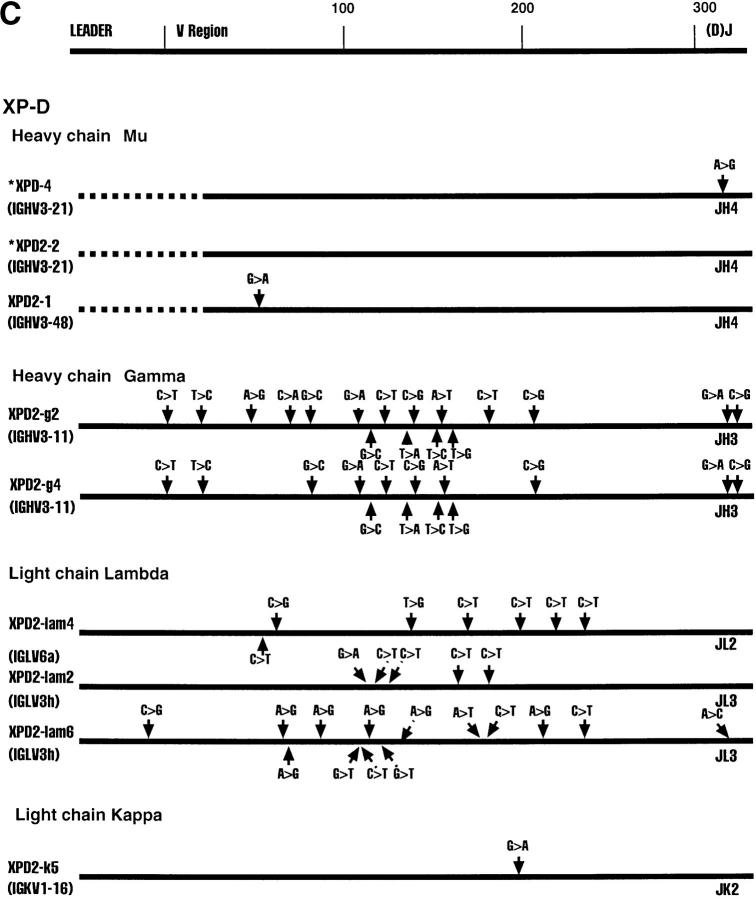
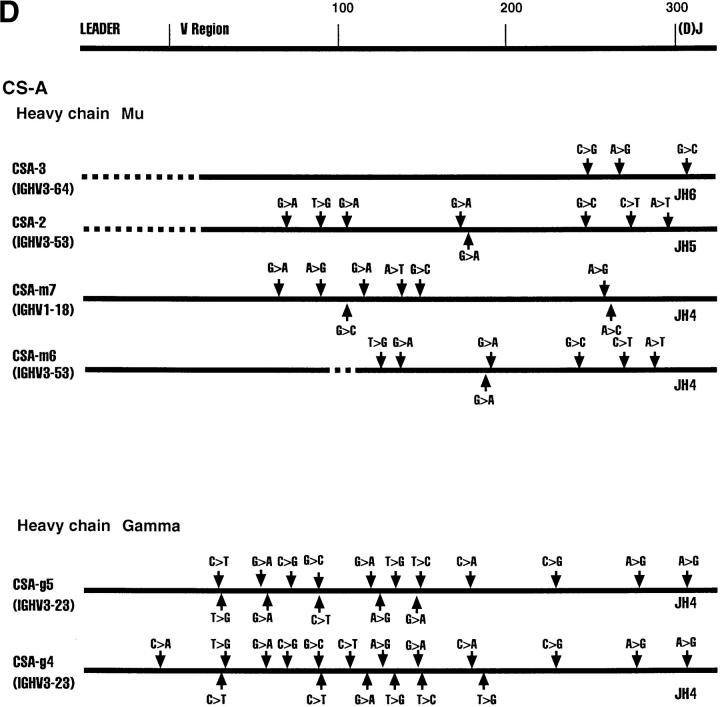
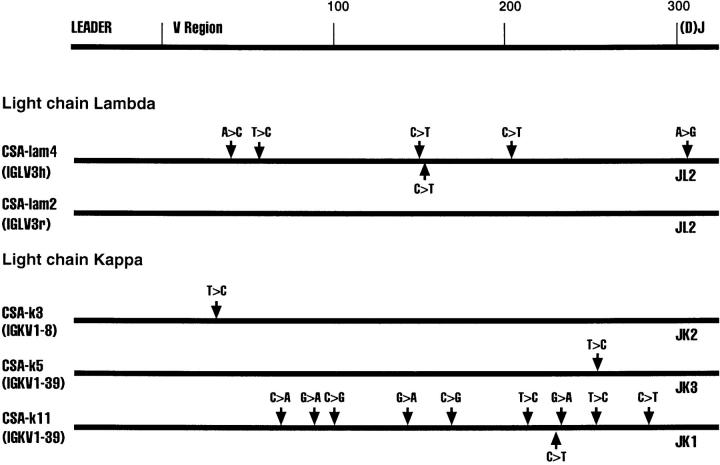
Fig. 1 shows various Ig clones obtained from each of the four cell lines and the region sequenced. We sequenced the D–J regions in the case of heavy chain genes and the J regions in the case of the light chain genes to confirm that individual clones are not represented in duplicates. Clones XPD4 and XPD2-2, for example, use the same V and J genes, but have distinctly different N regions and D regions (data not shown).
Figure 1.
Sequence alignments of heavy and light chain cDNAs from patients with XP-B, XP-D, XP-V and CS-A. On the top a general map of heavy and light chain genes is shown. The individual maps below show the approximate positions of nucleotide changes compared with germline V genes. Dotted lines represent unsequenced regions. As a control, ∼300 nucleotides of the constant regions of XPD-4, XPD2-1, XPD2-2, XPV-1, XPV-2, XPV-3, CSA-1, CSA-2, and CSA-3 were sequenced; no mutations were found. *These two clones have the same V and J, but different N and D sequences.
All Four Patients Show Evidence of Somatic Hypermutation of Their Heavy and Light Chain V Region Genes.
The sequence of each cDNA clone was aligned to germline V gene sequences. The differences between the cDNA sequence and the sequence of a germline gene that is the closest match were considered as mutations as outlined in Fig. 1. Table 2 shows that almost every sequence had mutations. The mutation frequency for individual clones ranges from 0–7.9% (Table 2). It is clear that all four patients from whom these cells are derived are capable of undergoing somatic hypermutation of Ig genes. The oldest patient, XP-B, had the highest mutation frequency, as is normally expected (9).
Table 2.
Summary of Somatic Point Mutations in XP and CS-A Cell Lines
| Cell line | Clone | Mutations* | Total bp* | Percentage of mutations | R/S‡ | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FR | CDR | |||||||||||
| XPB | -lam2 | 15 | 305 | 4.9 | 1.3 | 2.5 | ||||||
| XPB | -g1 | 28 | 475 | 5.9 | 1.2 | 2.0 | ||||||
| Total XP-B | 43 | 780 | 5.5 | 1.25 | 2.25 | |||||||
| XPD | -4 | 1 | 350 | 0.28 | ||||||||
| XPD | 2-2 | 0 | 348 | 0 | ||||||||
| XPD | 2-1 | 1 | 341 | 0.29 | 1.0 | |||||||
| XPD | 2-g2§ | 17 | 463 | 3.7 | 2.5 | 7.0 | ||||||
| XPD | 2-g4 | 14 | 474 | 2.9 | 4.0 | 3.5 | ||||||
| XPD | 2-lam4 | 7 | 342 | 2.0 | 0.25 | 2.0 | ||||||
| XPD | 2-lam2 | 5 | 318 | 1.6 | 0.25 | |||||||
| XPD | 2-lam6 | 14 | 308 | 4.5 | 1.0 | 4.0 | ||||||
| XPD | 2-k5 | 1 | 320 | 0.31 | >1.0 | |||||||
| Total XP-D | 60 | 3264 | 1.8 | 1.4 | 4.1 | |||||||
| XPV | -3 | 4 | 337 | 1.2 | >2.0 | 1.0 | ||||||
| XPV | -2 | 8 | 303 | 2.6 | 1.5 | >3.0 | ||||||
| XPV | -1 | 10 | 337 | 3.0 | 2.0 | 2.0 | ||||||
| XPV | -m1 | 11 | 413 | 2.6 | 0.7 | 2.0 | ||||||
| XPV | -g1 | 27 | 340 | 7.9 | 6.0 | 3.5 | ||||||
| XPV | -lam4 | 1 | 306 | 0.33 | <1.0 | |||||||
| XPV | -lam2‖ | 7 | 341 | 2.0 | 3.0 | >2.0 | ||||||
| XPV | -k5 | 5 | 347 | 1.4 | >1.0 | 0.3 | ||||||
| XPV | -k7 | 4 | 331 | 1.2 | >1.0 | >1.0 | ||||||
| Total XP-V | 77 | 3055 | 2.5 | 1.9 | 2.2 | |||||||
| CSA | -3 | 3 | 340 | 0.88 | >1.0 | |||||||
| CSA | -2 | 8 | 334 | 2.4 | 4.0 | >2.0 | ||||||
| CSA | -m7 | 8 | 445 | 1.8 | 1.0 | >4.0 | ||||||
| CSA | -m6 | 7 | 440 | 1.6 | 3.0 | >2.0 | ||||||
| CSA | -g5 | 16 | 472 | 3.4 | 2.5 | 7.0 | ||||||
| CSA | -g4 | 18 | 475 | 3.8 | 3.0 | 8.0 | ||||||
| CSA | -lam4 | 6 | 329 | 1.8 | 0.7 | |||||||
| CSA | -lam2 | 0 | 326 | 0 | ||||||||
| CSA | -k3‖ | 1 | 328 | 0.3 | >1.0 | |||||||
| CSA | -k5 | 1 | 330 | 0.3 | <1.0 | |||||||
| CSA | -k11 | 10 | 327 | 3.1 | 1.0 | 3.0 | ||||||
| Total CS-A | 78 | 4146 | 1.8 | 2.0 | 3.9 | |||||||
FR, framework region.
Only the base pairs sequenced in the V and J regions are included here.
Replacement/silent mutations in the V region; changes by V(D)J joining are excluded.
Stop codon due to mutation.
Out of frame: excluded from the total R/S (column 6, 7).
Discussion
This study shows that patients with defects in NER can somatically mutate Ig genes. As an aside, they must also be capable of Ig gene switch recombination, because IgG mRNAs were found in the B cells of all four patients.
The Base Changes Seen in This Study Are Due to Somatic Hypermutation.
The base pair changes seen in the cDNA sequences outlined in Fig. 1 are comparable to those seen in other studies of somatic hypermutation in human. They are considerably higher than the RT-PCR error rate of 10−4 bp (19). Although DNA repair in the cell cultures in this study is slightly compromised due to defects in components of NER, the mutation rate due to defective NER is at least five orders of magnitude lower than the mutation rate found here (20, 21). The fact that no mutations were found in the constant regions in this study also supports this. It also seems unlikely that the changes are due to polymorphism, because known polymorphism was taken into consideration and excluded when tabulating the mutations. Furthermore, there were very few changes compared with germline sequences in the leader region, the region that normally undergoes a low frequency of mutation, but where polymorphism would be as high as in the V region. Finally, there are cases where two sequences from one patient have exactly the same D and N sequences, suggesting that they arose from the same precursor cell, but where the V regions differ and thus apparently diverged during somatic mutation in different progeny cells.
Therefore, the mutations are most likely due to the somatic hypermutation process. It appears that the XP-B, XP-D, XP-V, and CS-A gene products are not required for somatic mutation of Ig genes.
It is possible that these genes involved in NER may somewhat alter the quality of the somatic point mutations. While, as is normal in somatic mutation, transitions were more frequent than transversions (Fig.1), subtle changes in unselectable nucleotides could not be determined. Almost all of the mRNAs were in an open translational reading frame (Table 2) and therefore likely selected for function. Selection is further supported by the higher R/S ratio in the CDRs than in the framework regions (Table 2). Therefore, an analysis of strand bias and hotspots would not be reliable. Such a study will now be carried out in mice that are mutant for certain NER genes.
DNA Repair and Somatic Hypermutation.
DNA repair coupled to transcription in eukaryotic cells was first proposed when it was observed that there is preferential repair of UV-induced cyclobutane pyrimidine dimers (CPD) in the transcribed strand of active genes relative to that in the nontranscribed strand (22, 23). In the model for transcription-coupled repair, the RNA polymerase complex, when stalled at a DNA lesion, such as CPD, recruits a DNA repair complex, such as the NER machinery, through binding to a transcription repair coupling factor (24, 25). Human genetic disorders like xeroderma pigmentosum (XP) and Cockayne's syndrome (CS) are attributed to defects in various components of the NER machinery (26). Several of the proteins involved in NER also play a role in transcription (8, 27). It appears that patients with defective NER genes who are not only deficient in NER, but also in transcription, exhibit symptoms of Cockayne disease (retarded growth, neurological abnormalities; reference 26). This is presumably because fetal and early postnatal development is especially sensitive to subtle defects in the transcription function. All of the NER-defective B cell lines chosen for this study were from patients who exhibited Cockayne symptoms (Table 1), because we were testing a model of transcription-coupled repair. In addition, these cells are completely or greatly deficient in the repair of UV-damaged DNA.
The XP-B gene encodes a 3′ → 5′ helicase, XP-D a 5′ → 3′ helicase. They are components of the transcription factor TFIIH and together are thought to form a bidirectional helicase (28). This helicase appears essential for the local opening of the DNA helix to allow loading of the RNA polymerase and initiation of transcription (28). This function is presumably weakened in the XP-B and XP-D patients with CS, but must still proceed sufficiently to allow overall transcription to occur. Given our findings here, these mutant proteins must also permit the loading of the postulated mutator factor and sufficient transcription to allow somatic mutation to proceed. XP-B and XP-D are also required for NER by opening the double helix around the lesion (8), again acting as components of TFIIH. It is not clear how their presence in the repair complex relates to their presence in the transcription complex (27). In any case, in the XP-B and XP-D patients whose cells we studied, NER is essentially absent. Finding somatically mutated Ig genes would suggest that classical NER is not required for somatic mutation, unless of course mutation-specific helicases are produced and can interact with the other components of NER. In another study, two siblings with XP-D without Cockayne syndrome were also found to exhibit normal levels of somatic hypermutation (14).
CS-A may be a transcription/repair-coupling factor (29). It interacts with the transcription factor TFIIH and is essential for NER of transcribed genes (30). Therefore, it was a prime candidate for a gene required for somatic mutation, if that involves NER coupled to transcription. However, CS-A does not seem to be essential for somatic mutation, suggesting once more that NER is not the basis for this process.
The XP-V gene has not been cloned, but XP-V is involved in the repair of DNA damage that interferes with DNA replication (postreplication repair) and perhaps NER (8). These patients are classified as XP, because of their photosensitivity and modest decrease in the repair of UV damage. The patient whose cells we studied had clinical symptoms of CS. However, somatic mutation of Ig genes appears normal.
From these data it appears likely that NER is not essential for somatic hypermutation, because defects in four genes required for NER allow normal levels of somatic mutation. Furthermore, in the analysis of a mouse mutant in the NER gene XP-C, normal somatic hypermutation was also found (31). A number of other genes known to be required for NER (XP-A, XP-E, XP-F, XP-G, and CS-B) have not been tested by us. We assume that their deletion would not eliminate somatic mutation either, because all the XP and CS proteins appear to be needed for NER. Thus, if the transcription coupled repair model is correct (7), another type of repair mechanism, perhaps a novel one, may be involved. These possibilities are currently being tested.
Acknowledgments
We thank V. Giudicelli for help with the alignments, B. Blomberg for suggestions concerning the amplification of human λ genes, and H. Zachau for hints about human κ genes. We are grateful to Dr. T. McKeithan for the human JH and BCR2 (control) probes.
This work was supported by National Institutes of Health grant GM38649 to U. Storb. N. Kim was supported by National Institues of Health predoctoral training grant GM07183.
Footnotes
Abbreviations used in this paper: C, constant; CPD, cyclobutane pyrimidine dimers; CS, Cockayne syndrome; J, joining; NER, nucleotide excision repair; RACE, rapid amplification of cDNA ends; V, variable; XP, xeroderma pigmentogum.
References
- 1.French D, Laskov R, Scharff M. The role of somatic hypermutation in the generation of antibody diversity. Science (Wash DC) 1989;244:1152–1157. doi: 10.1126/science.2658060. [DOI] [PubMed] [Google Scholar]
- 2.Storb, U., A. Peters, E. Klotz, N. Kim, H.M. Shen, K. Kage, and B. Rogerson. 1997. Somatic hypermutation of immunoglobulin genes is linked to transcription. Curr. Topics Microbiol. Immunol. In press. [DOI] [PubMed]
- 3.Storb U, Peters A, Klotz E, Rogerson B, Hackett J. The mechanism of somatic hypermutation studied with transgenic and transfected target genes. Semin Immunol. 1996;8:131–140. doi: 10.1006/smim.1996.0017. [DOI] [PubMed] [Google Scholar]
- 4.Storb U. Molecular mechanism of somatic hypermutation of immunoglobulin genes. Curr Opin Immunol. 1996;8:206–214. doi: 10.1016/s0952-7915(96)80059-8. [DOI] [PubMed] [Google Scholar]
- 5.Betz A, Milstein C, Gonzalez-Fernandes R, Pannell R, Larson T, Neuberger M. Elements regulating somatic hypermutation of an immunoglobulin K gene: critical role for the intron enhancer/matrix attachment region. Cell. 1994;77:239–248. doi: 10.1016/0092-8674(94)90316-6. [DOI] [PubMed] [Google Scholar]
- 6.Weber JS, Berry J, Manser T, Claflin JL. Position of the rearranged Vk and its 5′ flanking sequences determines the location of somatic mutations in the Jκ locus. J Immunol. 1991;146:3652–3655. [PubMed] [Google Scholar]
- 7.Peters A, Storb U. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity. 1996;4:57–65. doi: 10.1016/s1074-7613(00)80298-8. [DOI] [PubMed] [Google Scholar]
- 8.Friedberg, E., G. Walker, and W. Siede. 1995. DNA Repair and Mutagenesis. A.S.F. Microbiology, Washington, D.C.
- 9.Klein U, Kueppers R, Rajewsky K. Variable region gene analysis of B cell subsets derived from a 4-year-old child: somatically mutated memory B cells accumulate in the peripheral blood already at a young age. J Exp Med. 1994;180:1383–1393. doi: 10.1084/jem.180.4.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuppers R, Zhao M, Hansmann M, Rajewsky K. Tracing B cell development in human germinal centres by molecular analysis of single cells picked from histological sections. EMBO (Eur Mol Biol Organ) J. 1993;12:4955–4967. doi: 10.1002/j.1460-2075.1993.tb06189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hougs L, Barington T, Madsen HO, Ryder LP, Svejgaard A. Rapid analysis of rearranged kappa light chain genes of circulating polysaccharide-specific B lymphocytes by means of immunomagnetic beads and the polymerase chain reaction. Exp Clin Immunogenet. 1993;10:141–151. [PubMed] [Google Scholar]
- 12.Marks JD, Tristem M, Karpas A, Winter G. Oligonucleotide primers for polymerase chain reaction amplification of human immunoglobulin variable genes and design of family-specific oligonucleotide probes. Eur J Immunol. 1991;21:985–991. doi: 10.1002/eji.1830210419. [DOI] [PubMed] [Google Scholar]
- 13.Insel R, Varade W. Bias in somatic hypermutation of human VH genes. Int Immunol. 1994;6:1437–1443. doi: 10.1093/intimm/6.9.1437. [DOI] [PubMed] [Google Scholar]
- 14.Wagner S, Elvin J, Norris P, McGregor J, Neuberger M. Somatic hypermutation of Ig genes in patients with xeroderma pigmentosum (XP-D) Int Immunol. 1996;8:701–705. doi: 10.1093/intimm/8.5.701. [DOI] [PubMed] [Google Scholar]
- 15.Blomberg BB, Glozak MA, Donohoe ME. Regulation of human lambda light chain gene expression. Ann NY Acad Sci. 1995;764:84–98. [PubMed] [Google Scholar]
- 16.Maniatis, T., E. Fritsch, and J. Sambrook. 1984. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, New York.
- 17.Williams SC, Frippiat JP, Tomlinson IM, Ignativich O, Lefranc M-P, Winter G. Sequence and evolution of the human Vlambda repertoire. J Mol Biol. 1996;264:220–232. doi: 10.1006/jmbi.1996.0636. [DOI] [PubMed] [Google Scholar]
- 18.Thompson JD, Higgins DJ, Gibson TJ. Clustal W: improving the sensitivity of progressive multiple sequence alignments through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rogerson B. Somatic hypermutation of VHS107 genes is not associated with gene conversion among family members. Int Immunol. 1995;7:1225–1235. doi: 10.1093/intimm/7.8.1225. [DOI] [PubMed] [Google Scholar]
- 20.Norris P, Limb A, Hamblin A, Lehmann A, Arlett C, Cole J, Waught A, Hawk J. Immune function, mutant frequency, and cancer risk in the DNA repair defective genodermatoses xeroderma pigmentosum, Cockayne's syndrome, and trichothiodystrophy. J Invest Dermatol. 1990;94:94–100. doi: 10.1111/1523-1747.ep12873952. [DOI] [PubMed] [Google Scholar]
- 21.Norris P, Arlett C, Cole J, Lehmann A, Hawk J. Abnormal erythemal response and elevated T lymphocyte HRPT mutation frequency in Cockayne's syndrome. Brit J Dermatol. 1991;124:453–460. doi: 10.1111/j.1365-2133.1991.tb00625.x. [DOI] [PubMed] [Google Scholar]
- 22.Mellon I, Spivak G, Hanawalt P. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 23.Lommel L, Carswell-Crumpton C, Hanawalt PC. Preferential repair of the transcribed DNA strand in the dihydrofolate reductase gene throughout the cell cycle in UV-irradiated human cells. Mutation Res. 1995;336:181–192. doi: 10.1016/0921-8777(94)00055-b. [DOI] [PubMed] [Google Scholar]
- 24.Hanawalt PC. DNA repair comes of age. Mutation Res. 1995;336:101–113. doi: 10.1016/0921-8777(94)00061-a. [DOI] [PubMed] [Google Scholar]
- 25.Chalut C, Moncollin V, Egly JM. Transcription by RNA polymerase II: A process linked to DNA repair. BioEssays. 1994;16:651–655. doi: 10.1002/bies.950160910. [DOI] [PubMed] [Google Scholar]
- 26.Lambert W, Kuo H, Lambert M. Xeroderma pigmentosum. Dermatol Clinics. 1995;13:169–209. [PubMed] [Google Scholar]
- 27.Friedberg E. Relationships between DNA repair and transcription. Annu Rev Biochem. 1996;65:15–42. doi: 10.1146/annurev.bi.65.070196.000311. [DOI] [PubMed] [Google Scholar]
- 28.Hoeijmakers J, Egly J-M, Vermeulen W. TFIIH: a key component in multiple DNA transactions. Curr Opin Genet Dev. 1996;6:26–33. doi: 10.1016/s0959-437x(96)90006-4. [DOI] [PubMed] [Google Scholar]
- 29.Selby CP, Sancar A. Mechanism of transcription-repair coupling and mutation frequency decline. Microbiol Rev. 1994;58:317–329. doi: 10.1128/mr.58.3.317-329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henning K, Li L, Iyer N, McDaniel L, Reagan M, Legerski R, Schultz R, Stefanini M, Lehmann A, Mayne L, Friedberg E. The Cockayne syndrome group A gene encodes a WD repeat protein that interacts with CSB protein and a subunit of RNA polymerase II TFIIH. Cell. 1995;82:555–564. doi: 10.1016/0092-8674(95)90028-4. [DOI] [PubMed] [Google Scholar]
- 31.Shen, H.M., D.L. Cheo, E. Friedberg, and U. Storb. 1997. The inactivation of the XP-C gene does not affect somatic hypermutation or class switch recombination of immunoglobulin genes. Mol. Immunol. In press. [DOI] [PubMed]