Abstract
Although individual T lymphocytes have the potential to generate two distinct T cell receptor (TCR)-β chains, they usually express only one allele, a phenomenon termed allelic exclusion. Expression of a functional TCR-β chain during early T cell development leads to the formation of a pre-T cell receptor (pre-TCR) complex and, at the same developmental stage, arrest of further TCR-β rearrangements, suggesting a role of the pre-TCR in mediating allelic exclusion. To investigate the potential link between pre-TCR formation and inhibition of further TCR-β rearrangements, we have studied the efficiency of allelic exclusion in mice lacking the pre-TCR-α (pTα) chain, a core component of the pre-TCR. Staining of CD3+ thymocytes and lymph node cells with antibodies specific for Vβ6 or Vβ8 and a pool of antibodies specific for most other Vβ elements, did not reveal any violation of allelic exclusion at the level of cell surface expression. This was also true for pTα-deficient mice expressing a functionally rearranged TCR-β transgene. Interestingly, although the transgenic TCR-β chain significantly influenced thymocyte development even in the absence of pTα, it was not able to inhibit fully endogeneous TCR-β rearrangements either in total thymocytes or in sorted CD25+ pre-T cells of pTα−/− mice, clearly indicating an involvement of the pre-TCR in allelic exclusion.
Functional TCR genes are assembled by a program of somatic gene rearrangements from variable (Vβ)1 gene segments, diversity (Dβ) genes, and joining (Jβ) elements at the TCR-β loci and from Vα and Jα elements at the TCR-α loci. At the TCR-β locus, Dβ→ Jβ rearrangements precede Vβ→ DJβ rearrangements. Although this process of V(D)J recombination could theoretically give rise to cells with two in-frame TCR rearrangements at corresponding alleles, and thus two functional α or β TCR genes, virtually all T lymphocytes of the αβ lineage express only one particular TCR-β chain, a phenomenon referred to as allelic exclusion. Analysis of an increasing number of T cell clones and hybridomas has revealed that allelic exclusion at the TCR-β locus is largely due to the fact that αβ T cells carry as a rule only one productive TCR-β rearrangement, whereas the rearrangement on the other allele is either incomplete (DJβ) or out of frame (1). These findings are in line with the notion that a productive TCR-β rearrangement can somehow prevent further rearrangements at the TCR-β locus. Strong support for this hypothesis has been obtained in mice expressing productively rearranged TCR-β transgenes (2, 3), which enforce almost complete inhibition of endogeneous Vβ→ DJβ rearrangements, whereas Dβ→ Jβ rearrangements were essentially unimpaired. In contrast, no inhibition of endogeneous TCR-β rearrangements could be observed in mice expressing a nonproductive TCR-β transgene (3).
In mature T cells, the rearrangement status of the TCR-α locus differs from that of the TCR-β locus in that usually both alleles carry Vα→ Jα rearrangements and cells with two functional TCR-α alleles are easily detectable (1, 4, 5). In fact, in TCR-α transgenic mice there is no or only very inefficient inhibition of endogeneous Vα→ Jα rearrangements (6, 7, 8). Thus, it appears that rearrangements in the TCR-α locus continue on both alleles until a receptor is formed that can bind to thymic MHC molecules and induce positive selection, an event that leads to downregulation of RAG expression and complete termination of all TCR gene rearrangements (6, 9, 10, 11).
While, in general, TCR-α rearrangements occur relatively late during thymocyte development, primarily at the transition from the double-negative (DN) to the double-positive (DP) stage and during the DP stage itself (12, 13), TCR-β rearrangements are initiated and completed much earlier, namely at a CD4−8− (DN) stage defined by the expression of the IL-2 receptor α chain (CD25) (12, 14). Any model postulating a negative feedback of functional TCR-β chains on rearrangement at the second β allele therefore presumes a signaling function of TCR-β in the absence of TCR-α. A similar situation is encountered in B cells where IgH chains are thought to inhibit further rearrangements at the IgH locus, well before mature IgL chains become available. The discovery of the pre-B cell receptor (BCR) (15, 16) and pre-TCR (17, 18) provided likely candidates for the signaling machinery mediating allelic exclusion at the corresponding loci in the absence of mature light chains or TCRα chains, respectively, because these receptors consist, in the case of the pre-BCR, of a conventional IgH chain paired with surrogate light chains λ5 and VpreB (along with signal-transducing Igα [mb-1] and Igβ [B29] proteins) (19) and, in the case of the pre-TCR, of a conventional TCR-β chain disulfide-linked to the invariant pre-TCR-α (pTα) chain (in association with components of the CD3 complex) (20). Surprisingly, however, analysis of the limited number of mature B cells that develop in λ5-deficient and therefore pre-BCR–defective mice did not provide any evidence for violation of allelic exclusion (21). On the other hand, more recent experiments seem to indicate that allelic exclusion is not fully operating in the absence of λ5 when precursor rather than mature B cells are studied (22).
The role of the pre-TCR in allelic exclusion of the TCR-β locus has been investigated lately in mouse chimeras that were generated by injecting TCR-β–transgenic, pTα−/− embryonic stem (ES) cells into RAG−/− blastocysts (23). Analysis of ES cell–derived thymocytes in these chimeric mice revealed equivalent inhibition of endogeneous Vβ→ DJβ rearrangements in the presence and absence of the pTα chain and provided no evidence for the existence of T lymphocytes with more than one TCR-β chain (23). Here, we report on allelic exclusion in stable lines of pTα−/− and TCR-β–transgenic, pTα−/− mice rather than chimeric animals, which permitted a more rigorous analysis. Our results indicate an involvement of the pre-TCR in allelic exclusion at the TCR-β locus, but they also show that TCR-β signaling is not completely compromised in the absence of pTα, a finding that may explain why it has been difficult so far to demonstrate an effect of the pre-TCR, and by inference of the pre-BCR, on allelic exclusion when analysing pTα- (23) or λ5-deficient mice (21).
Materials and Methods
Mice.
C57BL6 (B6) mice, which were used in most experiments as wild-type controls, were purchased from IFFA-Credo (France). All other genetically modified mice (pTα−/−, TCR-β-transgenic, RAG-2−/−, and the respective intercross offspring) were bred and maintained in a specific pathogen-free facility of the Basel Institute for Immunology (Switzerland). All animals used were 6–12 wk of age. pTα-deficient mice were identified by Southern blotting of PstI-digested tail DNA using as a probe a genomic PstI–XhoI fragment (∼610 bp) corresponding to positions 5,432 to 6,046 of the pTα gene (numbering according to reference 24). TCR-β-transgenic mouse lines expressing functionally rearranged, Vβ8.2 transgenes have been described previously (2). In our experiments, we have used the line with ∼20 copies. Transgenic mice were identified by PCR with 1 μg of genomic tail DNA as template and primers specific for Vβ8.2 (5′-GCATGGGCTGAGGCTGATCCATTA-3′) and a region located immediately 3′ of the Jβ2 cluster (5′-TGAGAGCTGTCTCCTACTATCGATT-3′). Cycling conditions were as follows: 1 min at 94°C (denaturation); 30 s at 94°C; 1 min at 55°C; and 1 min 30 s at 72°C; 25 cycles. Amplification of a ∼920-bp fragment indicated the presence of TCR-β transgenes. Breeding stocks of RAG-2–deficient mice have been provided by Dr. F.W. Alt (Boston, MA). RAG-2−/− animals were identified based on the absence of B220+/surface Ig+ B lymphocytes as evidenced by double staining of peripheral blood cells with B220– PE and sheep anti-mouse Ig–FITC antibodies.
Antibodies and Flow Cytometry.
The following antibodies have been used (all purchased from PharMingen, San Diego, CA, unless stated otherwise): FITC anti–mouse CD3ε (2C11); biotin anti–mouse CD3ε (500A2); FITC anti–mouse CD4 (H129.19); R–PE anti–mouse CD4 (H129.19; GIBCO BRL, Gaithersburg, MD); FITC anti–mouse CD8α (53-6.7); R613 anti–mouse CD8α (53-6.7; GIBCO BRL); FITC anti–mouse CD11b (Mac1α) (M1/70); FITC anti–mouse CD19 (1D3); biotin anti–mouse CD25 (7D4); R–PE anti–mouse CD44 (IM7); PE anti–mouse γδ TCR (GL3); FITC anti–mouse Ly-6G (GR1) (RB6-8C5); R–PE anti–mouse CD45R/B220 (RA3-6B2); FITC anti–mouse Vβ6 (RR4-7); FITC anti–mouse Vβ8.1/8.2 (MR5-2); PE anti– mouse Vβ2 (B20.6); PE anti–mouse Vβ3 (KJ25); PE anti–mouse Vβ4 (KT4); PE anti–mouse Vβ5.1/5.2 (MR9-4); PE anti–mouse Vβ6 (RR4-7); PE anti–mouse Vβ7; PE anti–mouse Vβ8.1/8.2 (MR5-2); PE anti–mouse Vβ10b (B21.5); PE anti–mouse Vβ11 (RR3-15); PE anti–mouse Vβ13 (MR12-3); FITC sheep anti– mouse Ig F(ab′)2 fragment (Silenus, Australia). Streptavidin conjugates: Streptavidin–Tricolor (Caltag, CA) and streptavidin–APC (Molecular Probes, OR).
For flow cytometry, single cell suspensions from thymi and lymph nodes (axial, mesenteric, inguinal) were prepared in PBS containing 2% calf serum (CS). The number of viable cells was determined using a Coulter counter. Thymocytes were stained at 5 × 106 cells per ml in PBS, 2% CS containing the relevant antibodies at saturating concentrations. When pools of anti-Vβ reagents were used, it proved necessary to dialyze the antibody mix containing all first-step antibodies immediately before the staining, to eliminate the cytotoxicity of the concentrated reagents. Dialysis was performed at 4°C in ultra thimbles (Schleicher & Schuell UH 100/10; cutoff ∼10,000 MM) against two changes of PBS, 2% CS for 2 × 30 min. Phenotypes and proportions of thymocyte subsets were analyzed by three-color flow cytometry using a FACScan® (Beckton Dickinson, Mountain View, CA) and the Lysis II program. The data depicted in Figs. 2 and 3 were analyzed with the program Cellquest (Becton Dickinson). Dead cells were excluded from the analysis by forward- and side-scatter gating, when analyzing numerically small subpopulations also by addition of propidium iodine (PI) and gating on PI− cells. Four-color analyses and cell sorting were performed on a FACStar Plus® (Beckton Dickinson) equipped with a pulse processor for forward-scatter width (FSC-W) in order to gate out cell doublets.
Figure 2.
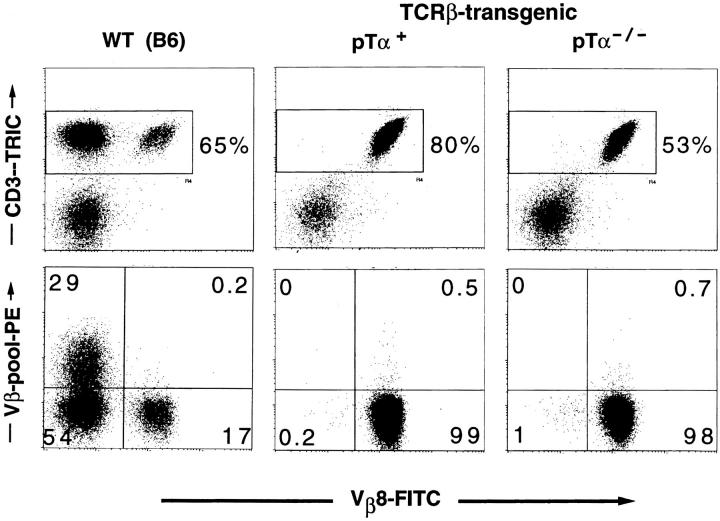
Vβ8.1/8.2 versus Vβ-pool staining of lymph node cells from TCR-β-transgenic mice expressing (pTα+) or lacking (pTα−/−) a functional pTα gene (four-color analysis). The transgene-encoded receptor contains the Vβ8.2 element. Staining and gating was done as described in Fig. 1.
Figure 3.
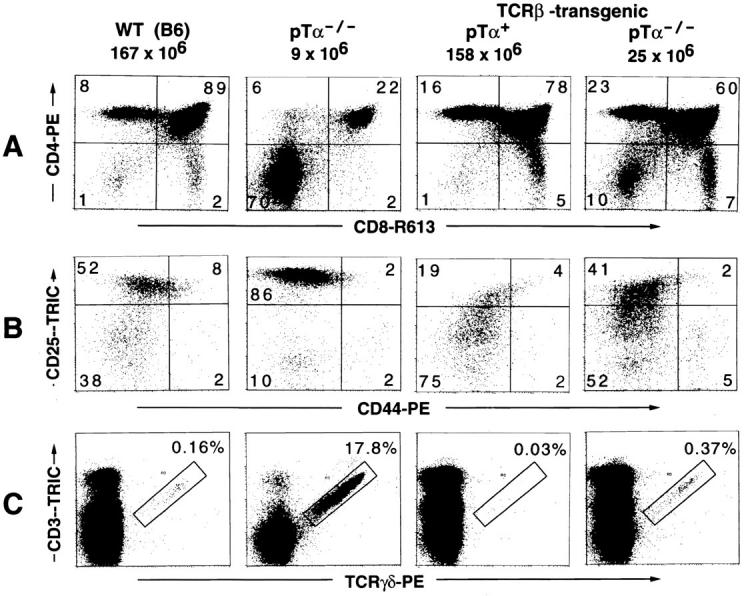
The effects of TCR-β transgenes on various aspects of thymic T cell development in the absence of pTα (the right most panel in A, B, and C). A TCR-β-transgenic mouse expressing functional pTα (TCR-β-transgenic, pTα+), a wild-type C57BL6 (WT [B6]), and a nontransgenic pTα-deficient (pTα−/−) mouse are included as controls. (A) CD4/CD8 profiles. The figures on top of each panel give the total number of thymocytes found in the particular mouse analyzed in this experiment. These values are very typical for mice with the respective genotype as seen in many similar experiments. (B) Analysis of triple-negative (CD3−CD4−CD8−) thymocytes for differential expression of the developmental markers CD25 and CD44. Thymocytes from three mice of the same genotype were pooled. CD4- and CD8-expressing cells (single positive and DP thymocytes) from B6 and TCR-β-transgenic, pTα+ mice were complement depleted (see Materials and Methods) before staining. For cytofluorometric analysis, thymocytes were incubated with an antibody mix containing CD4–FITC, CD8–FITC, CD3–FITC, GR1–FITC, MAC1–FITC, CD19–FITC (the latter three antibodies are specific for granulocytes, macrophages, and B cells, respectively), CD44–PE and CD25–biotin, and in a second step with streptavidin–Tricolor. FITC-positive cells were excluded from the analysis by electronic gating (data not shown). The cells shown in the four panels are thus highly enriched for immature triple-negative thymocytes. (C) The generation of γδ-expressing cells is suppressed in TCR-β-transgenic mice, even in the absence of pTα. Thymocytes were stained with a PE-conjugated TCR-γδ–specific antibody and CD3–biotin, in a second step with streptavidin–Tricolor. Only thymocytes that are positive for both CD3 and TCR-γδ were considered as genuine γδ-expressing cells (as defined by the rectangular gate). The data shown in A and C were obtained in the same experiment with the same mice.
Depletion of CD4 and CD8 Expressing Thymocytes.
Single-cell suspensions from thymi of three individual mice with the same genotype were pooled, resuspended in 8 ml serum-free DMEM medium, and incubated simultaneously with 1 ml of anti-CD8 (31M) and 1 ml of anti-CD4 (RL172.4) antibody supernatants on ice for 30 min. The cells were then washed in 10 ml DMEM, 2% CS and resuspended in 9 ml of the same medium. After adding 1 ml of freshly dissolved rabbit complement (Low-Tox-M, Cedarlane, Hornby, Canada), the suspension was incubated at 37°C for 40 min. Surviving cells were purified by Ficoll density–gradient centrifugation, washed and resuspended in PBS, 2% CS. The efficiency of CD4/CD8 depletion was usually more than 95% as determined by staining purified thymocytes with R–PE anti-mouse CD4 (H129.19) and R613 anti-mouse CD8α (53-6.7) antibodies, which bind epitopes distinct from those recognized by antibodies 31M and RL172.4.
Detection of Endogeneous TCR Rearrangements.
High molecular weight thymocyte DNA was prepared as follows: cells from two thymi (pTα+ mice) or three thymi (pTα−/− mice) of the same genotype were pooled and digested in proteinase K solution (50 μg/ml proteinase K, 0.5% SDS) at 50°C for 24 h. The DNA was phenol/chloroform extracted, precipitated with ethanol, recovered, and dissolved in TE buffer (10 mM Tris–HCl, pH 7.5, 1 mM EDTA) at a concentration of 100 μg/ml. PCR-based analysis of TCR-β chain rearrangements was performed using an assay originally described by van Meerwijk et al. (25) and modified by Anderson et al. (26). Vγ4–Jγ1 rearrangements (nomenclature according to reference 27) were detected in a similar fashion. Primers specific for Vβ4 (5′-CCTGATATGCGAACAGTATCTAGGC-3′), Vβ6 (5′-GAAGGCTATGATGCGTATCG-3′), Vβ11 (5′-TGCTGGTGTCATCCAAACACCTAG-3′), Vβ12 (5′-AGTTACCCAGACACCCAGACATGA-3′), and a region immediately downstream of the last Jβ segment in the Jβ2 gene cluster (5′-TGAGAGCTTGTCTCCTACTATCGATT-3′) were used to detect Vβ→ (D)Jβ rearrangements, and primers specific for Vγ4 (5′-TGTCCTTGCAACCCCTACCC-3′) and Jγ1 (5′-CAGAGGGAATTACTATGAGC-3′) were used to detect the respective rearrangement in the TCR-γ locus. Template DNA was used at three or more different concentrations in each experiment to ensure linearity of the PCR signal. As a control for the amount of template DNA, aliquots of a PCR mastermix were amplified with oligonucleotides specific for the IgM constant region (5′-CACTAGCCACACCCTTAGCAC-3′ and 5′-TGGCCATGGGCTGCCTAGCCCGGGACTT-3′). PCR amplifications were performed in 50 μl reaction buffer containing 10 mM Tris–HCl, pH 8.3, 50 mM KCl, 0.01% gelatin, 2.5 mM MgCl2, 0.2 mM of each dNTP (Pharmacia, Piscataway, NJ), 1 pmol of each primer, 0.5U Perfect Match PCR enhancer (Stratagene, La Jolla, CA), 1 U AmpliTaq DNA polymerase (Hoffmann-La Roche, Basel, Switzerland) and the indicated amount of template DNA. The amplification cycle consisted of 45 s at 94°C, 1 min 30 s at 62°C (Vβ→ [D]Jβ) or 50°C (Vγ4→ Jγ4), and 2 min at 72°C. The cycle was repeated 26 times. PCR products were separated on 1.2% agarose gels, blotted with a vacuum blotter (Bio-Rad; Richmond, CA) onto Hybond N+ nylon membrane (Amersham, Arlington Heights, IL) and detected by hybridization with end-labeled Jβ2.7-(5′-TATGAACAGTACTTCGGTCCC-3′), Jγ1-(5′-TGAATTCCTTCTGCAAATACCTTG-3′), or Cμ-specific (5′-CCTGCCCAGCACCATTTCCTTC-3′) probes, as appropriate.
Results
No Evidence for Cell Surface Expression of Two TCR-β Chains in pTα−/− Mice.
Assuming sufficient time for TCR-β rearrangements on both alleles, no feedback inhibition after a functional TCR-β chain has been generated and no selective disadvantage of cells with two functional TCR-β chains, one would expect that up to 1/5 (20%) of all TCR-β+ cells express two types of β chains, indicating the complete absence of allelic exclusion (see reference 4 for calculations). Any, even partial violation of these assumptions would reduce the percentage of double-expressing cells accordingly, albeit unpredictably. Given the low frequency of cells that express a particular Vβ element, it is clear that evidence for incomplete allelic exclusion is not easy to obtain by simple cell surface staining with antibodies specific for just two particular Vβ elements.
To alleviate this problem, we used a combination of antibodies directed against Vβ6 or Vβ8.1/8.2 elements on one hand and a pool of nine antibodies recognizing TCRs that contain most of the remaining Vβ elements on the other. Vβ6- or Vβ8.1/8.2-specific antibodies were chosen as individual antibodies, because they detect the two most frequent Vβ T cell populations on the B6 background, representing ∼5% and 15% of all TCR-αβ+ cells, respectively. To increase further the sensitivity of our analysis, we included as a third color a CD3ε-specific antibody, which allowed us to gate on CD3+ thymocytes/lymphocytes and exclude all other cells that do not express a TCR or only at low levels. Dead cells, which are known to absorb antibodies nonspecifically and which could therefore give rise to apparently double-expressing cells, were excluded not only by gating on live cells in a forward scatter/side scatter (FSC/SSC) analysis, but also by including PI staining as a fourth color and subsequent gating on PI− cells. By using the FSC pulse-width program on our flow cytometer, we could also efficiently exclude cell doublets, which otherwise would have raised the background of false TCR double-expressing cells.
Fig. 1 shows the result of a representative analysis using the Vβ8.1/8.2-specific antibody as the individual antibody and anti-Vβ2, 3, 4, 5.1/5.2, 6, 7, 9, 10b, and 13 as pooled antibodies. In the B6 control mouse, the frequency of CD3+ thymocytes and lymph node cells scoring positive for Vβ8.1/8.2 and one of the Vβs represented in the pool is extremely low (∼0.1–0.2%). This value may reflect the low proportion of T cells that genuinely express two β chains, violating allelic exclusion, as suggested by studies of human T lymphocytes (28, 29), or it could just indicate the level of nonspecific background staining in our experiment. More important in this context, however, no significant increase in the percentage of potentially double-expressing cells could be detected in the pTα−/− mouse, despite this very sensitive staining technique. These data clearly show that T cells and thymocytes expressing intermediate to high levels of the αβ TCR are allelically excluded even in pTα−/− mice, at least at the level of cell surface expression. Equivalent results were obtained in a similar staining with Vβ6 as the individual antibody versus a pool of antibodies including anti-Vβ8.1/8.2 (data not shown).
Figure 1.
Vβ8.1/8.2 versus Vβ-pool staining of thymocytes and lymph node cells from a wild-type C57BL6 (WT [B6]) and a pTα-deficient (pTα−/−) mouse (four-color analysis). Thymocytes or lymph node cells were stained with an antibody mix containing CD3–biotin, Vβ8.1/8.2– FITC, and nine distinct PE-conjugated anti-Vβ antibodies (anti-Vβ2, 3, 4, 5.1/5.2, 6, 7, 9, 10b, 13). CD3+ cells were detected with a streptavidin–Tricolor reagent. Before acquisition, PI was added. (Top) Dot blots of cells that have passed three gates: the FSC/SSC gate for live cells, the gate for PI− cells, and the pulse-width gate that eliminates cell doublets (data not shown). (Bottom) The cells depicted have passed in addition the gate for CD3high cells shown in the top.
Lack of Cell Surface Expression of Endogeneous TCR-β Chains in TCR-β–transgenic, pTα-deficient Mice.
To increase further the sensitivity of our anti-Vβ staining, we included TCR-β-transgenic mice in our analysis. As transgenic line, we used mice expressing a functional Vβ8.2 TCR-β transgene, previously shown to prevent expression of endogeneous TCR-β chains (2). As a consequence, essentially all mature T cells in these mice stain positive for Vβ8.2, while no other Vβ elements can be detected on the cell surface. To assess the efficiency of allelic exclusion in the absence of pTα, we crossed mice of the TCR-β–transgenic line with pTα-deficient animals and performed a four-color cytofluorometric analysis as described above, using Vβ8.1/8.2 to detect the transgenic TCR-β chain and all other available anti-Vβ reagents in a pool to detect expression of endogeneous TCR-β chains. Although the presence of the transgene allowed us to screen essentially all CD3+ cells for expression of two TCR-β chains rather than a fraction of cells expressing a particular Vβ element (i.e., Vβ8.1/8.2 or Vβ6) as above, we still found no evidence for cell surface expression of two distinct TCR-β chains, as endogeneous TCR-β chains could not be detected either on thymocytes (data not shown) or on mature T cells (Fig. 2) even in the absence of pTα. These data, in conjunction with those from nontransgenic pTα−/− mice, strongly suggest that functional TCR-β chains can regulate the expression of other TCR-β genes even in the absence of an intact pre-TCR.
A TCR-β Transgene Can Mediate Effects in Early T Cell Development Despite the Absence of pTα.
pTα-deficient mice exhibit a severe defect in early T cell development, which leads to a dramatic decrease in the relative proportion and absolute number of DP thymocytes and a concomitant, more than 90% reduction in total thymic cellularity (30). While analyzing Vβ surface expression in TCR-β-transgenic mice, we noticed that the presence of the TCR-β transgene on a pTα−/− background had a mild, but reproducible, effect on thymic cellularity, causing an up to threefold increase in the total number of thymocytes as compared with nontransgenic pTα−/− controls (Fig. 3; data not shown). This was usually accompanied by a reversion in the DP/DN thymocyte ratio (Fig. 3, compare the pTα−/− and the TCR-β–transgenic, pTα−/− panels), although the relative increase in the proportion of DP versus DN thymocytes varied considerably between individual TCR-β-transgenic, pTα−/− mice, possibly reflecting the weakness of the signal involved in this phenomenon.
The observation that a TCR-β transgene could influence thymic cellularity even in the absence of pTα, prompted us to analyze the effect of the β transgene on early T cell development in pTα-deficient mice. Early T cell development within the CD4−8− population can be subdivided into four discrete stages that are defined by the differential expression of CD44 and CD25 surface markers (31). In normal mice, TCR-β rearrangements take place at or shortly before the CD25+44−/low stage and only those thymocytes that manage to express a functional TCRβ chain can form a complete pre-TCR, which is required to trigger expansion and progression to the next developmental stage, defined as CD25−CD44−/low. In pTα-deficient mice, which cannot assemble an intact pre-TCR, this transition is severely impaired and relatively few CD25−CD44−/low cells are generated (Fig. 3 B, compare the two left panels). On the other hand, in pTα+ mice expressing a TCRβ transgene, essentially all CD25+ thymocytes can form a pre-TCR, resulting in a strong increase in the proportion of CD25−44−/low cells and a concomitant reduction in the percentage of CD25+CD44−/low thymocytes (Fig. 3 B, third panel from the left). Interestingly, analysis of TCR-β-transgenic mice lacking pTα revealed a marked down regulation of CD25 in comparison with nontransgenic pTα−/− mice and the appearance of a significant population of CD25−44−/low cells (compare second and fourth panel in Fig. 3 B). However, this effect of the TCR-β transgene was not as prominent as in the pTα+ background. These data indicate that the TCR-β transgene can actually influence the CD25+ compartment of CD4−8− thymocytes in the absence of pTα in a similar way as in pTα+ mice, although to a much lesser extent.
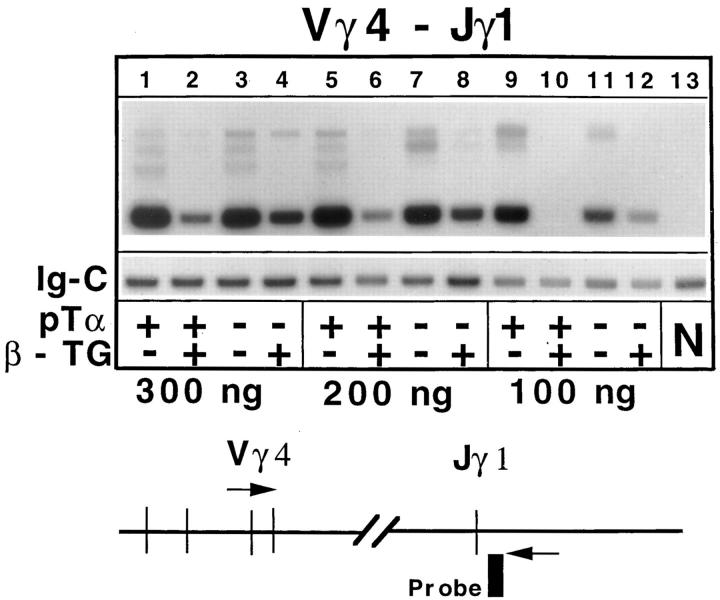
Previous experiments in normal (pTα+) mice have shown that a TCR-β transgene inhibits TCR-γ rearrangements and the generation of γδ expressing cells (32). Interestingly, the TCR-β transgene was able to suppress γ rearrangements, as measured by PCR with primers specific for Vγ4 and Jγ1, even in the absence of pTα (Fig. 4), although somewhat less efficiently than in the presence of pTα. The TCR-β transgene also mediated a strong reduction in the number of γδ cells in pTα-deficient mice (see Fig. 3 C). This effect appears particularly striking, when taking into account that the percentage and absolute number of γδ cells is strongly augmented in normal (nontransgenic) pTα−/− mice in comparison to wild-type littermates (30) (Fig. 3 C, compare the two left panels). The inhibition of TCR-γ rearrangements and the strong suppression of γδ T cell development illustrates once more that the TCR-β transgene is not innocuous despite the absence of pTα.
Figure 4.
Expression of TCR-β transgenes inhibits Vγ–Jγ rearrangements. Thymocyte DNA from four types of mice (pTα+ or pTα−/− mice with or without TCR-β transgenes [β-TG]) was amplified with primers specific for Vγ4 and Jγ1 (nomenclature according to reference 27). Three different amounts of template DNA (300 ng, 200 ng, 100 ng) were tested to ensure linearity of the signals. An aliquot of the PCR mastermix was amplified with primers specific for the constant region of IgM to provide an internal control for the amount of template used (Ig-C bands). Specific bands were detected with appropriate end-labeled oligonucleotide probes. N denotes a negative control (kidney DNA).
Introduction of a functionally rearranged TCRβ transgene into mice lacking one of the recombination activating genes (RAG-1 or RAG-2) leads to the induction of DP cells and the generation of essentially normal numbers of thymocytes (33), a result thought to reflect the physiological function of the pre-TCR. In this context, it was of interest to determine the potential effects of a TCR-β transgene on thymocyte development in RAG −/− mice lacking pTα. To this end, we crossed RAG-2 −/− mice carrying the TCR-β transgene with mice deficient for both RAG-2 and pTα, and analyzed thymi from littermates of the F2 intercross. Fig. 5 shows the result of a representative staining experiment. There was no difference between nontransgenic RAG-2 −/−, pTα+ and RAG-2 −/−, pTα−/− mice with respect to the total number of thymocytes (∼1–4 × 106) and the CD4/CD8 profiles (virtually complete absence of CD4+8+ cells) (Fig. 5, top right; data not shown). Introduction of a TCR-β transgene in RAG-2–deficient, pTα+ mice resulted in a dramatic increase in total thymocyte numbers and efficient generation of DP thymocytes, as reported previously (33). Surprisingly, expression of the TCR-β transgene in RAG-2 −/− mice lacking pTα resulted in the generation of a significant number of DP thymocytes as well, although the total thymic cellularity was raised only modestly (three- to fivefold over RAG-2 −/− controls in four independent experiments), which was also reflected by the limited reduction in the percentage of DN thymocytes in these RAG −/−, pTα−/−, TCR-β–transgenic mice (Fig. 5, bottom right). These findings are in full agreement with our observations regarding TCR-β–transgenic, pTα−/− mice with a wild-type RAG backgound, reported above. Taken together, the data indicate that a transgenic TCR-β chain can signal in the absence of pTα, promoting the differentiation of DN thymocytes into DP cells and inhibiting the generation of γδ-expressing cells, but that it cannot induce the massive cellular expansion, which normally accompanies the selection of DN CD25+44− thymocytes expressing a productive TCR-β chain, unless being associated with pTα.
Figure 5.

Expression of TCR-β transgenes in RAG-2–deficient mice in the absence of pTα induce differentiation, but no substantial proliferation of immature thymocytes. CD4 and CD8 staining of thymocytes isolated from a TCR-β–transgenic, RAG-deficient mouse (pTα+) and from a TCR-β–transgenic mouse that is RAG-deficient and in addition lacks pTα (pTα−/−). A wild-type C57BL6 (WT [B6]) and a RAG-deficient mouse are included as controls. The figures on top of each panel refer to the total number of thymocytes found in the respective animal. Similar results were obtained in four independent experiments. No differences were found between nontransgenic RAG −/− × pTα−/− and RAG−/− × pTα+ mice (data not shown).
Endogeneous Vβ→ (D)Jβ Rearrangements Are Less Efficiently Blocked in TCR-β–transgenic Mice Lacking pTα.
The expression of productively rearranged TCR-β transgenes in thymocytes of normal (pTα+) mice inhibits Vβ→ (D)Jβ rearrangements at endogeneous TCR-β loci almost completely, which is considered as a paradigm for allelic exclusion. To determine the impact of a transgenic TCR-β chain on TCR-β rearrangements in the absence of pTα, we compared the relative levels of endogeneous Vβ→ (D)Jβ rearrangements in thymocytes from TCR-β–transgenic and nontransgenic pTα+ and pTα−/− mice, using a sensitive DNA–PCR assay (see Materials and Methods; Fig. 6 A, legend). As reported previously (2, 25, 26), TCRβ-transgenic thymocytes of pTα+ mice contained nearly undetectable levels of endogeneous Vβ rearrangements (Fig. 6 A, lanes 2, 6, 10, 14, 18, 22, 26, 30, 34). Although the transgenic TCRβ chain was still able to inhibit endogeneous TCR-β rearrangements to a large degree even in the absence of pTα, the inhibition was significantly mitigated, as bands specific for endogeneous rearrangements involving all six functional Jβ2 segments were apparent at very substantial levels (Fig. 6 A, lanes 4, 8, 12, 16, 20, 24, 28, 32, 36). Similar results were obtained with two additional Vβ primers (Vβ12, Vβ14; data not shown), clearly indicating an involvement of pTα in the inhibition process.
Figure 6.
Inefficient inhibition of Vβ→ (D)Jβ rearrangements in TCR-β–transgenic mice lacking pTα. PCR-based analysis of genomic DNA isolated from unfractionated thymocytes of nontransgenic and TCR-β–transgenic pTα+ and pTα−/− mice, as indicated in the panel below the autoradiographs (β-TG, β-transgenic). Primers complementary to one of several Vβ elements were used in combination with a reverse primer positioned immediately downstream of the Jβ2 gene cluster, allowing amplification of rearranged, but not germline Vβ segments, as shown in the diagram and described previously (25, 26). Specific PCR products corresponding to rearrangements that involve each of the six functional Jβ2 elements were identified by Southern blotting with a probe hybridizing to a region immediately upstream of the Jβ2 primer-annealing site. The numbers below the panels refers to the amount of template DNA used. Ig-C indicates bands corresponding to the IgM constant region, an internal control to estimate the actual amount of template used.
Because the proportion of various thymocyte subpopulations is markedly altered in pTα-deficient mice, owing to the severe block in αβ T cell development (30), we were concerned that our results might have been influenced inappropriately by differences in the cellular composition of thymi from TCR-β–transgenic pTα+ and pTα−/− mice, respectively, although it was difficult to envisage specifically how this could have mimicked enhanced endogeneous rearrangements in the absence of pTα. Nevertheless, to exclude such a possibility completely and to allow a comparison of equivalent thymic subpopulations, we verified our PCR analysis of endogeneous Vβ→ (D)Jβ rearrangements with DNA from sorted CD25+CD44−/low DN thymocytes, which represent the developmental stage at which TCR-β rearrangements predominantly occur (12, 14). Fig. 7 shows that the result of this analysis was exactly the same as with unfractionated thymocytes; although the transgenic TCR-β chain could inhibit endogeneous Vβ→ (D)Jβ rearrangements in the absence of pTα (Fig. 7, compare lanes 4, 8, 12 with lanes 3, 7, 11), the degree of inhibition was less pronounced than in CD25+ thymocytes expressing pTα. Taken together, our data clearly indicate that the pTα chain contributes to the inhibition of endogeneous Vβ rearrangements by productive TCR-β transgenes.
Figure 7.
Analysis of genomic DNA isolated from sorted CD25+ triple-negative (CD3−4−8−) thymocytes. (A) Gates used to isolate CD25+CD44−/low triple-negative thymocytes. Thymocytes from three mice of the same genotype were pooled and stained as described in Fig. 3 B. FITC-positive cells (macrophages, granulocytes, B cells, CD4-, and CD8-expressing cells) were eliminated by electronic gating (data not shown). The panels representing the B6 and the TCR-β-transgenic, pTα+ mice have a much higher density of dots, because CD4- and CD8-expressing thymocytes from these mice (but not from pTα−/− animals) had been depleted with complement before staining, so that triple-negative thymocytes were already strongly enriched. The lower panels show the purity of the populations after sorting. (B) PCR-based analysis of genomic DNA from sorted CD25+ triple-negative thymocytes. For details, see legend to Fig. 6. In this particular experiment, Vβ11 rearrangements involving all Jβ2 elements, except Jβ2.6, were not appropriately amplified at the highest template concentration (lane 4). This phenomenon is due to excess template DNA and is not related to the mouse genotype, because a failure to amplify Vβ→ Jβ rearrangements that correspond to larger PCR bands than Vβ→ Jβ2.6 rearrangements has also been observed in a few other experiments at the highest DNA concentration when the template DNA was derived from wild-type or nontransgenic pTα−/− mice.
Discussion
The data reported here demonstrate that expression of a functional TCR-β chain efficiently prevents the expression of a second functional chain on the cell surface of thymocytes and lymph node T cells in both nontransgenic and TCR-β–transgenic mice even in the absence of pTα. At first glance, this result seems to suggest that an intact pre-TCR is not required for mediating allelic exclusion. However, impaired cell surface expression of two functional TCR-β chains in pTα−/− mice does not automatically exclude a role for the pre-TCR in implementing allelic exclusion in normal mice. Recent data indicate that TCRα chains can, at least in part, substitute for pre–TCR-α chains, because they induce the generation of almost normal numbers of thymocytes in pTα-deficient mice, when expressed at a relatively early developmental stage (34, 35). Moreover, a recent analysis of pTα−/− × TCR-α−/− and pTα−/− × TCR-δ−/− mice has provided strong evidence that a significant proportion of the αβ thymocytes that develop in pTα−/− mice are actually derived from precursors that have been selected for further development based on the presence of a functional TCR-β chain and early expression of a conventional TCR-α chain (35). The underlying assumption that some Vα→ Jα rearrangements and TCR-α expression can occur already in the CD25+ DN subpopulation is supported by a PCR-based analysis of TCR-α rearrangements in sorted CD25+ thymocytes from normal mice (36). If early expression of TCRα chains in a few CD25+ pre-T cells was responsible for the generation of most mature αβ T cells in pTα−/− mice, the early formation of an αβ TCR in these cells might as well be accountable for the allelic exclusion of TCR-β chains that is observed despite the absence of pTα.
A priori, it would also be possible that neither the TCR-α nor the pTα chain are involved in mediating allelic exclusion and that TCR-β chains can signal independently, even in the absence of these two partner chains. Our analysis of TCR-β-transgenic, pTα−/− mice clearly demonstrates that TCR-β chains can signal without being associated with a pTα chain, resulting in marked effects on early T cell development; for instance, a small increase in the number of DP thymocytes, partial downregulation of CD25 in the pre-T cell population, significant inhibition of Vγ→ Jγ and Vβ→ (D)Jβ rearrangements, suppression of the generation of γδ-expressing cells and, most striking, induction of DP thymocyte formation in RAG −/− × pTα−/− mice. These effects of a transgenic TCR-β chain in the absence of pTα could be due to the association of TCR-β chains with some other proteins, like TCR-α, TCR-δ (37), or the hypothetical VpreT subunit (20), giving rise to a signaling complex that can assume part but not all of pre-TCR function. The idea that the transgenic TCR-β chain mediates its effects in the absence of pTα via a signal-transducing complex including CD3 components is consistent with the observation that a functionally rearranged TCR-β transgene can no longer inhibit Vβ→ (D)Jβ rearrangements in CD3-deficient mice (B. Malissen, personal communication).
The question arises whether the observed effects of the transgenic TCR-β chain in the absence of pTα have some physiological relevance or whether they are solely due to a peculiarity of the transgenic system; for instance, expression of the TCR-β transgene at inappropriately high levels. In this context, it should be mentioned that the TCR-β chains in our TCR-β–transgenic mice can be expressed on thymocytes in gpi-linked form, a feature not found in nontransgenic animals (38). If the significant inhibition of endogeneous Vβ→ (D)Jβ rearrangements in pTα-deficient mice was due to a transgenic artifact, one might anticipate a variation in the degree of inhibition when analyzing different TCR-β–transgenic lines. This may explain, why Xu et al. (23) have seen an equally strong inhibition of endogeneous TCR-β rearrangements in both pTα+ and pTα−/− thymocytes, while in our experiments the potentially artifactual, transgene-specific effects of the TCR-β chain may be fortuitously less pronounced, allowing us to detect significant differences in the absence and presence of pTα. Moreover, the use of a stable line of transgenic mice allowed us to compare endogeneous rearrangements within the same transgenic background, which was not possible in the chimeric mice of Xu et al. (23), because they were generated from pTα+ and pTα−/− ES cell clones that had been transfected individually with the respective TCR-β transgene, almost certainly giving rise to pTα+ and pTα−/− animals with distinct transgene copy numbers and/or insertion sites. Whatever the reason for the discrepant results may be, our observation that the inhibition of endogeneous rearrangements at the TCR-β locus is less profound in the absence of pTα, despite the capacity of the transgenic β chain to signal in part independently of pTα, clearly indicates a role of pTα and the pre-TCR in the regulation of Vβ→ (D)Jβ rearrangements. The importance of an intact pre-TCR for allelic exclusion at the level of TCR gene rearrangements therefore may be even more obvious in normal (nontransgenic) mice, in which a newly formed, functional TCR-β chain is by definition expressed at physiological levels and therefore possibly less prone to pTα-independent signal transduction. This will be assessed by comparing the rearrangement status at the TCR-β locus in a statistically significant number of sorted DN CD25+ thymocytes from pTα-deficient mice and wild-type littermates at the single cell level.
Acknowledgments
We thank C. Laplace for skillful and efficient technical support; M. Dessing and A. Pickert for expert assistance with cell sorting and four-color cytofluorometric analysis; W. Metzger, E. Wagner, and B. Aschwanden for their care in maintaining the mouse colonies; J. Hatton and R. Schulze for oligonucleotide synthesis; B. Pfeiffer and H. Spalinger for photography, and T. Rolink and S. Gilfillan for critical reading of the manuscript. A. Krotkova is affiliated with the Vavilov Institute of General Genetics, Russian Academy of Sciences, Moscow. H. von Boehmer is supported by the Human Frontier Science Foundation. The Basel Institute for Immunology was founded and is supported by F. Hoffmann-La Roche, Basel.
Footnotes
Abbreviations used in this paper: BCR, B cell receptor; CS, calf serum; D, diversity; DN, double-negative; DP, double-positive; ES, embryonic stem; FSC, forward scatter; J, joining; PI, propidium iodine; pTα, pre-TCRα; SP, single-positive; SSC, side scatter; V, variable.
References
- 1.Malissen M, Trucy J, Jouvin-Marche E, Cazenave P-A, Scollay R, Malissen B. Regulation of TCRα and β gene allelic exclusion during T-cell development. Immunol Today. 1992;13:315–322. doi: 10.1016/0167-5699(92)90044-8. [DOI] [PubMed] [Google Scholar]
- 2.Uematsu Y, Ryser S, Dembiç Z, Borgulya P, Krimpenfort P, Berns A, von Boehmer H, Steinmetz M. In transgenic mice the introduced functional T cell receptor β gene prevents expression of endogeneous β genes. Cell. 1988;52:831–841. doi: 10.1016/0092-8674(88)90425-4. [DOI] [PubMed] [Google Scholar]
- 3.Krimpenfort P, Ossendorp F, Borst J, Melief C, Berns A. T cell depletion in transgenic mice carrying a mutant gene for TCRβ. Nature (Lond) 1989;341:742–746. doi: 10.1038/341742a0. [DOI] [PubMed] [Google Scholar]
- 4.Casanova JL, Romero P, Widman C, Kourilsky P, Maryanski JL. T cell receptor genes in a series of class I major histocompatibility complex–restricted cytotoxic T lymphocyte clones specific for a Plasmodium berghei nonapeptide: implications for T cell allelic exclusion and antigen-specific repertoire. J Exp Med. 1991;174:1371–1383. doi: 10.1084/jem.174.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Padovan E, Casorati G, Dellabona P, Meyer S, Brockhaus M, Lanzavecchia A. Expression of two T cell receptor α chains: dual receptor T cells. Science (Wash DC) 1993;262:422–424. doi: 10.1126/science.8211163. [DOI] [PubMed] [Google Scholar]
- 6.Borgulya P, Kishi H, Uematsu Y, von Boehmer H. Exclusion and inclusion of α and β T cell receptor alleles. Cell. 1992;69:529–537. doi: 10.1016/0092-8674(92)90453-j. [DOI] [PubMed] [Google Scholar]
- 7.Hardardottir F, Baron JL, Janeway CA., Jr T cells with two functional antigen-specific receptors. Proc Natl Acad Sci USA. 1995;92:354–358. doi: 10.1073/pnas.92.2.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heath WR, Miller JFAP. Expression of two α chains on the surface of T cells in T cell receptor transgenic mice. J Exp Med. 1995;178:1807–1811. doi: 10.1084/jem.178.5.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brändle D, Müller C, Rülicke T, Hengartner H, Pircher H. Engagement of the T cell receptor during positive selection in the thymus down-regulates RAG-1 expression. Proc Natl Acad Sci USA. 1992;89:9529–9533. doi: 10.1073/pnas.89.20.9529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petrie HT, Livak F, Schatz DG, Strasser A, Crisp IN, Shortman K. Multiple rearrangements in T cell receptor α chain genes maximize the production of useful thymocytes. J Exp Med. 1993;178:615–622. doi: 10.1084/jem.178.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kouskoff V, Vonesch J-L, Benoist C, Mathis D. The influence of positive selection on RAG expression in thymocytes. Eur J Immunol. 1995;25:54–58. doi: 10.1002/eji.1830250111. [DOI] [PubMed] [Google Scholar]
- 12.Petrie HT, Livak F, Burtrum D, Mazel S. T cell receptor gene recombination patterns and mechanisms: cell death, rescue, and T cell production. J Exp Med. 1995;182:121–127. doi: 10.1084/jem.182.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson A, de Villartay J-P, MacDonald HR. T cell receptor δ gene rearrangement and T early α (TEA) expression in immature αβ lineage thymocytes: implications for αβ/γδ lineage commitment. Immunity. 1996;4:37–45. doi: 10.1016/s1074-7613(00)80296-4. [DOI] [PubMed] [Google Scholar]
- 14.Godfrey DI, Kennedy J, Mombaerts P, Tonegawa S, Zlotnik A. Onset of TCR-β gene rearrangement and role of TCR-β expression during CD3−CD4−CD8−thymocyte differentiation. J Immunol. 1994;152:4783–4792. [PubMed] [Google Scholar]
- 15.Sakaguchi N, Berger CN, Melchers F. Isolation of a cDNA copy of an RNA species expressed in murine pre-B cells. EMBO (Eur Mol Biol Organ) J. 1986;5:2139–2147. doi: 10.1002/j.1460-2075.1986.tb04477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kudo A, Melchers F. A second gene, VpreBin the λ5 locus of the mouse which appears to be selectively expressed in pre-B lymphocytes. EMBO (Eur Mol Biol Organ) J. 1987;6:2267–2272. doi: 10.1002/j.1460-2075.1987.tb02500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groettrup M, Ungewiss K, Azogui O, Palacios R, Owen MJ, Hayday AC, von Boehmer H. A novel disulfide-linked heterodimer on pre-T cells consists of the T cell receptor β chain and a 33 kD glycoprotein. Cell. 1993;75:283–294. doi: 10.1016/0092-8674(93)80070-u. [DOI] [PubMed] [Google Scholar]
- 18.Saint-Ruf C, Ungewiss K, Groettrup M, Bruno L, Fehling HJ, von Boehmer H. Analysis and expression of a cloned pre-T cell receptor gene. Science (Wash DC) 1994;266:1208–1212. doi: 10.1126/science.7973703. [DOI] [PubMed] [Google Scholar]
- 19.Melchers F, Karasuyama H, Haasner D, Bauer S, Kudo A, Sakaguchi N, Jameson B, Rolink A. The surrogate light chain in B cell development. Immunol Today. 1993;14:60–68. doi: 10.1016/0167-5699(93)90060-X. [DOI] [PubMed] [Google Scholar]
- 20.von Boehmer H, Fehling HJ. Structure and function of the pre-T cell receptor. Annu Rev Immunol. 1997;15:433–452. doi: 10.1146/annurev.immunol.15.1.433. [DOI] [PubMed] [Google Scholar]
- 21.Kitamura D, Kudo A, Schaal S, Müller W, Melchers F, Rajewsky K. A critical role for the λ5 protein in B cell development. Cell. 1992;69:823–831. doi: 10.1016/0092-8674(92)90293-l. [DOI] [PubMed] [Google Scholar]
- 22.Löffert D, Ehlich A, Müller W, Rajewsky K. Surrogate light chain expression is required to establish immunoglobulin heavy chain allelic exclusion during early B cell development. Immunity. 1996;4:133–144. doi: 10.1016/s1074-7613(00)80678-0. [DOI] [PubMed] [Google Scholar]
- 23.Xu Y, Davidson L, Alt FW, Baltimore D. Function of the pre-T-cell receptor α chain in T-cell development and allelic exclusion at the T-cell receptor β locus. Proc Natl Acad Sci USA. 1996;93:2169–2173. doi: 10.1073/pnas.93.5.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fehling HJ, Laplace C, Mattei M-G, Saint-Ruf C, von Boehmer H. Genomic structure and chromosomal location of the mouse pre-T-cell receptor alpha gene. Immunogenetics. 1995;42:275–281. doi: 10.1007/BF00176445. [DOI] [PubMed] [Google Scholar]
- 25.van Meerwijk JPM, Blüthmann H, Steinmetz H. T-cell specific rearrangement of T-cell receptor β transgenes in mice. EMBO (Eur Mol Biol Organ) J. 1990;9:1057–1062. doi: 10.1002/j.1460-2075.1990.tb08210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson, S.J., K.M. Abraham, T. Nakayama, A. Singer, and R.M. Perlmutter. 1992. Inhibition of T-cell receptor β-chain gene rearrangement by overexpression of the non-receptor protein tyrosine kinase p56lck. EMBO (Eur. Mol. Biol. Organ.) J. 11:4877–4886. [DOI] [PMC free article] [PubMed]
- 27.Itohara S, Mombaerts P, Lafaille J, Iacomini J, Nelson A, Clarke AK, Hooper ML, Farr A, Tonegawa S. T cell receptor δ gene mutant mice: independent generation of αβ T cells and programmed rearrangements of γδ TCR genes. Cell. 1993;72:337–348. doi: 10.1016/0092-8674(93)90112-4. [DOI] [PubMed] [Google Scholar]
- 28.Davodeau F, Peyrat MA, Romagné F, Necker A, Hallet MM, Vié H, Bonneville M. Dual T cell receptor β chain expression on human T lymphocytes. J Exp Med. 1995;181:1391–1398. doi: 10.1084/jem.181.4.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Padovan E, Giachino C, Cella M, Valitutti S, Acuto O, Lanzavecchia A. Normal T lymphocytes can express two different T cell receptor β chains: implications for the mechanism of allelic exclusion. J Exp Med. 1995;181:1587–1591. doi: 10.1084/jem.181.4.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fehling HJ, Krotkova A, Saint-Ruf C, von Boehmer H. Crucial role of the pre-T-cell receptor α gene in development of αβ but not γδ T cells. Nature (Lond) 1995;375:795–798. doi: 10.1038/375795a0. [DOI] [PubMed] [Google Scholar]
- 31.Fehling HJ, von Boehmer H. Early αβ T cell development in the thymus of normal and genetically altered mice. Curr Opin Immunol. 1997;9:263–275. doi: 10.1016/s0952-7915(97)80146-x. [DOI] [PubMed] [Google Scholar]
- 32.von Boehmer H, Bonneville M, Ishida I, Ryser S, Lincoln G, Smith RT, Kishi H, Scott B, Kisielow P, Tonegawa S. Early expression of a T-cell receptor β-chain transgene suppresses rearrangement of the Vγ4 gene segment. Proc Natl Acad Sci USA. 1988;85:9729–9732. doi: 10.1073/pnas.85.24.9729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shinkai Y, Koyasu S, Nakayama K, Murphy KM, Loh DY, Reinherz EL, Alt FW. Restoration of T cell development in RAG-2–deficient mice by functional TCR transgenes. Science (Wash DC) 1993;259:822–825. doi: 10.1126/science.8430336. [DOI] [PubMed] [Google Scholar]
- 34.Bruno L, Fehling HJ, von Boehmer H. The αβ T cell receptor can replace the γδ receptor in the development of γδ lineage cells. Immunity. 1996;5:343–352. doi: 10.1016/s1074-7613(00)80260-5. [DOI] [PubMed] [Google Scholar]
- 35.Buer J, Aifantis I, DiSanto JP, Fehling HJ, von Boehmer H. Role of different T cell receptors in the development of pre-T cells. J Exp Med. 1997;185:1541–1547. doi: 10.1084/jem.185.9.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mertsching E, Wilson A, MacDonald HR, Ceredig R. T cell receptor α gene rearrangement and transcription in adult thymic γδ cells. Eur J Immunol. 1997;27:389–396. doi: 10.1002/eji.1830270208. [DOI] [PubMed] [Google Scholar]
- 37.Hochstenbach F, Brenner MB. T-cell receptor δ-chain can substitute for α to form a βδ heterodimer. Nature (Lond) 1989;340:562–565. doi: 10.1038/340562a0. [DOI] [PubMed] [Google Scholar]
- 38.Groettrup M, von Boehmer H. T cell receptor β chain dimers on immature thymocytes from normal mice. Eur J Immunol. 1993;23:1393–1396. doi: 10.1002/eji.1830230633. [DOI] [PubMed] [Google Scholar]