Abstract
The early growth response (Egr)-1 is a zinc finger–containing transcription factor belonging to the immediate–early genes. Its expression in CD4/CD8 double negative (DN) immature thymocytes suggests that Egr-1 expression may be involved in early thymocyte development. In transgenic mice overexpressing Egr-1 in a recombinase-activating gene–deficient background, thymocytes bypassed the block at the CD25+CD44− DN stage and matured to the immature CD8 single-positive (ISP) cell stage, but not further to the CD4/CD8 double-positive (DP) cell stage. When these mice were irradiated, thymocytes did develop to the DP stage, suggesting transcriptional induction of additional genes by irradiation that are required to promote thymocyte development from the ISP to the DP stage. These results provide genetic evidence for two distinct steps during early thymocyte development from the CD25+CD44− DN to the DP stage. The first step, from the CD25+CD44− DN to the ISP stage, can be entirely promoted by overexpression of Egr-1.
Intrathymic α/βT cell development can be grossly separated into three phases based on the configuration of TCR genes and the expression of cell-surface markers including the CD4 and CD8 coreceptors. In the earliest phase, thymocytes are characterized by TCR loci in germline configuration and thus, no surface expression of any TCR protein. The most immature cells identified in this first phase are present in small numbers and express c-kit and low levels of CD4 (1). Thymocyte maturation is accompanied by loss of CD4 and onset of CD44 expression, immediately followed by CD25 expression (the CD25+CD44+ CD4/CD8 double-negative [DN]1 stage) and finally, down-regulation of CD44 and c-kit expression (2). This final maturation stage of the first phase is defined as the CD25+CD44− DN stage, during which rearrangement of TCR-β gene begins and functional TCR-β chains are expressed on the cell surface associated with the pre–TCR-α chain (3) as a pre-TCR complex. Several cytokine and cytokine receptor–deficient mice have implied that this first phase of thymocyte development appears entirely dependent on growth factors. IL-7/IL-7 receptor (4, 5), IL-2 receptor–γ (common γ chain) (6–8), and c-kit (9) –deficient mice revealed a mild, but not complete, block of early thymocyte development. More recently, Rodewald et al. reported a complete block before TCR-β gene rearrangement in c-kit/common γ chain double mutant mice (10).
The second phase of thymocyte development begins with expression of the pre-TCR complex. Several mutant mice have revealed that expression of the pre-TCR complex plays a key role during subsequent development. Recombinase-activating gene deficient (RAG−/−) mice in which TCR-β genes cannot be rearranged and hence no pre-TCR complex is expressed on the cell surface, have complete block of thymocyte development at the CD25+CD44− stage (11, 12). In addition, TCR-β−/− (13) and pre–TCR-α−/− mice (3) show greatly reduced numbers of CD4+CD8+ double-positive (DP) cells. This first checkpoint regulated by the pre-TCR complex is termed β selection (14). When the pre-TCR complex is expressed and its signaling is initiated, thymocytes lose CD25 expression and, in mice, most start to express CD8 molecules. After this immature CD8 single-positive (ISP) stage (15–18), thymocytes begin to express CD4 molecules and become CD4/CD8 DP cells. During this process, TCR-α genes undergo rearrangement, and the TCRα/TCRβ (TCR-α/β) complex is expressed on the cell surface. The second checkpoint for the final steps of thymocyte maturation is positive/negative selection, which depends on the interaction between TCR- α/β and MHC complexes, but is also influenced by the CD4 and CD8 coreceptors. Cells surviving selection can achieve development to mature to CD4 or CD8 single-positive cells, exit the thymus, and enter the periphery. Although these developmental processes are well described phenotypically, the molecular mechanisms involved in developmental control is still unclear.
The molecular events during the second phase of thymocyte development between the two checkpoints for β selection and TCR-α/β selection are elusive because a possible ligand for the pre-TCR complex, which plays a key role in this period, has not been identified. Although a recent report suggested CD81, a transmembrane 4 superfamily protein, as a potential candidate for the pre-TCR ligand (19), this is unlikely because CD81-deficient mice (20, 21) have normal thymocyte development. Alternatively, this issue has been studied intensively using RAG−/− mice. Several experimental manipulations of the mutant mice either by irradiation (22, 23), CD3 ligation (24, 25) as well as introduction of activated lck (26) or activated ras (27) transgenes effectively bypass β selection and promote thymocyte development to the DP stage in the absence of pre-TCR expression. All of these stimuli somehow compensate for pre-TCR– mediated signaling. Because all of these manipulations can activate the Ras protein (28), a ras-related pathway could play an important role in β selection as well as in further developmental progression to the DP stage. Downstream of Ras, however, multiple pathways may be involved which have not yet been dissected. Intracellular signals during this rapid developmental process must be differentially controlled, and rapidly responding transcriptional regulators able to elicit a cascade of changes in gene expression should be necessary.
Immediate–early genes, expression of which is rapidly induced after cell-surface receptor ligation without de novo protein synthesis (29), are strong candidates for such rapid response mediators. Many of these genes encode transcription factors such as c-jun and c-fos, as well as the zinc finger–containing early growth response (Egr) family members (30, 31). Of these Egr family members, early growth response (Egr)-1, also known as krox-24, NGFI-A, zif-268, and Tis-8, has been studied most intensively for its involvement in T cell function and activation, because of its rapid expression after TCR ligation (32–35). However, Egr-1 is expressed not only in mature T cells, but also in thymocytes. Shao et al. recently reported the expression of Egr-1 in DN immature thymocytes, suggesting a potential involvement in β selection (36). Nevertheless, very little functional information is available concerning the possible involvement of Egr-1 in thymocyte development.
In this report, a potential role for Egr-1 expression in thymocyte development is described. First, endogenous Egr-1 expression in DN thymocytes from wild-type (C57Bl/6) mice was dissected in detail. Second, by generating RAG-2−/− transgenic mice that overexpress Egr-1 in immature thymocytes, the functional effects of Egr-1 expression on β selection as well as further development of thymocytes were evaluated.
Materials and Methods
Mice.
All mice used here were maintained in the specific pathogen-free facility of the Basel Institute for Immunology. Matched sets of littermates with transgene-positive and -negative genotypes were used in all experiments.
Generation of Transgenic Mice.
A λ clone containing a 16-kb genomic DNA fragment including the whole Egr-1 sequence (clone GE-1) was isolated by screening a genomic DNA library from 129/SVJ mice (Stratagene Corp., La Jolla, CA) with an Egr-1 cDNA fragment cloned into the EcoRI site of pBluescript (32P-labeled Egr-1 cDNA fragment: positions 121–1,970, numbering according to ref. 33). A 2.3-kb ApaI–BglII fragment was subcloned and then the 5′ end ApaI site was modified to a BamHI site by T4 DNA polymerase and a BamHI linker. The BamHI– BglII fragment was cloned into the BamHI site of the plck-human growth hormone (hGH) expression vector (37). The complete plasmid (plck-Egr) was digested with NotI and the transgene fragment no longer containing vector sequence was purified by Geneclean II (BIO101, Vista, CA). DNA was microinjected into fertilized eggs of C57Bl/6 × DBA2 F2 mice (BDF2). Resulting founders were screened for transgene by Southern blot and PCR. Three independent founders carried the transgene. All three mice were backcrossed within RAG-2−/− background.
Northern Blot Analysis.
Total RNA was isolated from thymus using RNAzolB solution (Tel-Test. Inc., Friendswood, TX) either from transgenic mice or negative littermates with a RAG-2+/− background. 15 μg total RNA was denatured, electrophoresed on 1% agarose gel, blotted on nylon membrane, and then hybridized with 32P-labeled Egr-1 cDNA fragment.
Reverse Transcriptase PCR.
Thymocytes were stained for CD4, CD8, CD25, and CD44, and then appropriate cell populations were sorted by FACstar® (Becton Dickinson, Mountain View, CA). From 50,000 cells of each population, total RNA was isolated by RNAzolB solution (Tel-Test, Inc.), and cDNA was synthesized by reverse transcriptase with random-hexamer primers. PCR was performed with titrated amounts of this cDNA as template, using a set of primers: 5′-TGCTCCTGGCTTTTGGCCTGCTCTG (positions 26–50 of the hGH cDNA sequence, from ATG, the start codon) and 5′-GTTGTGTGAGTTTGTGTCGAACTT (positions 534–511) for transgene expression, 5′-GCAGATCTCTGACCCGTTCGG (positions 300–320 of the mouse Egr-1 cDNA, according to ref. 33) and 5′-CCGAGTCGTTTGGCTGGGATA (positions 630–610) for Egr-1 expression, 5′-ATGGATGACGATATCGCTGCG (positions 80–101, regarding ref. 38), and 5′-CATGTTCAATGGGGTACTTCA (positions 300–280) for β actin expression. PCR reactions were performed as described; 10% of each reaction was analyzed on a Southern blot with either an hGH, Egr-1, or β actin cDNA probe.
Serological Reagents and Flow Cytometry.
Reagents used for staining T cells and subsets thereof were as described (39–41, and references therein). Thymus, lymph node, and spleen cells were stained with saturating levels of mAbs and analyzed using a FACScan® cytometer.
Cell Cycling Analysis.
100,000 ISP thymocytes were sorted and fixed in 70% ethanol at 4°C overnight. Cells were collected by centrifugation and then resuspended in 500 μl of RNase A (0.5 mg/ml in 0.1 M Tris, pH 7.5, 0.1 M NaCl; Sigma Chemical Co., St. Louis, MO) and incubated for 30 min at 37°C. 500 μl of pepsin (1 mg/ml in 0.4% HCl; Sigma Chemical Co.) was added, mixed well, and incubated for 15 min at 37°C. 1 ml of ethidium bromide (0.02 mg/ml in 0.2 M Tris, pH 8.5, 0.5% BSA) was then added, mixed well, and incubated for another 15 min at room temperature with protection from light. After the incubation, the sample was analyzed directly using a FACScan® with the double discriminating module.
TCR Rearrangement Assay.
100,000 thymocytes were sorted from various populations. Cells were digested in cell lysis buffer; 1× SSC, 10 mM Tris, pH 7.4, 1 mM EDTA, 0.5% SDS containing protenase K (Sigma Chemical Co.) at 0.1 mg/ml in a 55°C water bath with vigorous shaking for 2 h. Samples were treated with phenol/chloroform solution once and then ethanol precipitated. DNA corresponding to 5,000 cells was used as template for PCR reactions. PCR reactions were performed for 25, 30, 35, and 40 cycles under the following conditions: 94°C for 1 min, 60°C for 1.5 min, 72°C for 1.5 min in a total 20 μl of volume. Primers used for Vα rearrangement detection were 5′-CAGAAGGTGCAGCAGAGCCCAGAA (Vα5H), 5′-ACTGTCTCTGAAGGAGCCTCTCTG (Vα2C), 5′-ACCCAGACAGAAGGCCTGGTCACT (VαF3), and 5′-GACCCTATTACTCACATACTTGGCTTG (VαTT11); and for Vβ rearrangement detection were, 5′-CCTGATTGGTCAGGAAGGGC (Vβ6), 5′-TCCCTGATGGGTAGAAGGCC (Vβ8), and 5′-TAACACGAGGAGCCGAGTGC (Jβ2.5), as described (42). 10% of each reaction was analyzed on a Southern blot with a 32P-labeled internal oligo as a probe, for Vα, 5′-GAAAGCAGAGTCCCAATTCCAAAG, for Vβ, 5′-CTGGCCCAAAGTACTGGGTG. As a control, the β actin gene was amplified from each template using primers 5′-TTGAGACCTTCAACACCCCAG (positions 450–471, according to ref. 37) and 5′-CGAAGTCTAGAGCAACATAGC (positions 750–730), and then hybridized with a β actin cDNA fragment.
Results
Endogenous Egr-1 Expression in DN Thymocytes of Wild-type Mice.
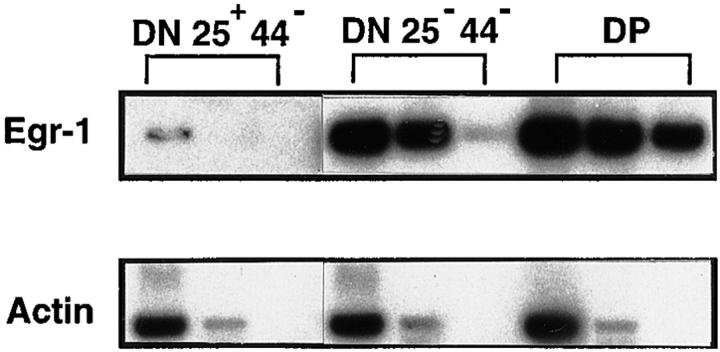
Shao et al. recently described Egr-1 expression in DN thymocytes of mice using intracellular fluorocytometry and suggested the possible involvement of Egr-1 expression during β selection in immature thymocytes (36). Because the pre-TCR–expressing population in DN thymocytes can be distinguished by CD25/CD44 expression, endogenous Egr-1 expression was dissected in DN thymocytes from wild-type C57Bl/6 (B6) mice by semiquantitative reverse transcriptase PCR (RT-PCR), using sorted CD25+CD44− cells that precede pre-TCR expression, and CD25−CD44− cells that express the pre-TCR complex. As shown in Fig. 1, much higher endogenous Egr-1 expression was detected in CD25−CD44− cells than in CD25+CD44− cells. Egr-1 was expressed at even higher levels in DP cells than in CD25− CD44− DN cells, consistent with the recent report (36).
Figure 1.
Increased endogenous Egr-1 expression in CD25−CD44− DN thymocytes of wild-type (C57Bl/6) mice. CD25+CD44− DN, CD25− CD44− DN, and DP thymocytes were sorted from C57Bl/6 (B6) mice, total RNA was isolated, and the expression of Egr-1 gene was assessed by semiquantitative RT-PCR. The amount of template cDNA for PCR corresponds to 5 × 104, 5 × 103, or 103 cells from the left lane to the right in each type of cell.
Generation of Transgenic Mice Overexpressing Egr-1 in Thymocytes with a RAG-2−/− Background.
Pre-TCR–mediated signals are necessary to promote development from the CD25+CD44− to the CD25−CD44− stage as revealed in several mutant mice. For instance, in RAG-2−/− mice lacking pre-TCR expression, thymocyte development is blocked at the CD25+CD44− stage (12). The increased expression of Egr-1 seen in CD25−CD44− DN thymocytes (Fig. 1) might reflect induction of Egr-1 expression by pre-TCR engagement, which may help to promote development of DN thymocytes from the CD25+CD44− to the CD25−CD44− stage. To test this hypothesis, transgenic mice designed to overexpress Egr-1 in immature thymocytes were generated and bred to RAG-2−/− mice (12).
The Lck proximal promoter was used to limit transgene expression primarily to immature thymocytes (37, 43). A mouse genomic DNA fragment of the Egr-1 gene, including coding exons and intron 1 but no 3′ noncoding sequence was used to insure efficient and stable expression of transcripts (44, 45). Three independent lines of transgenic founder mice were obtained. Phenotypes of these original transgenic mice will be detailed elsewhere (Miyazaki, T., and U. Müller, manuscript in preparation). All three lines were backcrossed to RAG-2−/− mice. All progeny carrying the transgene were of normal size and appeared healthy. As observed for many other transgenes driven by this promoter (43), transgene expression in the thymus was detected at levels far exceeding the endogenous Egr-1 when analyzed by Northern blotting using thymic RNA from transgenic and negative littermate mice, both of which had a RAG-2+/− background (data not shown). All three lines showed essentially the same expression levels.
Thymocyte Development.
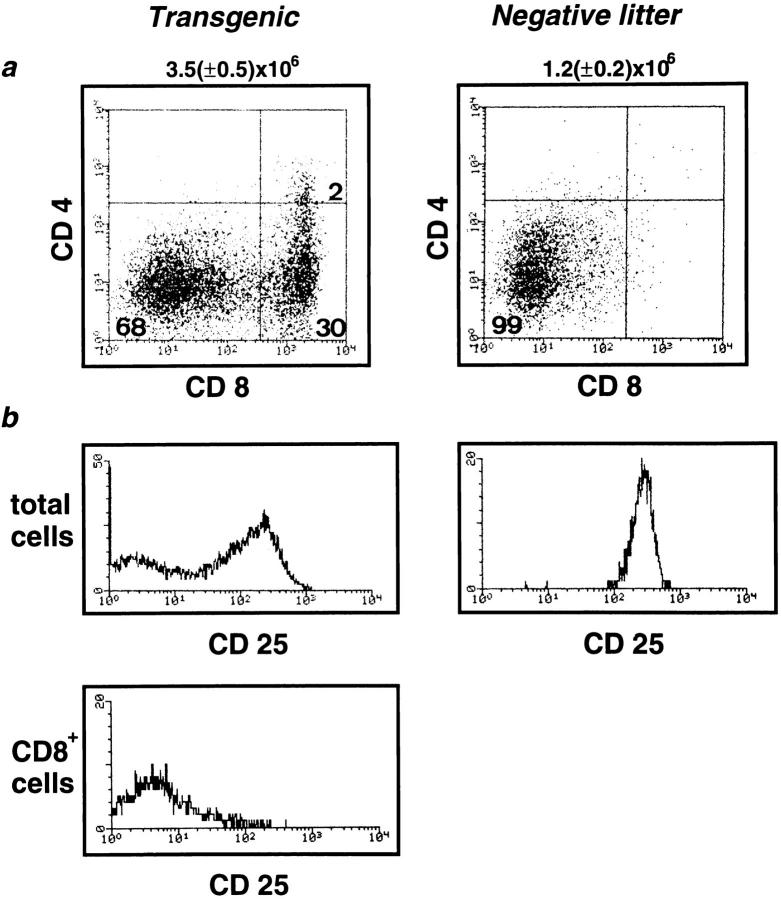
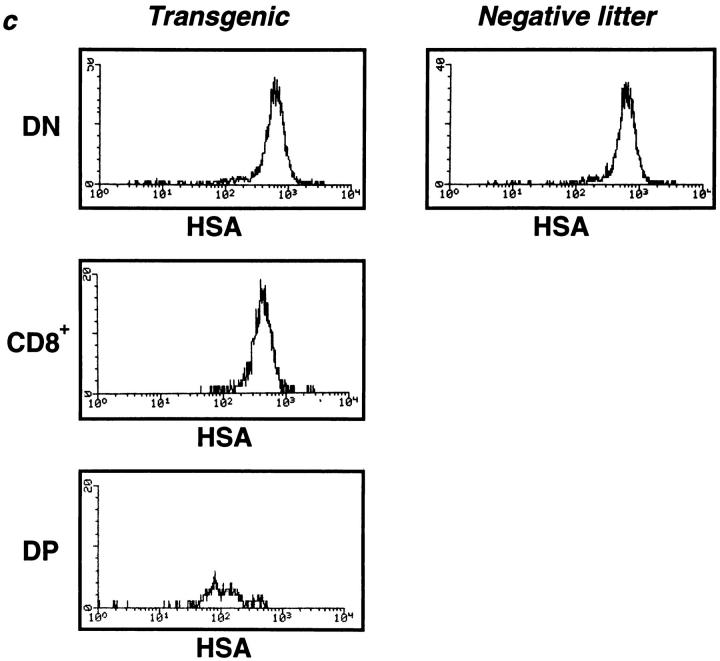
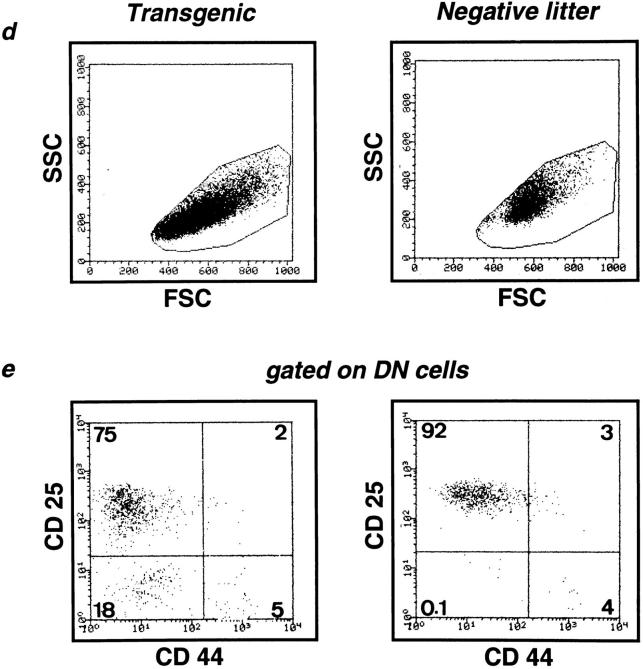
The thymi of Egr-1 transgenic mice with a RAG-2−/− background (Egr-1 Tg-RAG−) were significantly larger than those of RAG-2−/− negative littermates (NL-RAG−). The absolute number of thymocytes was 3.5 (± 0.5) × 106 in Egr-1 Tg-RAG− (n = 10), compared to 1.2 (± 0.3) × 106 in NL-RAG− (n = 10). Typical staining profiles of thymocytes are presented in Fig. 2 a. The most impressive aberration in the transgenic thymi was the presence of a large population (30%) of CD4−CD8+ cells. These CD8+ cells are CD25-negative (Fig. 2 b) and display lower heat-stable antigen (HSA) levels than those on DN cells, but higher levels than those on DP cells (Fig. 2 c), indicating that they are ISP transitional stage thymocytes (15–18). Consistently, size profiles of total thymocytes show that smaller cells appeared (Fig. 2 d) and a significant population (18%) of CD25−CD44− DN cells was detected in Egr-1 Tg-RAG− thymi (Fig. 2 e). Thus, overexpression of Egr-1 promoted thymocyte development from the DN to the ISP stage, overcoming the β block in the RAG-2−/− background. In accordance with this, small numbers of ISP cells appeared to express CD4 also, falling into the CD4/ CD8-DP quadrant (Fig. 2 a). These cells form, however, a very small (2%), indistinct population, possibly representing the intermediate stage cells between the ISP and the DP. Neither mature single-positive thymocytes nor peripheral T cells were detected in either Egr-1 Tg-RAG− or NL-RAG− mice. ISP cells isolated from Egr-1 Tg-RAG− could not develop to the DP stage spontaneously in in vitro culture for 12–24 h (data presented in following section as Fig. 3 d).
Figure 2.
Development of ISP cells induced by Egr-1 expression in RAG-2−/− mice. Thymocyte suspensions from 6–8-wk-old transgenic mice or negative littermates, both with a RAG-2−/− background, were stained and analyzed by fluorocytometry. (a) CD4/CD8 profiles. Numbers indicate relative percentages of positive cells within a quadrant. The average of the total thymocyte numbers of 10 of each type of mice are indicated above the profiles. (b) CD25 expression of total thymocytes from transgenic or negative littermates (top) or CD8+ cells from transgenic mice. (c) HSA histograms. (d) Side scatter (SSC)/forward scatter (FSC) profiles indicating cell size. Only living cells were gated. (e) CD25/CD44 profiles of DN cells. Thymocytes were stained with anti-CD25–Tricolor, anti-CD44–FITC, anti-CD4–PE, and anti-CD8–PE. PE-negative cells (CD4/CD8 DN cells) were gated and analyzed for CD25/CD44 expression. Numbers indicate relative percentages of positive cells within a quadrant.
Figure 3.
Development of DP cells induced by irradiation. Both transgenic mice and negative littermates with a RAG-2−/− background were sublethaly irradiated (600 rads). 4 wk after irradiation, their thymocyte suspensions were stained and analyzed by a fluorocytometer. (a) CD4/ CD8 profiles. Numbers indicate relative percentages of positive cells within a quadrant. The average of the total thymocyte numbers of seven of each type of mice are indicated above the profiles. (b) CD25 expression on total thymocytes. (c) Peanut-agglutinin binding capacity of each subpopulation. (d) In vitro culture of ISP cells. 50,000 ISP cells were sorted from irradiated or nonirradiated transgeic mice with a RAG-2−/− background and cultured in 200 μl of complete culture medium for 12–24 h. After the culture period, cells were stained for CD4 and CD8, and analyzed by fluorocytometer. This experiment was repeated three times and the results were reproducible. A representative result is shown.
DN and ISP cells from transgenic thymi were sorted, and transgene expression in each population was confirmed by RT-PCR. To avoid amplification of endogenous Egr-1 transcripts, a set of primers specific for the hGH gene sequence present in transgene transcripts was used. Transgene expression was detected in either CD25+CD44−, CD25− CD44−, or ISP cells (data not shown).
Irradiation-induced DP Cell Development in Egr-1 Tg Mice with a RAG-2−/− Background.
As discussed above, once the β block is bypassed in a RAG−/− thymus, either by irradiation, CD3-ligation, or by expression of activated lck or ras transgenes, thymocytes do mature to the DP stage, whereas in Egr-1 Tg-RAG− mice, the vast majority of thymocytes develop to the ISP stage, but not to the DP stage. Two potential explanations can be considered for this difference. First, overexpression of Egr-1 is sufficient to drive development of DN thymocytes to the ISP stage, but other gene(s) might be required to promote ISP cells to the DP stage. Second, overexpression of Egr-1 at ISP stage might interfere with further development of ISP cells towards the DP stage.
To investigate this question, both Egr-1 Tg-RAG− and NL-RAG− mice were sublethally irradiated (600 rads) and their thymi were examined. As observed previously (22), the thymic cellularity in both Tg and negative littermates was strikingly decreased 2–3 d after irradiation. Most of the thymocytes were dead when examined by trypan blue exclusion (data not shown), and very few living ISP cells were detected in irradiated Tg thymi by fluorocytometric analysis (data not shown). 4 wk after irradiation, however, thymi were significantly enlarged as compared to nonirradiated thymi both in Tg and negative littermates, and the absolute numbers of thymocytes were 1.8 (± 0.3) × 107 in Tg versus 1.0 (± 0.2) × 107 in negative littermates (n = 7 each). As shown in Fig. 3 a, in both types of mice, an obvious DP population appeared which was 1.2 (± 0.2) × 107 cells in Tg and 0.8 (± 0.1) × 107 cells in negative littermates. Large numbers of total thymocytes both in Tg and negative littermates did not express CD25 as shown in Fig. 3 b. These results indicate that overexpression of Egr-1 does not block thymocyte development from the ISP to the DP stage and, therefore, perhaps additional gene(s) induced by irradiation are required to drive ISP cells to the DP stage.
In the thymus of Egr-1 Tg- RAG− 4-wk after irradiation, a substantial population (20%) of CD4−/lowCD8+ cells was detected. This population differs from the ISP cells in nonirradiated Egr-1 Tg- RAG− thymi, since the ISP cells disappeared soon after irradiation. However, these cells are apparently repopulated ISP and ISP/DP intermediate stage cells because their peanut agglutinin–binding capacity which is reduced as thymocytes mature, is even higher than that of DP cells (Fig. 3 c), indicating that these CD4−/low CD8+ cells are more immature than DP cells. This is also supported by the observation that these CD4−/lowCD8+ cells bear higher levels of HSA than do DP cells, but lower levels than do DN cells (data not shown). In addition, ISP cells isolated from irradiated Egr-1 Tg-RAG− mice spontaneously mature to the DP stage in 12–24 h of culture (Fig. 3 d), indicating these immature cells have the potential to mature into DP cells, in sharp contrast to ISP cells from nonirradiated Egr-1 Tg-RAG− mice which do not develop to the DP cell stage in identical culture conditions (Fig. 3 d).
In both types of mice, no mature CD4+ or CD8+ cells were detected, which is consistent with previous reports regarding irradiated RAG−/− mice (22, 23). TCR Vα and Vβ rearrangements were examined in each sorted population and, as expected (22, 23), no rearrangement could be detected in any thymocyte population, either from irradiated transgenic or negative littermates with a RAG-2−/− background (data not shown).
Cell Cycling of ISP Cells in Egr-1 Tg with a RAG+ Background.
Seeing a significant population of ISP (and ISP/ DP intermediate) cells in Egr-1 Tg-RAG− mice even after irradiation, which have the potential to further develop to the DP stage, I wondered if overexpression of Egr-1 might lead the ISP cells to be hyperproliferative, resulting in enlargement of this population in the profiles. To asses this question, the cell cycle of ISP cells was determined. To compare the fraction of cycling cells between Egr-1 overexpressing ISP and normal ISP cells, RAG+ background Tg (Egr-1 Tg-RAG+) and NL-RAG+ mice were used. In RAG+ background animals, ISP cells can be distinguished as CD8+ TCRlow. These cells were sorted from Egr-1 Tg-RAG+ and NL-RAG+ mice, and their cell cycling condition was examined by DNA content .
As shown in Fig. 4, essentially the same percentage of ISP cells from transgenic and negative littermate mice were in S or G2/M phase, indicating that cell cycling was not altered by Egr-1 overexpression.
Figure 4.
Normal cell cycling in ISP cells of transgenic mice. ISP (CD8+TCRlow) thymocytes from transgenic mice or negative littermates with a RAG-2+/+ background were sorted and their DNA content was determined. The high peaks at ∼75 of relative intensity represent the G1 phase DNA and the other smear part at ∼200 represents the S or G2/M phase DNA. The amount of S or G2/M phase DNA was essentially same in transgenic mice and negative littermate controls.
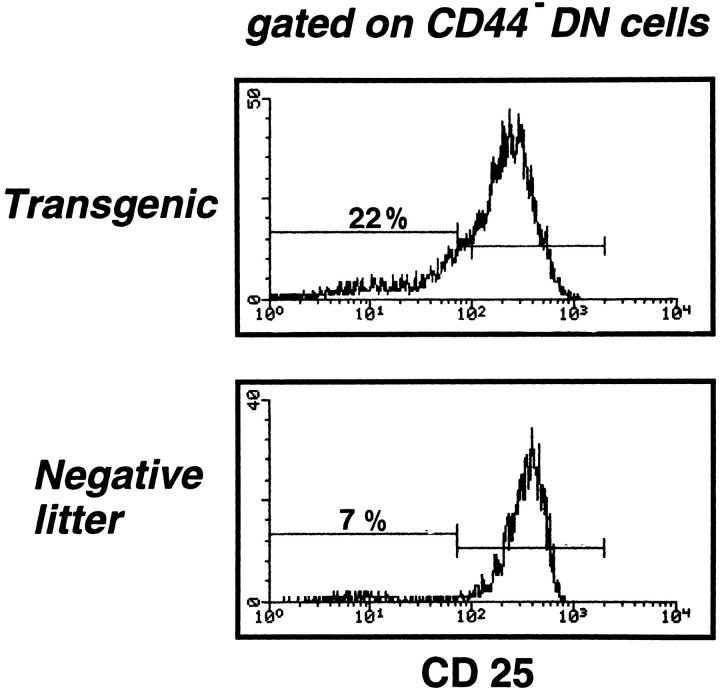
More Efficient Maturation from the DN to the ISP Stage in Irradiated Egr-1 Tg Mice.
An alternative explanation for the large population of ISP (and ISP/DP intermediate) cells in irradiated Egr-1 Tg-RAG− thymi is that the high level of Egr-1 expression might boost the thymocyte maturation from the DN to the ISP stage. This question was assessed by comparing the CD25/CD44 profiles of DN cells in both types of mice 4 wk after irradiation. Fig. 5 shows histograms of CD25 expression in CD44− DN thymocytes. In irradiated NL-RAG− thymi, two clear populations (CD25− and CD25+ ) could be identified and seven (± 2.0; n = 8) percent of CD44− DN cells exhibited the CD25− phenotype. In irradiated Egr-1 Tg-RAG− thymi, however, CD25− and CD25+ populations were not clearly separated, and a large population of intermediate (CD25low CD44−) cells bearing broadly differing levels of CD25 were observed. Using the same gate as for the negative littermate cells, CD25− cells were 22 (± 1.6; n = 8) percent of CD44− DN cells. In addition, the relative peak intensity of the CD25+ population was 1.7 times higher in negative littermates than in Tg mice, indicating that many of the CD25+CD44− cells had shifted to the CD25−CD44− stage. This shift was detected even in nonirradiated Egr-1 Tg-RAG− DN cells (see Fig. 2 e). These results suggest that the maturation may be boosted in Egr-1 Tg DN thymocytes. An increase in the number of CD4−CD8low in irradiated Tg thymi (6 [± 0.8] × 105 in Tg versus 1.7 [± 0.5] × 104 in negative littermate thymus [n = 8 each]), might also reflect the boost also during maturation from the CD25−CD44− DN to the ISP cell stage.
Figure 5.
More CD25−/lowCD44− DN cells in irradiated transgenic thymus than in irradiated negative littermate thymus. Thymocyte suspensions from either irradiated transgenic or irradiated negative littermate mice 4 wk after irradiation were stained for CD4, CD8, CD25, and CD44 and analyzed by fluorocytometry. CD44− DN cells were gated and their CD25 expression is presented. Percentages of the CD25-negative population (the gate for which was created using the negative littermate's histogram as a standard) are indicated.
Discussion
Egr-1 Overexpression Is Sufficient to Promote Development of CD25+CD44− DN Cells to the CD25−CD44− DN and Further to the ISP Stage.
As previously discussed, pre-TCR independent development of thymocytes in RAG−/− mice has been achieved by various experimental manipulations, including irradiation, CD3 ligation, and introduction of several transgenes. These stimuli are thought to compensate for signaling through the pre-TCR complex, though the precise molecular mechanisms are unknown. Because all of these manipulations can activate Ras (28), activation events downstream of the Ras pathway might be strong candidates for key players in promoting thymocyte development beyond β selection. We are, however, entirely ignorant of the gene regulation/transcription involved in this maturation control. In this report, an immediate–early gene, Egr-1, is presented as a primary transcription factor, overexpression of which does initiate thymocyte development in RAG-2−/− mice.
The initial observation that endogenous Egr-1 expression is far upregulated in CD25−CD44− DN cells in comparison to the immediately preceding CD25+CD44− DN population, suggested that Egr-1 might be induced after ligation of the pre-TCR complex and a set of genes which are transcriptionally regulated by Egr-1 could possibly play key roles in promoting CD25+CD44− DN cells to develop to the CD25−CD44− stage. This idea was well supported by transgenic mice overexpressing Egr-1 with a RAG-2−/− background in which CD25+CD44− DN cells achieved maturation to the CD25−CD44− and further to the ISP stage. Because several reports have indicated that Egr-1 expression is upregulated by irradiation (46) and ras activation (47, 48), the observation in these transgenic mice is consistent with the previously reported ras-related promotion of thymocyte development in RAG−/− mice.
Despite these results which implicate involvement of Egr-1 in early thymocyte development, Egr-1−/− mice showed no defect in thymocyte development (49; my unpublished results). Potential compensation for the role of Egr-1 by other Egr family members, such as Egr-2, -3, and -4, which are reported to be expressed in the thymus (36), could explain normal thymocyte development in Egr-1−/− mice. Because the sequence of the zinc finger DNA–binding site is highly conserved in all members of the Egr family (29), the set of expressed genes that are required to promote DN thymocyte development could be induced by other members of the Egr family, even in the absence of Egr-1.
Thus, although expression of Egr-1 is not necessary for β selection, overexpression allows thymocytes to overcome β selection and mature to the ISP stage.
Two Distinct Steps during Thymocyte Maturation between the DN and ISP Stages.
To our surprise, though thymocytes overexpressing Egr-1 bypassed the β block and developed to the ISP stage in a RAG−/− background, these cells could not further mature to the DP stage. All other manipulations reported that induce thymocyte development in RAG−/− mice drive thymocyte maturation past the β block to the DP stage. However, irradiation did promote maturation of DP cells in Egr-1 Tg-RAG−, demonstrating that overexpression of Egr-1 itself does not impair thymocyte maturation from the ISP to the DP stage. Thus, transcriptional regulation by Egr-1 is sufficient to promote maturation of thymocytes from the CD25+CD44− DN to the ISP stage, but additional regulations that can be induced by irradiation are necessary to drive ISP cells to the DP stage. Several immediate–early genes other than Egr-1, such as c-jun and c-fos, are upregulated by irradiation (28, 46). In addition, p53 expression, which can rescue both the rearrangement of the TCR-β locus and thymocyte development to the DP stage in scid mutant mice is also induced by irradiation (50), though induction of rearrangement is clearly not a factor in the RAG-2−/− background animals. On the other hand, null-mutant mice of an high-mobility group box protein family member, T cell factor 1, revealed a maturation block of thymocytes at the ISP stage, and this protein appeared indispensable for thymocyte development from the ISP to the DP stage (51). Though it is not known whether T cell factor 1 is induced by irradiation, it is a potential candidate as a key player in this developmental process. Further studies are required to identify the responsible gene(s) expression for thymocyte maturation from the ISP to the DP stage.
One might argue that the signals mediated by TCR-α/β instead of pre-TCR, might be responsible for the development from ISP to DP cells, because the rearrangement of α chain becomes active and low levels of TCR-α/β expression can be detected on the cell surface at the ISP stage. It appears unlikely, however, because thymocytes do develop to the DP stage even in TCR-α−/− mice (13).
Overexpression of Egr-1 Might Boost Thymocyte Maturation.
The presence of a large population of ISP (and ISP/ DP intermediate) cells in Egr-1 Tg-RAG− mice after irradiation revealed an unexpected effect of Egr-1 overexpression on immature thymocytes. These cells have the potential to develop to the DP stage, and it is unlikely that there is a distinct population that is resistant to induction of maturation by irradiation. In addition, this phenotype was not due to the hyperproliferation of ISP cells because cell cycling was not affected by overexpression of Egr-1. The shifted balance of CD25+CD44− versus CD25−CD44− DN cell populations and increased number of CD4−CD8low cells in irradiated Tg thymi suggest that overexpression of Egr-1 may boost thymocyte maturation from CD25+ CD44− DN to the ISP stages, resulting in an enlarged ISP population in the thymic profiles. The precise mechanism of this boost effect by Egr-1 overexpression is unclear. However, not only CD25 downregulation and CD8 upregulation are affected, because these changes were accompanied by reduction of cell size and HSA downregulation. In addition, Egr-1–binding domains have not been identified either in CD8 or CD25 gene promoter regions. Recently, Eibel and colleagues have generated transgenic mice which overexpress Egr-1 in B cell lineage under the control of immunogloblin promoter/enhancer. In these transgenic mice, B cell maturation seems to be advanced (Eibel, H., personal communication). This result appears analogous with the boosted thymocyte maturation in our transgenic mice.
Perspective.
Newly generated Egr-1 transgenic mice with a RAG-2−/− background provide several novel insights, two of which are worth reemphasizing. First, overexpression of Egr-1 is sufficient to promote the development of CD25+CD44− DN thymocytes to the ISP stage, beyond β selection. Second, there are two distinct steps regulating thymocyte maturation between the DN and DP stages, the turning point of which appears to be at the ISP stage. Egr-1 transgenic mice may provide a model for further dissection of the gene regulation involved in early thymocyte development.
Footnotes
The author thanks U. Müller for microinjection; Drs. K.S. Campbell, M. Colonna, S. Gilfillan, H.J. Fehling, and M. Bogue (Baylor College of Medicine, Waco, TX) for critical reading of the manuscript; E. Wagner, W. Metzger, and R. Zedi for help with the mice; and M. Dessing for help with the cell sorting and cell cycle analysis.
Basel Institute for Immunology was founded and is supported by F. Hoffmann-La Roche Ltd. (Basel, Switzerland).
Abbreviations used in this paper: DN, double negative; DP, double-positive; Egr, early growth response; hGH, human growth hormone; HSA, heat-stable antigen; ISP, immature CD8 single-positive; NL, negative littermates; RAG, recombinase-activating gene; RT-PCR, reverse transcriptase PCR; Tg, transgenic.
References
- 1.Malissen B, Malissen M. Functions of TCR and pre-TCR subunits: lessons from gene ablation. Curr Opin Immunol. 1996;8:383–393. doi: 10.1016/s0952-7915(96)80129-4. [DOI] [PubMed] [Google Scholar]
- 2.Godfrey DI, Zlotnik A, Suda T. Phenotypic and functional characterization of c-kit expression during intrathymic T cell development. J Immunol. 1992;149:2281–2285. [PubMed] [Google Scholar]
- 3.Fehling HJ, Krotkova A, Saint-Ruf C, von Boehmer H. Crucial role of the pre–T-cell receptor α gene in development of α/β but not γ/δ cells. Nature (Lond) 1995;375:795–798. doi: 10.1038/375795a0. [DOI] [PubMed] [Google Scholar]
- 4.Ware CB, Meyer JD, Davidson BL. Early lymphocyte expansion is severely impaired in interleukin 7 receptor–deficient mice. J Exp Med. 1994;180:1955–1960. doi: 10.1084/jem.180.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Freeden-Jeffry U, Vieira P, Lucian LA, McNeil T, Burdach SE, Murray R. Lymphopenia in interleukin (IL) 7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J Exp Med. 1995;181:1519–1526. doi: 10.1084/jem.181.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao X, Shores EW, Hu-Li J, Anver MR, Kelsall BL, Russell SM, Drago J, Noguchi M, Grinberg A, Bloom ET, et al. Defective lymphoid development in mice lacking expression of the common cytokine receptor γ chain. Immunity. 1995;2:223–238. doi: 10.1016/1074-7613(95)90047-0. [DOI] [PubMed] [Google Scholar]
- 7.DiSanto JP, Muller W, Guy-Grand D, Fisher A, Rajewsky K. Lymphoid development in mice with a targeted deletion of the interleukin 2 receptor γ chain. Proc Natl Acad Sci USA. 1995;92:377–381. doi: 10.1073/pnas.92.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohbo K, Suda T, Hashiyama M, Mantani A, Ikebe M, Miyakawa K, Moriyama M, Nakamura M, Katsuki M, Takahashi K, et al. Modulation of hematopoiesis in mice with a truncated mutant of the interleukin-2 receptor γ chain. Blood. 1996;87:956–967. [PubMed] [Google Scholar]
- 9.Rodewald HR, Kretzschmar K, Swat W, Takeda S. Intrathymically expressed c-kit ligand (stem cell factor) is a major factor driving expansion of very immature thymocytes in vivo. Immunity. 1995;3:313–319. doi: 10.1016/1074-7613(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 10.Rodewald HR, Ogawa M, Haller C, Waskow C, DiSanto JP. Pro-thymocyte expansion by c-kit and the common cytokine receptor γ chain is essential for repertoire formation. Immunity. 1997;6:265–272. doi: 10.1016/s1074-7613(00)80329-5. [DOI] [PubMed] [Google Scholar]
- 11.Mombaerts P, Lacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1 deficient mice have no mature β and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 12.Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, Mendlsohn M, Charron V, Datta M, Yung F, Stall AM, Alt FW. Rag-2 deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 13.Mombaerts P, Clarke AR, Rudnicki MA, Lacomini J, Itohara S, Lafaille JJ, Wang L, Ichikawa Y, Jaenisch R, Hooper ML, Tonegawa S. Mutations in T-cell antigen receptor genes α and β block thymocyte development at different stages. Nature (Lond) 1992;360:225–231. doi: 10.1038/360225a0. [DOI] [PubMed] [Google Scholar]
- 14.Godfrey DI, Zlotnik A. Control points in early T-cell development. Immunol Today. 1993;14:547–553. doi: 10.1016/0167-5699(93)90186-O. [DOI] [PubMed] [Google Scholar]
- 15.Paterson DJ, Williams AF. An intermediate cell in thymocyte differenciation that expresses CD8 but not CD4 antigen. J Exp Med. 1987;166:1603–1608. doi: 10.1084/jem.166.5.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crispe IN, Bevan MJ. Expression and functional significance of the J11d marker on mouse thymocytes. J Immunol. 1987;138:2013–2018. [PubMed] [Google Scholar]
- 17.Shorman K, Wilson A, Egerton M, Perse M, Scollay R. Immature CD4−CD8+murine thymocytes. Cell Immunol. 1988;113:462–479. doi: 10.1016/0008-8749(88)90042-1. [DOI] [PubMed] [Google Scholar]
- 18.MacDonald HR, Budd RC, Howe RC. A CD3− subset of CD4−CD8+ thymocytes: a rapidly cycling intermediate in the generation of CD4+CD8+cells. Eur J Immunol. 1988;18:519–523. doi: 10.1002/eji.1830180405. [DOI] [PubMed] [Google Scholar]
- 19.Boismenu R, Rhen M, Fischer WH, Havran WL. Role of CD81 in early T cell development. Science (Wash DC) 1996;271:198–200. doi: 10.1126/science.271.5246.198. [DOI] [PubMed] [Google Scholar]
- 20.Miyazaki T, Müller U, Campbell KS. Normal develpment but differencially altered proliferative responses of lymphocytes in mice lackind CD81. EMBO (Eur Mol Biol Organ) J. 1997;16:4217–4225. doi: 10.1093/emboj/16.14.4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maecker HT, Levy S. Normal lymphocyte development but delayed humoral immune response in CD81-null mice. J Exp Med. 1997;185:1505–1510. doi: 10.1084/jem.185.8.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuniga-Phücker JC, Jiang D, Scwartzberg PL, Lenard MJ. Sublethal γ-radiation induces differenciation of CD4−/CD8− into CD4+/CD8+thymocytes without T cell receptor β rearrangement in recombinase activation gene 2 −/− mice. J Exp Med. 1994;180:1517–1521. doi: 10.1084/jem.180.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guidos CJ, Williams CJ, Wu GE, Paige CJ, Danska JS. Development of CD4+CD8+thymocytes in RAG-deficient mice through a T cell receptor β chain–independent pathway. J Exp Med. 1995;181:1187–1195. doi: 10.1084/jem.181.3.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shinkai Y, Alt FW. CD3ε-mediated signals rescue the development of CD4+CD8+ thymocytes in RAG-2−/−mice in the absence of TCR β chain expression. Int Immunol. 1994;6:995–1001. doi: 10.1093/intimm/6.7.995. [DOI] [PubMed] [Google Scholar]
- 25.Jacobs H, Vandeputte D, Tolkamp L, de Vries E, Borst J, Berns A. CD3 components at the surface of pto-T cells can mediate pre–T cell development in vivo. . Eur J Immunol. 1994;24:934–939. doi: 10.1002/eji.1830240423. [DOI] [PubMed] [Google Scholar]
- 26.Mombaerts P, Anderson SJ, Perlmutter RM, Mak TW, Tonegawa S. An activated lck transgene promotes thymocyte development in RAG-1 mutant mice. Immunity. 1994;1:261–267. doi: 10.1016/1074-7613(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 27.Swat W, Shinkai Y, Cheng H-L, Davidson L, Alt FW. Activated Ras signals differentiation and expansion of CD4+CD8+thymocytes. Proc Natl Acad Sci USA. 1996;93:4683–4687. doi: 10.1073/pnas.93.10.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robey E, Allison JP. T-cell activation: integration of signals from the antigen receptor and costimulatory molecules. Immunol Today. 1995;16:306–313. doi: 10.1016/0167-5699(95)80140-5. [DOI] [PubMed] [Google Scholar]
- 29.Kelly K, Siebenlist U. Immediate-early genes induced by antigen receptor stimulation. Curr Opin Immunol. 1995;7:327–332. doi: 10.1016/0952-7915(95)80106-5. [DOI] [PubMed] [Google Scholar]
- 30.Milbrandt J. Nerve growth factor induces a gene homologus to the glucocorticoid receptor gene. Neuron. 1988;3:183–188. doi: 10.1016/0896-6273(88)90138-9. [DOI] [PubMed] [Google Scholar]
- 31.Hazel TG, Nathans D, Lau LF. A gene inducible by serum growth factors encodes a member of the steroid and thyroid hormone receptor superfamily. Proc Natl Acad Sci USA. 1988;85:8444–8448. doi: 10.1073/pnas.85.22.8444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Milbrandt J. A nerve growth factor–induced gene encodes a possible transcriptional regulatory factor. Science (Wash DC) 1987;238:797–799. doi: 10.1126/science.3672127. [DOI] [PubMed] [Google Scholar]
- 33.Sukhatme VP, Cao X, Chang LC, Tsai-Morris CH, Stamenkovich D, Ferrira PCP, Cohen DR, Edwards SA, Shows TB, Curran T, et al. A zinc finger–encoding gene coregulated with c-fos during growth and differentiation, and after cellular depolarization. Cell. 1988;53:37–43. doi: 10.1016/0092-8674(88)90485-0. [DOI] [PubMed] [Google Scholar]
- 34.Christy BA, Lau LF, Nathans D. A gene activated in mouse 3T3 cells by serum growth factors encodes a protein with “zinc finger” sequences. Proc Natl Acad Sci USA. 1988;85:7857–7861. doi: 10.1073/pnas.85.21.7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perez-Castillo A, Pipaon C, Garcia I, Alemany S. NGFI-A gene expression is necessary for T lymphocyte proliferation. J Biol Chem. 1993;268:19445–19450. [PubMed] [Google Scholar]
- 36.Sha H, Kono DH, Chen L-Y, Rubin EM, Kaye J. Induction of the early growth response (Egr) family of transcription factors during thymic selection. J Exp Med. 1997;185:731–744. doi: 10.1084/jem.185.4.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allen JA, Forbush KA, Perlmutter RM. Functional dissection of the lck proximal promoter. Mol Cell Biol. 1992;12:2758–2768. doi: 10.1128/mcb.12.6.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tokunaga K, Taniguchi H, Yoda K, Shimizu M, Sakiyama S. Nucleotide sequence of a full-length cDNA for mouse cytoskeltal beta-actin mRNA. Nucleic Acids Res. 1986;14:2829. doi: 10.1093/nar/14.6.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tourne S, Nakano N, Viville S, Benoist C, Mathis D. The influence of invariant chain on the positive selection of single TCR specificities-an argument for the role of peptide selection of CD4+T cells. Eur J Immunol. 1995;25:1851–1856. doi: 10.1002/eji.1830250709. [DOI] [PubMed] [Google Scholar]
- 40.Chan SH, Cosgrove D, Walzinger C, Benoist C, Mathis D. Another view of the selective model of thymocyte selection. Cell. 1993;73:225–236. doi: 10.1016/0092-8674(93)90225-f. [DOI] [PubMed] [Google Scholar]
- 41.Miyazaki T, Wolf P, Tourne S, Walzinger C, Dierich A, Barois N, Ploegh H, Benoist C, Mathis D. Mice lacking H2-M complexes, enigmatic elements of the MHC class II peptide-loading pathway. Cell. 1996;84:531–541. doi: 10.1016/s0092-8674(00)81029-6. [DOI] [PubMed] [Google Scholar]
- 42.Mertsching E, Burget C, Ceredig R. IL-7 transgenic mice: analysis of the role of IL-7 in the differentiation of thymocytes in vivo and in vitro. Int Immunol. 1995;7:401–414. doi: 10.1093/intimm/7.3.401. [DOI] [PubMed] [Google Scholar]
- 43.Calnan BJ, Szychowski S, Chan FKM, Cado D, Winoto A. A role for the orphan steroid receptor Nur77 in apoptosis accompanying antigen-induced negative selection. Immunity. 1995;3:273–282. doi: 10.1016/1074-7613(95)90113-2. [DOI] [PubMed] [Google Scholar]
- 44.Brinster RL, Allen JM, Behringer RR, Gelinas RE, Palmiter RD. Introns increase transcriptional efficiency in transgenic mice. Proc Natl Acad Sci USA. 1988;85:836–840. doi: 10.1073/pnas.85.3.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruther U, Barber C, Komitowski D, Muller R, Wagner EF. Deregulated c-fos expression interferes with normal bone development in transgenic mice. Nature (Lond) 1987;325:412–416. doi: 10.1038/325412a0. [DOI] [PubMed] [Google Scholar]
- 46.Prasad AV, Mohan N, Chandrasekar B, Melts ML. Induction of transcription of “immediate early genes” by low-dose ionizing radiation. Radiat Res. 1995;143:263–272. [PubMed] [Google Scholar]
- 47.Huang R-P, Adamson ED. A biological role for Egr-1 in cell survival following ultra-violet irradiation. Oncogene. 1995;10:467–475. [PubMed] [Google Scholar]
- 48.McMahon SB, Monroe JG. Activation of the p21raspathway couples antigen receptor stimulation to induction of the primary response gene egr-1 in B lymphocytes. J Exp Med. 1995;181:417–422. doi: 10.1084/jem.181.1.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee SL, Tourtellotte LC, Wesselschmidt RL, Milbrandt J. Growth and differentiation proceeds normally in cells deficient in the immediate early gene NGFI-A. J Biol Chem. 1995;270:9971–9977. doi: 10.1074/jbc.270.17.9971. [DOI] [PubMed] [Google Scholar]
- 50.Bogue MA, Zhu C, Aguilar-Cordova E, Donehower LA, Roth DB. p53 is required for both radiation-induced differenciation and rescue of V(D)J rearrangement in scid mouse thymocytes. Genes Dev. 1996;10:553–565. doi: 10.1101/gad.10.5.553. [DOI] [PubMed] [Google Scholar]
- 51.Verbeek S, Izon D, Hofhuis F, Robanus-Maandag E, te Riele H, van de Wetering M, Oostermegel M, Wilson A, MacDonald HR, Clevers H. An HMG-box– containing T-cell factor required for thymocyte differentiation. Nature (Lond) 1995;374:70–74. doi: 10.1038/374070a0. [DOI] [PubMed] [Google Scholar]