Abstract
Dendritic cells are potent antigen-presenting cells involved in the initiation of immune responses. The trafficking of these cells to tissues and lymph nodes is mediated by members of the chemokine family. Recently, a novel CC chemokine known as MIP-3α or liver and activation-regulated chemokine has been identified from the EMBL/GenBank/DDBJ expressed sequence tag database. In the present study, we have shown that the messenger RNA for MIP-3α is expressed predominantly in inflamed and mucosal tissues. MIP-3α produced either synthetically or by human embryonic kidney 293 cells is chemotactic for CD34+-derived dendritic cells and T cells, but is inactive on monocytes and neutrophils. MIP-3α was unable to displace the binding of specific CC or CXC chemokines to stable cell lines expressing their respective high affinity receptors, namely CCR1–5 and CXCR1 and CXCR2, suggesting that MIP-3α acts through a novel CC chemokine receptor. Therefore, we used degenerate oligonucleotide-based reverse transcriptase PCR to identify candidate MIP-3α receptors in lung dendritic cells. Our results show that the orphan receptor known as GCY-4, CKRL-3, or STRL-22 is a specific receptor for MIP-3α, and that its activation leads to pertussis toxin–sensitive and phospholipase C–dependent intracellular Ca2+ mobilization when it is expressed in HEK 293 cells.
Dendritic cells (DCs)1 play an essential role in the induction of immune responses (1). DC precursors are thought to originate in the bone marrow and subsequently migrate into the peripheral tissues and lymph nodes. The main function of these cells is to sample and process incoming antigenic material, which is then presented to naive T cells, either locally or within regional lymph nodes, resulting in T cell activation and the generation of immune responses necessary to clear the infection. The trafficking of DCs from blood to tissues and then to lymph nodes is mediated in part by members of the chemokine superfamily.
Chemokines are small molecular weight (8–10-kD) proteins, most of which contain four conserved cysteine residues in their primary amino acid sequence. There are two major groups: the CXC chemokines in which the two NH2-terminal cysteines are separated by a single amino acid, or the CC chemokines in which the two NH2-terminal cysteines are adjacent (2). A third type of chemokine represented by lymphotactin, contains only two of the four conserved cysteines (3). Recently, the prototype of a fourth class of chemokines has been described in which the first two of the four conserved cysteines are separated by three amino acids (CX3C) (4).
Chemokines were originally identified through their chemoattractant effects on different leukocyte populations, but other approaches have included the selective cloning of secreted proteins using the signal sequence trap (5, 6) or by subtractive cloning methods (7). In addition to this, a number of putative chemokine sequences have recently been reported in expressed sequence tag (EST) databases (8). About 30 chemokines are known to date and this number is likely to increase as more sequence information is obtained from genome sequencing projects.
The specific effects of chemokines on target cell types are mediated by a family of G protein–coupled seven-transmembrane receptors. 10 human chemokine receptors have been identified so far; 4 of which are specific for CXC chemokines (CXCR1–4) (9–12); 5 of which are specific for CC chemokines (CCR1–5) (13–17); and the Duffy antigen receptor (DARC), which binds both CC and CXC chemokines (18). All of these receptors bind more than one chemokine at high affinity with the exception of CXCR1, which is specific for IL-8, and CXCR4, which so far appears to be specific for stromal cell-derived factor-1 (SDF-1). In addition, distinct chemokines appear to act on more than one receptor type. However, there is increasing evidence to suggest that this redundancy does not occur in vivo (19). Despite the overlapping ligand specificities shown by chemokine receptors, the number of chemokines identified far exceeds the number of known receptors. Degenerate oligonucleotide–based PCR cloning strategies used to identify chemokine receptor cDNAs have also resulted in the cloning of numerous orphan receptors (20). Although these orphan receptors show significant levels of identity (30–50%) at the amino acid level to the known chemokine receptors, their natural ligands remain to be identified. It is more than likely that the ligands will turn out to be many of the newly identified EST-derived chemokines, but progress in this area is slow due to the limited availability of these proteins for testing.
To appreciate fully the role of chemokines in diverse biological processes and the mechanisms by which they control the recruitment of immune cells necessitates the identification and characterization of their specific receptors. We and others have recently identified a novel CC chemokine from the EMBL/GenBank/DDBJ EST database known as MIP-3α or liver and activation-regulated chemokine (LARC) (21, 22). Here, we show that synthetic or recombinantly expressed MIP-3α is a potent chemoattractant for certain DCs and T cells. MIP-3α was unable to bind to any of the known CC chemokine receptors (CCR1–5) or to CXCR1 or CXCR2, suggesting the existence of a specific receptor for this chemokine. Therefore, we used degenerate oligonucleotide–based RT-PCR to identify candidate receptors in cells that responded to MIP-3α. As chemokine receptor activation typically leads to pertussis toxin–sensitive phospholipase C activation resulting in calcium mobilization in most systems (23), we used a fluorescent imaging plate reader to measure changes in intracellular cytosolic free calcium concentration ([Ca2+]i) in human embryonic kidney (HEK) 293 cells transiently expressing high levels of orphan chemokine-like receptors to identify the putative MIP-3α receptor. Here, we show that an orphan receptor, formerly known as GCY4, STRL-22 (24), or CKR-L3 (25), is a specific receptor for the novel CC chemokine, MIP-3α, and its activation by MIP-3α leads to both calcium influx and mobilization in cells in which it is expressed.
Materials and Methods
Materials.
Restriction enzymes and DNA-modifying enzymes were purchased from New England Biolabs (Beverly, MA) unless otherwise stated. AmplitTaqTM was from Perkin-Elmer Cetus (Norwalk, CT). AMV reverse transcriptase was from Promega (Madison, WI). All cell culture reagents were from GIBCO BRL Life Sciences (Paisley, UK). MIP-1α, RANTES, and IL-8 were expressed in Escherichia coli and prepared as described earlier (26, 27). MCP-1 was obtained using similar methodology. SDF-1α was prepared at Glaxo, Inc. (Research Triangle Park, NC). Synthetic MIP-3α, MIP-5, and the NH2-terminal chemokine domain of the CX3C chemokine also known as fractalkine (4) were chemically synthesized according to published methods (28) using the sequences discussed in Wells and Peitsch (8). All other recombinant chemokines used in this study were purchased from R&D Systems, Inc. (Minneapolis, MN) or Peprotech, Inc. (Rocky Hill, NJ). Radiolabeled [125I] MIP-3α was prepared by Amersham (Cardiff, UK) to a specific activity of 2,000 Ci/mmol. Fluo-3 AM was purchased from Molecular Probes, Inc. (Eugene, OR). Oligonucleotide primers were synthesized by Microsynth (Balgach, Switzerland).
Purification of Leukocyte Populations.
Leukocyte populations used for chemotaxis assays or for RT-PCR experiments were purified from the buffy coats of normal healthy blood donors after centrifugation through Ficoll–Paque (Pharmacia Biotech, Uppsala, Sweden). The PBMCs were subfractionated into T cells and monocytes after rossetting by sheep RBCs. CD4 and CD8 T cell subpopulations were further purified using T cell purification columns (R&D Systems). NK cell populations were purified as described previously (29). Eosinophils were purified from the fresh blood of asymptomatic hypereosinophilic donors by negative selection from the granulocyte pellet using anti-CD16 monoclonal antibodies (30). The purity of each cell population was determined by FACS®, and only populations showing >99% purity were used for experiments, except for monocytes that were routinely >80% pure. Peripheral blood monocyte–derived DCs were prepared as described previously (31). CD34+-derived DCs were obtained by CD34+ selection from PBMCs using CD34+ columns (Minimacs, Milteny Biotec, Bergisch Gladbach, Germany) as described (32). In brief, CD34+ cells were cultured at 5 × 104 cells/ml in RPMI-1640 containing 10% FCS supplemented with growth factors. During the first week, the following factors were used: stem cell factor (50 ng/ml) (Amgen, Thousand Oaks, CA) and GM-CSF (50 ng/ml) (Sandoz, Basel, Switzerland). During the second week, cells were maintained in GM-CSF and TNF (10 ng/ml) (Basf Knoll, Ludwigschaffen, Germany). Cells (70–80% CD1a+, MHC class II+, and CD80+) were used after 12–15 d of culture. Lung macrophages and lung DCs were purified from resected human lung using the method of Aubry et al. (33). In brief, lung tissue was dissociated with DNAse I and collagenase type I (Sigma Chemical Co., St. Louis, MO). The cell suspension was subjected to centrifugation through Ficoll and the resultant mononuclear cell layer was subsequently washed twice and plated at a concentration of 2 × 106 cells/ml on 10-cm-diam petri dishes. After incubation at 37°C for 1 h, nonadherent cells were removed by washing with HBSS. Cells adhering to the plastic were incubated overnight at 37°C. Loosely adherent mononuclear cells (LAMCs) were recovered by washing the monolayers four times with HBSS. LAMCs were then sorted by FACS®. Macrophages were sorted based on their autofluorescence. The remaining population was then sorted using four-color fluorescence, selecting for the CD40+, CD3−, CD14−, CD20−, and CD56− population representing lung DCs. The lung DCs (>95% purity) express high levels of HLA class II, CD80, and CD40 and are able to induce a strong mixed lymphocyte reaction.
Preparation of Total RNA and RT-PCR.
Total RNA was prepared from leukocytes using the TrizolTM reagent (Life Sciences, Paisley, Scotland) according to the instructions of the manufacturer. Reverse transcriptase reactions were performed on 3 μg of total RNA using an oligo dT primer with a reverse transcription kit (Promega) in a final volume of 20 μl. 2-μl aliquots of each reverse transcriptase reaction were then subjected to 40 cycles of PCR in a 50 μl reaction mixture containing 10 mM Tris–HCl buffer, pH 8.3, 50 mM KCl, 1.5 mM MgCl2, 0.2 mM dNTPS, 2.5 U of AmpliTaq, and 1 μM each of forward and reverse primer in a Crocodile III thermal cycler (Appligene, Strasbourg, France) or a Perkin-Elmer DNA thermal cycler. As control, PCR reactions were performed on RNA samples that had been incubated in the absence of reverse transcriptase. As a check for the quality of the cDNA used in each PCR reaction, we used glyceraldehyde 3-phosphate dehydrogenase primers as described previously (16). The identity of PCR products that corresponded to the predicted size was verified by sequencing, using the same primers as for the PCR reaction, after gel purification using a Wizard PCR preps kit (Promega).
Cloning of MIP-3α.
The full coding sequence of MIP-3α was cloned by RT-PCR using specific primers (5′ forward CGG GAT CCA CCA TGT GCT GTA CCA AGA GTT TG) and (5′ reverse CGG AAT TCC AGT TTT TAC ATG TTC TTG AC) based on the sequence deposited in the EMBL/GenBank/ DDBJ EST database (accession number D31065) for 40 cycles of PCR (95°C for 1 min, 37°C for 1 min, and 72°C for 1 min). The cDNA template used in the PCR reaction was either 2 μl of 16 different Quick Clone cDNAs (Clontech, Palo Alto, CA) or was generated by reverse transcription of isolated leukocyte RNAs (as described above). PCR products corresponding to the predicted size of 306 bp were gel purified and subcloned as BamHI–EcoRI fragments into the mammalian cell expression vector pcDNA3.1(+) (Invitrogen, San Diego, CA) and sequenced using T7 and pcDNA3AS primers. Two of the resultant clones, MIP-3α–11, which contained the full error-free coding sequence of MIP-3α, and MIP-3α–16, which contained a 3-bp deletion in the putative signal peptide coding sequence, were used in subsequent experiments.
Analysis of MIP-3α Messenger RNA Expression.
Multiple tissue Northern blots (Clontech) and Northern Territory blots (Invitrogen) were probed with a 306-bp BamHI–EcoRI insert from MIP-3α–11 according to the instructions of the manufacturer. [α-32P]dCTP-labeled probes were generated using an Amersham Rediprime kit.
Degenerate Oligonucleotide PCR to Identify Chemokine Receptors Expressed in Human Lung DCs.
Total RNA was isolated from 5 × 105 FACS® purified human lung DCs using Trizol™. Single-stranded cDNA was prepared from the resultant RNA using the Promega reverse transcription system. One-tenth of the reaction mixture was subjected to 40 cycles of PCR (95°C for 1 min, 37°C for 1 min, and 72°C for 1 min) using 3 μM of each degenerate oligonucleotide primer (5′ forward GAY MGI TAY YTI GCI ATH GTX CA and 5′ reverse RMR TAI ADI AII GGR TTI AXR CA ) in a Perkin-Elmer DNA thermal cycler. PCR reaction products were visualized on 1% agarose gels containing 0.5 μg/ml ethidium bromide. Reaction products migrating at the predicted size (500–550 bp) were gel purified, subcloned into pBluescript II KS(−) and sequenced on an Applied Biosystems ABI 377 machine using T3 and T7 primers as described elsewhere (16).
Cloning of Full-Length DCCR2.
The full coding sequence of DC chemokine receptor 2 (DCCR2) was cloned from human lung DCs by RT-PCR essentially as described above using 1 μM of specific primers (based on the sequence of full-length cDNA deposited in the EMBL/Genbank/DDBJ database under accession number U45984) (5′ forward TCA ATG AAT TTC AGC GAT GTT TTC G) and (5′ reverse CTA TCA CAT AGT GAA GGA CGA CGC) using 40 cycles of PCR (95°C for 2 min, 55°C for 2 min, and 72°C for 2 min). PCR products corresponding to the predicted size of 1.1 kb were gel purified and subcloned into the EcoRV site of the mammalian cell expression vector pcDNA3.1(+) and sequenced using T7 and pcDNA3AS primers. One of the resultant clones, DCCR2–10, which was identical to the U45984 coding sequence (also known as GCY-4), was subsequently used for expression studies.
Transient Expression of DCCR2 in HEK 293 Cells.
HEK 293 cells were maintained in DMEM–F12 medium containing 10% heat-inactivated FCS, 2 mM glutamine, and 100 U/ml penicillin–streptomycin. 24 h before transfection cells were plated at 2 × 106 cells per 10-cm-diam petri dish or at 105 cells per well in 96-well black-walled microtiter plates (Polyfiltronics Group, Inc., Rockland, MA) precoated with 0.1% gelatin and 10 μg/ml poly l-lysine (Sigma Chemical Co.). Cells were transfected using a calcium phosphate transfection kit (Life Sciences) according to the instructions of the manufacturer. Cytosolic free calcium concentration ([Ca2+]i) and radioligand binding were measured 36–72 h after transfection.
Transient Expression of MIP-3α by HEK 293 Cells.
HEK 293 cells plated in 10-cm diameter petri dishes were transfected as described above with pcDNA3.1(+) MIP-3α or pcDNA3.1(+) chloramphenicol acetyl transferase (CAT) as control except that calcium phosphate precipitate containing medium was replaced with serum-free medium 16 h after transfection. Conditioned medium was collected 48 h later. Cell debris was removed by centrifugation at 2,000 rpm for 5 min and diluted medium was used directly in chemotaxis assays.
For verification of the NH2-terminal amino acid sequence of mature MIP-3α, conditioned medium was concentrated 10-fold using Centriprep-3 ultrafiltration units (Millipore Corp., Milford, MA) and subjected to reverse phase HPLC using an Aquapore RP-300 7 μm column (0.2 × 22 cm) equilibrated in 0.1% trifluoroacetic acid. Proteins were eluted with a linear gradient of 22.5–45% acetonitrile in 0.1% trifluoroacetic acid and analyzed by 4–20% SDS-PAGE. Bands migrating at the predicted molecular mass for chemokines (8 kD) were purified and sequenced on a (494 Procise; Applied Biosystems) protein sequencer.
Calcium Fluorometry.
Cytosolic-free calcium determinations were conducted in monolayers of HEK 293 cells transiently expressing the DCCR2 receptor by adapting the method of Capponi et al. (34). In brief, black-walled 96-well microtiter plates containing confluent monolayers of transfected cells were washed with 150 μl/well serum-free DMEM–F12 medium containing 2 mM glutamine and 100 U/ml penicillin–streptomycin (serum-free medium). The washes were removed, and the monolayers were incubated at 37°C in a water-saturated air–CO2 atmosphere (19:1) for 60 min with 100 μl/well serum-free medium containing 1 μM cyclosporin A, 1 μM probenicid, and 4 μM Fluo-3AM previously dissolved in 20% pluronic acid (1 mg/ml stock). After the loading procedure, the monolayers were washed four times at room temperature with 150 μl/well 10 mM Hepes buffer, pH 7.4, containing 145 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM glucose, 1 μM cyclosporin A, and 1 μM probenicid (assay buffer), after which 180 μl of the same buffer was added to each well, and the plates were used immediately. Chemokine-induced calcium mobilization and influx in DCCR2 receptor–expressing HEK 293 cells was monitored at room temperature by means of a fluorescent imaging plate reader (Molecular Devices Ltd., Menlo Park, CA). The excitation wavelength of 488 nm was supplied by a 3W argon laser (Coherent) set at 0.8– 1.2 W according to the level of cell confluency. Fluorescent emission was measured at 512 nM. All assays were conducted at a mean fluorescent baseline signal of 3.0–3.5 × 104 fluorescent cpm per well and per plate. Lens aperture was maintained constant at f/1.4, and shutter speed was set at 0.3–0.4 s. A total of 120 sampling points were collected for each well over 6–8-min periods. Chemokine experiments were run by adding 20 μl of each chemokine at 10-fold the desired final concentration to each well, and monitoring Fluo-3 fluorescence for a further 6 min. For antagonist assays, 20 μl of each chemokine or pharmacological agent of interest was added at 10-fold the desired final concentration to each well, followed by a 5-min incubation period after which 20 μl of synthetic MIP-3α was added to each sample (final [MIP-3α] = 100 nM), and fluorescence was monitored for an additional 3 min. Sample injection speed was set at 175 μl/min, and pipettor height at 180 μl. Under these conditions, maximal signal amplitude was an additional 0.5–1.0 × 104 fluorescent counts upon addition of 100 nM synthetic MIP-3α to wells containing only vehicle. Experiments with pertussis toxin–treated cells were conducted by preincubating DCCR2-expressing cells with 500 nM pertussis toxin as previously described (35). For statistical analysis of fluorometry experiments, Student–Fisher unpaired bilateral t tests and analysis of variance using the Scheffe F test criterion for unbalanced groups were used, where applicable. The data represent the means ± SEM of at least four experiments performed in single determinations.
Chemotaxis Assays.
Chemotaxis assays were performed in 48-well micro chemotaxis chambers (Neuroprobe, Inc., Cabin John, MD). Neutrophils, monocytes, and DCs were suspended at 106 cells/ml, and T cells at 2 × 106 cells/ml, in RPMI medium containing 2 mM glutamine and 25 mM Hepes, and applied to the top wells of the chamber. Standard 5-μm pore filters for monocytes, T cells, and DCs and 3-μm filters for neutrophils separated the cells from the bottom chamber, which contained dilutions of conditioned medium from cells transfected with either pcDNA3.1(+) MIP-3α–11, MIP-3α–16 or pcDNA3.1(+) CAT alone (mock transfected) or synthetic MIP-3α, at concentrations of 10−10–10−6 M. Chambers were incubated at 37°C for 20 min (neutrophils), 60 min (monocytes and T cells), or 90 min (DCs), after which time cells that had migrated onto the underside of the filter were stained using Diff-Quik (Baxter Scientific, Inc., Unterschliesshe, Germany) and counted using an image analyzer or five high power (×100) oil immersion fields. Human recombinant MCP-1, MCP-3, and IL-8 were used as positive controls in the chemotaxis assays at a concentration of 100 nM. Each data point was performed in triplicate and the results are representative of at least three independent experiments using leukocytes from different donors.
Radioligand Competition Binding Analysis.
Binding assays were performed in triplicate on 105 HEK 293 cells transfected with DCCR2/pcDNA3.1(+) or with pcDNA3.1/CAT (as control), in Millipore 96-well filtration plates as described previously (26). Results are representative of at least three different experiments. The data obtained were curve-fitted to obtain the IC50 values using GraFit version 3.01 software (Erithicus Software Ltd., Staines, UK) according to a modified four-parameter logistic equation where F = B/Bmaxapp = 1 / (1 + [L]n / IC50) + background, where B = cpm bound, Bmaxapp = cpm bound in the absence of competing ligand, [L] is the concentration of competing ligand, and n is the slope of the curve.
Results
Cloning of a Novel CC Chemokine, MIP-3α.
A search of the human EST databases revealed the presence of several novel chemokine-like sequences (8). The full coding sequence of one of the EST sequences that appeared to encode a novel CC chemokine was cloned by reverse transcriptase PCR using primers based on the EMBL/GenBank/ DDBJ database accession number, D31065. Of the 16 cDNA libraries and 8 leukocyte cDNAs tested, D31065 cDNA was only detected in lung macrophages, lung DCs, and eosinophils. Sequencing of one of the resultant clones (clone 11) from the macrophage PCR products revealed that it was identical to the recently described MIP-3α and LARC sequences (Fig. 1). In addition to the D31065 sequence, we also detected a population of PCR products that contained an in-frame, three nucleotide deletion in the putative signal sequence coding region. As the resultant amino acid deletion may effect the position of cleavage of the signal peptide, we expressed both forms of MIP-3α, MIP-3α clone 11 (wild type), and MIP-3α clone 16 (mutant) in HEK 293 cells. MIP-3α protein was purified from conditioned medium 48 h after transfection. Amino acid sequence analysis demonstrated that the NH2 terminus of wild-type MIP-3α occurred at Ala 27. By contrast, the NH2 terminus of the mutant MIP-3α was one amino shorter starting at Ser 27.
Figure 1.
Amino acid sequence of MIP-3α (clone 11). The signal peptide is underlined. The amino acid deleted in MIP-3α (clone 16) is italicized. Arrows denote the NH2-terminal amino acid of the mature protein (a), synthetic MIP-3α (b), wild-type MIP-3α (clone 11), and (c) mutant MIP-3α (clone 16).
Analysis of MIP-3α Messenger RNA Expression in Human Tissues and Leukocytes.
The expression of MIP-3α messenger RNA (mRNA) was analyzed in human multiple tissue Northern blots (Fig. 2 A). MIP-3α mRNA was highly expressed in lung and in inflammed tissues such as tonsil and appendix. Weaker expression was also detected, predominantly at mucosal sites and in lymphatic tissues: placenta, gall bladder, stomach, thymus, prostrate, testis, small intestine, and colon. RT-PCR analysis of distinct leukocyte populations indicated MIP-3α mRNA present in lung macrophages, DCs, and peripheral blood eosinophils (Fig. 2 B).
Figure 2.


(A) Northern blot analysis of MIP-3α expression in human tissues. (a) Clontech multiple tissue Northern blots (1 μg poly A+ mRNA/lane); (b) Invitrogen Northern territory blots (20 μg total RNA/ lane). (B) RT-PCR analysis of MIP-3α expression in leukocytes. (Top) MIP-3α; (bottom) glyceraldehyde 3-phosphate dehydrogenase control. Molecular weight markers (1-kb ladder) are shown on the left. Lane 1, lung macrophages; lane 2, T cells; lane 3, monocytes; lane 4, neutrophils; lane 5, eosinophils; lane 6, peripheral blood monocyte–derived DCs; lane 7, NK cells (IL-2–stimulated); lane 8, lung DCs.
Chemoattractant Activity of MIP-3α.
Synthetic MIP-3α was tested at concentrations of 10−6 to 10−10 M in chemotaxis assays to characterize the leukocyte populations that responded to this chemokine (Fig. 3). MIP-3α was a potent chemoattractant for T lymphocytes and CD34+-derived DCs, which migrated with typical bell-shaped dose– response curves. The maximal chemotactic activity on T cells was observed at 10−9 M, whereas on CD34+ DCs the potency was 12.5-fold lower. No chemotactic activity was observed with peripheral blood monocyte–derived DCs, monocytes, or neutrophils. We also tested the chemotactic activity of conditioned medium from HEK 293 cells transfected with wild-type MIP-3α (MIP-3α clone 11) or mutant MIP-3α (MIP-3α clone 16). The level of expression of both proteins (estimated from the HPLC peak height) was similar and ∼0.01–0.1 μg/ml. Whereas the media from both transfectants was chemotactic when compared with conditioned media from mock transfectants, the MIP-3α wild type appeared to be at least 2.5-fold more efficacious than the mutant form.
Figure 3.
Chemotaxis of leukocytes in response to synthetic MIP-3α. (a) T cells; (b) monocytes; (c) neutrophils. The response to MIP-3α is indicated by the open circles. As controls for the chemotaxis we used 100 nM each of MCP-1 for T cells and monocytes and IL-8 for neutrophils (closed circles). (d) CD34+ DCs (closed circles) and peripheral blood monocyte–derived DCs (open circles). (e) Chemotaxis of T cells in response to conditioned medium from MIP-3α clone 11 transfectants (open circles); MIP-3α clone 16 transfectants (closed circles); and mock transfectants (open squares).
Identification of Candidate MIP-3a Receptors Expressed on T Cells and DCs by Degenerate Oligonucleotide RT-PCR.
MIP-3α was unable to displace the binding of specific chemokine ligands to Chinese hamster ovary or HEK 293 cells stably expressing the known chemokine receptors CCR1–5 and CXCR1 and CXCR2 (data not shown). Therefore, we used a degenerate oligonucleotide PCR strategy to identify other candidate chemokine receptor–like cDNAs expressed in these cells. Degenerate oligonucleotide primers based on the conserved amino acid sequence found in the second intracellular loop and the seventh transmembrane domain of the known human chemokine receptors were used in RT-PCR experiments on human lung DCs. PCR products corresponding to the predicted size of ∼500–550 bp were gel purified, subcloned, and sequenced. The majority of the resultant clones corresponded to known chemokine receptors CCR1–5 and CXCR1–4. In addition, we also detected several cDNAs encoding orphan chemokine receptor–like sequences.
One of the orphan receptor sequences that we identified in DCs, which we have called DCCR2, had earlier been identified as GCY4, CKR-L3 (25), or STRL22 (24). RT-PCR on RNA extracted from distinct human leukocyte populations indicated that DCCR2 mRNA expression was restricted to T cells, CD34+ DCs, and lung DCs but not on peripheral blood monocyte–derived DCs (Fig. 4). Full-length DCCR2 was subsequently cloned from human lung DCs by RT-PCR using specific primers based on the full-length sequence given in the EMBL/GenBank/DDBJ database under accession number U48494. The resultant 1.1-kb cDNA was subcloned into the mammalian cell expression vector pcDNA3.1 and sequenced. One of the clones, DCCR2–10, had an identical sequence to that deposited under U48494. The phylogenetic relationship of DCCR2 with other chemokine receptors is shown in Fig. 5. DCCR2 clusters with the CXC chemokine receptor family being most closely related to another orphan receptor, EBI-1 (36) (42% amino acid identity), which just before submission of this manuscript, was reported to be CCR7 (37).
Figure 4.
RT-PCR analysis of DCCR2 expression in leukocytes. Molecular weight markers are shown on the left. Lane 1, lung DCs; lane 2, peripheral blood monocyte–derived DCs; lane 3, CD34+ DCs; lane 4, CD4 T cells; lane 5, CD8 T cells.
Figure 5.
Dendrogram showing the relationship of DCCR2 with known chemokine receptors and a number of orphan receptors. The EMBL/GenBank/ DDBJ accession numbers for the receptors are CCR1, P32246; CCR2, P41597; CCR3, P51677; CCR4, P51679; CCR5, P51681; V28 (orphan receptor), U28934; U95626 (orphan receptor); TER1 (CCR8), U45983; CXCR1, P25024; CXCR2, P25025; CXCR3, P49682; CXCR4, Q28474; CCR7/EBI1, P32248; DCCR2 is GCY-4, U45984; BLR1 (orphan receptor), X68149; and DARC, Q16570.
Functional Expression of DCCR2 in HEK 293 Cells.
It has previously been shown that chemokine receptors typically couple to phospholipase C (PLC) through a pertussis toxin–sensitive G protein–dependent mechanism, which subsequently leads to increased phosphatidyl inositol 4, 5, bisphosphate hydrolysis, and inositol 1, 4, 5 trisphosphate (IP3)–dependent calcium mobilization (23). In view of this, we attempted to identify the putative ligand(s) for DCCR2 by monitoring intracellular free calcium levels in monolayers of transiently transfected HEK 293 cells exposed to a battery of 18 different human CC and CXC chemokines. Of all the chemokines tested at 250 nM–1 μM concentrations (MCP-1, MCP-3, MIP-1α, MIP-1β, MIP-3α, MIP-5, RANTES, eotaxin, HCC-1, I-309, fractalkine, lymphotactin, SDF-1α, SDF-1β, IL-8, NAP-2, GROα, and IP-10), only MIP-3α induced a significant calcium response in these preparations. As shown in Fig. 6, incubation of DCCR2 expressing HEK 293 cells with 100 nM MIP-3α promoted a rapid, transient increase in cytosolic-free calcium, a response that was maximal within 25–30 s of application (from 2,230 ± 489 cpm to 7,541 ± 1,237 cpm at 25 s; n = 8, P <0.05 versus basal), and was sustained for up to 5 min thereafter (4,234 ± 869 cpm at 3.5 min, n = 8). Furthermore, the response was dose dependent, exhibiting a half-maximal effective concentration (EC50) of 100–200 nM.
Figure 6.
Effect of MIP-3α on intracellular free calcium in DCCR2 receptor–expressing HEK 293 cells. HEK 293 cells transiently expressing the DCCR2 receptor were loaded with Fluo-3AM and exposed to either vehicle (buffer) or to 50 nM–1 μM concentrations of MIP-3α. Calcium fluorometry was conducted as described in Materials and Methods. Tracings represent the means of eight experiments conducted in single determinations.
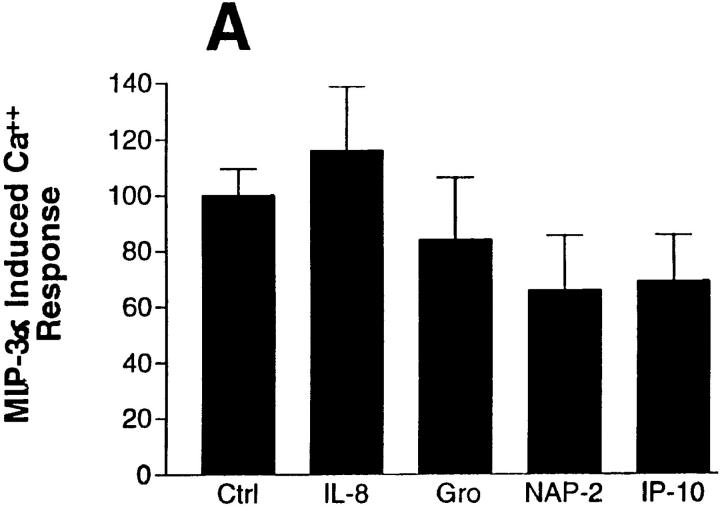
In view of the observations that many CC chemokines bind to more than one chemokine receptor in vitro, but that only MIP-3α promotes significant increases in cytosolic-free calcium in our preparations, we further attempted to detect chemokine–DCCR2 interactions by submitting transfected HEK 293 cells to desensitization protocols in which the cells were exposed for 5 min to 500 nM to 1 μM concentrations of 14 CC and CXC chemokines before being challenged with 100 nM MIP-3α. As shown in Fig. 7 A, preexposure of cells to 1 μM concentrations of IL-8, GROα, NAP-2, or IP-10 had no significant effect on the calcium signal induced by 100 nM MIP-3α (n = 4). Preincubation with 500 nM to 1 μM concentrations of nine human CC chemokines (MCP-1, MIP-1α, MIP-1β, MIP-5, RANTES, eotaxin, HCC-1, I-309, and fractalkine), also had no effect significant on MIP-3α–induced calcium responses (n = 4; see Fig. 7 B), although preincubation with both 1 μM RANTES and 1 μM HCC-1 did decrease the MIP-3α–induced calcium response in two out of four experiments. Indeed, only preincubation with 1 μM MIP-3α led to a significant decrease in MIP-3α–induced calcium responses. The effect appeared to correspond to near maximal receptor desensitization (97 ± 12% inhibition of calcium response; n = 4; see Fig. 7 B), and was dose dependent, with MIP-3α exhibiting a half-maximal inhibiting concentration (IC50) of 1–10 nM (n = 4; see Fig. 7 C).
Figure 7.
Effect of preincubation with chemokines on MIP-3α–induced calcium responses in DCCR2 receptor–expressing cells. HEK 293 cells transiently expressing DCCR2 were loaded with Fluo-3AM, and exposed for 5 min to either 1 μM IL-8, 1 μM GROα, 1 μM NAP-2, or 500 nM IP-10 (A); 1 μM RANTES, 1 μM MIP-1α, 1 μM MIP-1β, 1 μM MIP-3α, 1 μM MIP-5, 1 μM MCP-1, 1 μM eotaxin, 1 μM fractalkine, 1 μM HCC-1, or 1 μM I-309 (B), or to increasing concentrations of MIP-3α (C), followed by addition of 100 nM MIP-3α. Calcium fluorometry was conducted as described in Materials and Methods. Results represent the mean calcium signal amplitudes of four separate experiments conducted in single determinations. Asterisk, P <0.05 versus effect of 100 nM MIP-3a preceded by vehicle pretreatment, or 1 nM MIP-3α pretreatment for C. The value of 100 = normalized response induced by 100 nM MIP-1α.
We also investigated the cell signaling pathways underlying MIP-3α–induced calcium responses in DCCR2– expressing HEK 293 cells by monitoring the effect of MIP-3α on cytosolic free calcium in pertussis toxin–treated monolayers, a procedure that leads to the inhibition of G protein effector coupling by inactivational ADP–ribosylation. As shown in Fig. 8 A, a 24-h pretreatment of DCCR2 receptor–expressing cells with 500 nM pertussis toxin led to a marked decrease in the calcium responses induced by either 100 or 500 nM MIP-3α (74% and 61% inhibition, respectively; n = 4, P <0.05), a result suggesting that the G protein–dependent activation of a PLC is implicated in MIP-3α–induced calcium responses in this system. Consistent with this last hypothesis, MIP-3α–induced calcium responses were only partially inhibited when studied in the presence of 1.6 mM extracellular EGTA (30% inhibition; n = 4, P <0.05; see Fig. 8 B), a result suggesting that a substantial portion of the MIP-3α–induced calcium response is derived from the IP3-mediated mobilization of calcium from intracellular stores. In agreement with this possibility, MIP-3α–induced calcium responses were reduced in the presence of 10 μM of the selective PLC inhibitor, U-73122 (38), as well as in cells pretreated with 1 μM 4b PMA, a widely used experimental condition known to activate protein kinase C, and consequently inhibit PLC through a negative feedback process (39). Consistent with these last results, preincubation of DCCR2 receptor– expressing cells with the Ca2+–ATPase inhibitors thapsigargin and cyclopiazonate (40, 41) promoted a significant inhibition of the calcium response induced by MIP-3α (55 and 60% inhibition at 1 and 120 μM, respectively; n = 4, P <0.05; see Fig. 8 D).
Figure 8.
Effect of pertussis toxin treatment, EGTA, PLC inhibition, and Ca2+–ATPase inhibition on MIP-3α–induced calcium responses in DCCR2-expressing HEK 293 cells. (A) HEK 293 cells transiently expressing the DCCR2 were treated overnight with vehicle (−Ptx) or 500 nM pertussis toxin (+Ptx), loaded with Fluo-3AM, and incubated with increasing concentrations of MIP-3α. Calcium fluorometry was conducted as described in Materials and Methods. (B) Cells were stimulated with 100 nM MIP-3α in the presence (plus) and absence (minus) of 1.6 mM extracellular EGTA. (C) Cells were exposed for 5 min to 10 μM U-73122 (U73) or 1 μM PMA before addition of 100 nM MIP-3α. (D) Cells were preincubated for 5 min with increasing concentrations of thapsigargin (closed squares) or cyclopiazonate (open circles) before addition of 100 nM MIP-3α. Results in A, C, and D represent the mean maximal calcium signal amplitudes of four separate experiments conducted in single determinations. Asterisk, P <0.05 versus corresponding control. B shows averaged tracings from four experiments. The value of 100 = normalized response induced by 100 nM MIP-1α.
Binding Studies.
We also tested the ability of the DCCR2-transfected HEK 293 cells to bind to radiolabeled synthetic MIP-3α. In competition binding assays, synthetic MIP-3α was able to displace [I125]-MIP-3α with an IC50 value of ∼12 nM (Fig. 9).
Figure 9.
Competition binding of [I125] MIP-3α to HEK 293 cells transiently transfected with DCCR2. Binding assays were performed as described in Materials and Methods. Open circles denote binding to pcDNA3.1(+) DCCR2 transfectants; closed circles denote binding to pcDNA3.1(+) CAT transfectants.
Discussion
Chemokines play an important role in the trafficking of immune cells around the body and in diverse physiological processes such as inflammation, infection, hematopoeisis, and development (42, 43). Their actions are mediated through a family of highly homologous, G protein–coupled, seven-transmembrane receptors. Here, we report the cloning and functional characterization of a novel chemokine receptor from lung DCs that is highly specific for the recently described CC chemokine, MIP-3α.
MIP-3α was first identified in EST databases (8, 21, 22). We have shown by Northern blot analysis that the mRNA for this chemokine is highly expressed in potentially inflamed tissues such as tonsil and appendix, and in lymphoid and mucosal tissues, particularly in lung. Despite the earlier identification of LARC as a liver chemokine (22), we found only weak, barely detectable expression in liver on two independent Northern blots, suggesting that the expression may vary among individuals, possibly depending on immune status. The specific cell types that express MIP-3α appear to be macrophages, eosinophils, and DCs, as shown by RT-PCR experiments. These cell populations are known to be well represented at mucosal sites. In lung, macrophages are found in large numbers both within the airway lumen and the lung interstitium. They are the first line of defense against invading organisms and allergens and play an important role in innate and acquired immunity (44, 45). Lung DCs reside within the airway epithelium and are the most potent APCs known (46, 47). These cells uptake, process, and present foreign antigen or allergens to T cells either in situ or at local lymph nodes, resulting in T cell activation and proliferation. Eosinophils are not usually found in large numbers in normal human lung. However, they do occur at other mucosal sites, notably in the gastro– intestinal tract of normal individuals, where they are thought to play a major role in host defense against mucosal pathogens (48). Thus, the finding that MIP-3α mRNA is constitutively expressed at high levels in these cell types suggest that MIP-3α may play an important role in the rapid recruitment of inflammatory cells to potential sites of infection.
To identify the specific cell populations which responded to MIP-3α, and for characterization of the MIP-3α receptor, we used chemically synthesized protein. Synthetic MIP-3α was chemotactic for T cells but not for monocytes and neutrophils. The maximal chemotactic effect was observed at around 10−9 M. MIP-3α was also chemotactic for CD34+-derived DCs but was an order of magnitude less potent than on T cells. Peripheral blood monocyte–derived DCs did not migrate in response to MIP-3α. Differences in chemokine responsiveness of CD34+ derived DCs and peripheral blood monocyte–derived DCs have been reported in the literature (31, 49). These differences in the chemotactic activity may be a consequence of different degrees of maturation of the DCs or reflect important functional differences between DCs obtained from distinct sources. We have preliminary data to show that MIP-3α is chemotactic on lung DCs between 10−7 and 10−9 M but due to the difficulties in obtaining sufficient numbers of lung DCs to do statistically significant numbers of chemotaxis assays, we are unable to present this data at present.
We and others have shown that MIP-3α does not act through any of the known CC chemokine receptors or through CXCR1 or CXCR2 (CXCR3, CXCR4, and DARC were not tested). We have cloned a number of orphan chemokine receptor–like molecules from lung DCs in addition to the published chemokine receptors. One of these orphans, DCCR2, variously known as GCY-4, STRL-22, or CKRL-3 was shown by RT-PCR to be expressed in T cells and certain DC populations, an mRNA expression pattern consistent the MIP-3α responsiveness in chemotaxis assays.
We also demonstrated that synthetic MIP-3α is able to mobilize intracellular Ca2+ in HEK 293 cells transiently expressing DCCR2. The maximal effect of MIP-3α was observed at 1 μM and could not be agonized or antagonized by at least 18 other human chemokines tested, thus confirming the specificity of this receptor. In common with other CC chemokine receptors, DCCR2-dependent calcium responses appear to result from both EDTA-sensitive calcium influx on the one hand, and pertussis toxin–sensitive PLC activation on the other. Furthermore, the results obtained in the presence of U-73122 and in PMA-treated cells suggest that IP3-mediated calcium mobilization is at the basis of this second element of the response, particularly as depletion of intracellular calcium stores with either thapsigargin or cyclopiazonate also inhibit the MIP-3α– induced increases in cytosolic free calcium. Interestingly, the EC50 of the Ca2+ response was much higher than the concentration of MIP-3α required to produce a maximal chemotactic effect in T cells and CD34+ derived DCs, and the IC50 of radiolabeled MIP-3α binding to HEK 293 cells transiently expressing DCCR2. This may be due to the fact that different concentrations of chemokine may be required for different physiological functions as has been observed for other chemokines (50). Earlier studies have shown that modifications at the NH2 terminus of some chemokines can alter their activity; for example, the addition of methionine to the NH2 terminus of RANTES changes it from an agonist of CC chemokine receptors, to an antagonist, without changing its receptor binding properties (51). While it is clear that the NH2 terminus of the synthetic MIP-3α used in this study contains an additional three amino acids compared with the mature protein expressed in HEK 293 cells, our results would indicate that this modification is only likely to change the binding affinity and potency in different functional assays rather than receptor specificity. In support of this, recent studies on mature LARC produced in insect cells indicated that it failed to bind to any of the known chemokine receptors (22) suggesting that the synthetic MIP-3α used here behaves in a manner analogous to the recombinantly expressed protein. However, it remains to be seen whether there are other as yet uncharacterized orphan receptors that bind MIP-3α or, indeed, whether other ligands for DCCR2 will be found amongst the ever-increasing number of chemokines.
DCCR2 shows several features that make it distinct from other CC chemokine receptors: the gene for DCCR2 resides on chromosome 6q27 (24); all of the other CC chemokine receptors identified to date have been localized on chromosome 3p21–24 (52); so far, it appears to be highly selective for a single ligand (MIP-3α), unlike the other CC chemokine receptors, which are activated by at least two to three chemokine ligands; and the constitutive mRNA expression of DCCR2 seems to be highly restricted to T cells and DCs. The finding of both ligand and receptor in DCs is perhaps also indicative of an autocrine loop existing in these cells, suggesting that the ligand, MIP-3α, may have functions in addition to being a chemoattractant.
The primary role of DCs is to take up and process foreign antigens, which are then presented to naive T cells, thus initiating the immune cascade. During inflammation, the numbers of DCs resident in tissues increases and there is also increased trafficking of DCs from the tissue to the local lymph nodes (53). This ensures rapid resolution of the infection or inflammation. However, excessive local T cell activation by DCs may be an important contributory factor in allergic diseases such as asthma. As DCCR2 shows relatively restricted expression to T cells and DCs, and is highly specific for MIP-3α, this ligand–receptor pair is likely to be an important therapeutic target for modulation of the immune response.
Acknowledgments
The authors thank Dr. J.-P. Aubry for FACS®; F. Borlat, L. Menoud, and Y. Cambet for technical assistance; D. Bertschy and M. Huguenin for DNA sequencing; E. Magenat for protein sequencing; and C. Hebert for photography.
Footnotes
Abbreviations used in this paper: CAT, cloramphenicol acetyl transferase; DARC, Duffy antigen receptor; DCs, dendritic cells; DCCR2, dendritic cell chemokine receptor 2; EST, expressed sequence tag; HEK, human embryonic kidney; LAMCs, loosely adherent mononuclear cells; LARC, liver and activation-regulated chemokine; mRNA, messenger RNA; PLC, phospholipase C; RT, reverse transcriptase; SDF-1, stromal cell– derived factor-1.
References
- 1.Steinman RM. The dendritic cell system and its role in immunogenicity. Annu Rev Immunol. 1991;9:271–296. doi: 10.1146/annurev.iy.09.040191.001415. [DOI] [PubMed] [Google Scholar]
- 2.Schall, T.J. 1994. The Chemokines. In The Cytokine Handbook. A. Thompson, editor. Academic Press, New York. 419–460.
- 3.Kelner GS, Kennedy J, Bacon KB, Kleyensteuber S, Largaespada DA, Jenkins NA, Copeland NG, Bazan JF, Moore KW, Schall TJ. Lymphotactin: a cytokine that represents a new class of chemokine. Science (Wash DC) 1994;266:1395–1399. doi: 10.1126/science.7973732. [DOI] [PubMed] [Google Scholar]
- 4.Bazan JF, Bacon KB, Hardiman G, Wang W, Soo K, Rossi D, Greaves DR, Zlotnik A, Schall TJ. A new class of membrane-bound chemokine with a CX3C motif. Nature (Lond) 1997;385:640–644. doi: 10.1038/385640a0. [DOI] [PubMed] [Google Scholar]
- 5.Tashiro K, Tada H, Heilker R, Shirozu M, Nakano T, Honjo T. Signal sequence trap: a cloning strategy for secreted proteins and type I membrane proteins. Science (Wash DC) 1993;261:600–603. doi: 10.1126/science.8342023. [DOI] [PubMed] [Google Scholar]
- 6.Hieshima K, Imai T, Opdenakker G, Van Damme J, Kusuda J, Tei H, Sakaki Y, Takatsuki K, Miura R, Yoshie O, Nomiyama H. Molecular cloning of a novel human CC chemokine liver and activation-regulated chemokine (LARC) expressed in liver. Chemotactic activity for lymphocytes and gene localization on chromosome 2. J Biol Chem. 1997;272:5846–5853. doi: 10.1074/jbc.272.9.5846. [DOI] [PubMed] [Google Scholar]
- 7.Schall TJ, Jongstra J, Dyer BJ, Jorgensen J, Clayberger C, Davis MM, Krensky AM. A human T cell–specific molecule is a member of a new gene family. J Immunol. 1988;141:1018–1025. [PubMed] [Google Scholar]
- 8.Wells TNC, Peitsch MC. The chemokine information source: identification and characterisation of novel chemokines from expressed sequence tags using the world wide web. J Leukocyte Biol. 1997;61:545–550. doi: 10.1002/jlb.61.5.545. [DOI] [PubMed] [Google Scholar]
- 9.Holmes WE, Lee J, Kuang WJ, Rice GC, Wood WI. Structure and functional expression of a human interleukin-8 receptor. Science (Wash DC) 1991;253:1278–1280. [Google Scholar]
- 10.Murphy PM, Tiffany HL. Cloning of complementary DNA encoding a functional human interleukin-8 receptor. Science (Wash DC) 1991;253:1280–1283. doi: 10.1126/science.1891716. [DOI] [PubMed] [Google Scholar]
- 11.Loetscher M, Gerber B, Loetscher P, Jones SA, Piali L, Clark-Lewis I, Baggiolini M, Moser B. Chemokine receptor specific for IP-10 and MIG: structure, function, and expression in activated T-lymphocytes. J Exp Med. 1996;184:963–969. doi: 10.1084/jem.184.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagasawa T, Nakajima T, Tachibana K, Iizasa H, Bleul CC, Yoshie O, Matsushima K, Yoshida N, Springer TA, Kishimoto T. Molecular cloning and characterization of a murine pre-B-cell growth-stimulating factor/stromal cell–derived factor 1 receptor, a murine homolog of the human immunodeficiency virus 1 entry co-receptor fusin. Proc Natl Acad Sci USA. 1996;93:14726–14729. doi: 10.1073/pnas.93.25.14726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neote K, DiGregorio D, Mak JY, Horuk R, Schall TJ. Molecular cloning, functional expression, and signaling characteristics of a C–C chemokine receptor. Cell. 1993;72:415–425. doi: 10.1016/0092-8674(93)90118-a. [DOI] [PubMed] [Google Scholar]
- 14.Charo IF, Myers SJ, Herman A, Franci C, Connolly AJ, Coughlin SR. Molecular cloning and functional expression of two monocyte chemoattractant protein 1 receptors reveals alternative splicing of the carboxy-terminal tails. Proc Natl Acad Sci USA. 1994;91:2752–2756. doi: 10.1073/pnas.91.7.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ponath PD, Qin S, Post TW, Wang J, Wu L, Gerard NP, Newman W, Gerard C, Mackay CR. Molecular cloning and characterization of a human eotaxin receptor expressed selectively on eosinophils. J Exp Med. 1996;183:2437–2448. doi: 10.1084/jem.183.6.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Power CA, Meyer A, Nemeth K, Bacon KB, Hoogewerf AJ, Proudfoot AE, Wells TN. Molecular cloning and functional expression of a novel CC chemokine receptor cDNA from a human basophilic cell line. J Biol Chem. 1995;270:19495–19500. doi: 10.1074/jbc.270.33.19495. [DOI] [PubMed] [Google Scholar]
- 17.Samson M, Labbe O, Mollereau C, Vassart G, Parmentier M. Molecular cloning and functional expression of a new human CC–chemokine receptor gene. Biochemistry. 1996;35:3362–3367. doi: 10.1021/bi952950g. [DOI] [PubMed] [Google Scholar]
- 18.Chaudhuri A, Zbrzezna V, Polyakova J, Pogo AO, Hesselgesser J, Horuk R. Expression of the Duffy antigen in K562 cells. Evidence that it is the human erythrocyte chemokine receptor. J Biol Chem. 1994;269:7835–7838. [PubMed] [Google Scholar]
- 19.Cook DN, Beck MA, Coffman TM, Kirby SL, Sheridan JF, Pragnell IB, Smithies O. Requirement of MIP-1 alpha for an inflammatory response to viral infection. Science (Wash DC) 1995;269:1583–1585. doi: 10.1126/science.7667639. [DOI] [PubMed] [Google Scholar]
- 20.Power CA, Wells TNC. Cloning and characterization of human chemokine receptors. Trends Pharmacol Sci. 1996;17:209–213. doi: 10.1016/0165-6147(96)10019-5. [DOI] [PubMed] [Google Scholar]
- 21.Rossi D, Vicari A, Bacon-Franz K, McClanahan TK, Zlotnik A. Identification through bioinformatics of two new macrophage proinflammatory human chemokines MIP-3α and MIP-3β. J Immunol. 1997;158:1033–1036. [PubMed] [Google Scholar]
- 22.Hieshima K, Imai T, Opendakker G, Van Damme J, Kusuday J, Tei H, Sakaki Y, Takatsuki K, Miura R, Yoshie O, Nomiyama H. Molecular cloning of a novel human CC chemokine liver and activation-regulated chemokine (LARC) expressed in liver. J Biol Chem. 1997;272:5846–5853. doi: 10.1074/jbc.272.9.5846. [DOI] [PubMed] [Google Scholar]
- 23.Howard OM, Ben-Baruch A, Oppenheim JJ. Chemokines: progress toward identifying molecular targets for therapeutic agents. Trends Biotechnology. 1996;14:46–51. doi: 10.1016/0167-7799(96)80920-6. [DOI] [PubMed] [Google Scholar]
- 24.Liao F, Lee H, Farber JM. Cloning of STRL22, a new human gene encoding a G protein–coupled receptor related to chemokine receptors and located on chromosome 6q27. Genomics. 1997;40:175–180. doi: 10.1006/geno.1996.4544. [DOI] [PubMed] [Google Scholar]
- 25.Zaballos A, Varona R, Gutierrez J, Lind P, Marquez G. Molecular cloning and RNA expression of two new human chemokine receptor–like genes. Biochem Biophys Res Commun. 1996;227:846–853. doi: 10.1006/bbrc.1996.1595. [DOI] [PubMed] [Google Scholar]
- 26.Proudfoot AE, Power CA, Hoogewerf A, Montjovent MO, Borlat F, Wells TNC. Characterisation of the RANTES/MIP-1 alpha receptor (CC CKR-1) stably transfected in HEK 293 cells and the recombinant ligands. FEBS Lett. 1995;376:19–23. doi: 10.1016/0014-5793(95)01235-x. [DOI] [PubMed] [Google Scholar]
- 27.Alouani S, Gaertner HF, Mermod JJ, Power CA, Bacon KB, Wells TN, Proudfoot AE. A fluorescent interleukin-8 receptor probe produced by targeted labeling at the amino terminus. Eur J Biochem. 1995;227:328–334. doi: 10.1111/j.1432-1033.1995.tb20393.x. [DOI] [PubMed] [Google Scholar]
- 28.Clark-Lewis I, Moser B, Walz A, Baggiolini M, Scott GJ, Aebersold R. Chemical synthesis, purification, and characterization of two inflammatory proteins, neutrophil activating peptide 1 (interleukin-8) and neutrophil activating peptide. Biochemistry. 1991;30:3128–3135. doi: 10.1021/bi00226a021. [DOI] [PubMed] [Google Scholar]
- 29.Mainiero F, Gismondi A, Milella M, Morrone S, Palmieri G, Piccoli M, Frati L, Santoni A. Long term activation of natural killer cells results in modulation of beta-1 integrin expression and function. J Immunol. 1994;152:446–454. [PubMed] [Google Scholar]
- 30.Hansel TT, De Vries IJ, Iff T, Rihs S, Wandzilak M, Betz S, Blaser K, Walker C. An improved immunomagnetic procedure for the isolation of highly purified human blood eosinophils. J Immunol Methods. 1991;145:105–110. doi: 10.1016/0022-1759(91)90315-7. [DOI] [PubMed] [Google Scholar]
- 31.Sozzani S, Sallusto F, Luini W, Zhou D, Piemonti L, Allavena P, Van Damme J, Valitutti S, Lanzavecchia A, Mantovani A. Migration of dendritic cells in response to formyl peptides, C5a, and a distinct set of chemokines. J Immunol. 1995;155:3292–3295. [PubMed] [Google Scholar]
- 32.Romani N, Gruner S, Brang D, Kampgen E, Lenz A, Trockenbacher B, Konwalinka G, Fritsch PO, Steinman RM, Schuler G. Proliferating dendritic cell progenitors in human blood. J Exp Med. 1994;180:83–93. doi: 10.1084/jem.180.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aubry, J.-P., S. Rison, C. Power, and J.-Y. Bonnefoy. 1997. Purification of human lung dendritic cells by four-color flow cytometry. In Immunology Methods Manual. I. Lefkovits, editor. Academic Press, London. 1447–1454.
- 34.Capponi AM, Lew PD, Vallotton MB. Cytosolic free calcium levels in monolayers of cultured rat aortic smooth muscle cells. Effects of angiotensin II and vasopressin. J Biol Chem. 1985;260:7836–7842. [PubMed] [Google Scholar]
- 35.Eistetter HR, Church DJ, Mills A, Godfrey PP, Capponi AM, Brewster R, Schulz M-F, Kawashima E, Arkinstall SJ. Recombinant bovine neurokinin-2 receptor stably expressed in Chinese hamster ovary cells couples to multiple signal transduction pathways. Cell Regulation. 1991;2:767–779. doi: 10.1091/mbc.2.10.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Birkenbach M, Josefsen K, Yalamanchili R, Lenoir G, Kieff E. Epstein–Barr virus–induced genes: first lymphocyte-specific G protein–coupled peptide receptors. J Virol. 1993;67:2209–2220. doi: 10.1128/jvi.67.4.2209-2220.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshida R, Imai T, Hieshima K, Kusuda J, Baba M, Kitaura M, Nishimura M, Kakisaki M, Nomiyama H, Yoshie O. Molecular cloning of a novel human CC chemokine, EBI1-ligand chemokine that is a specific functional ligand for EBI1, CCR7. J Biol Chem. 1997;272:13803–13809. doi: 10.1074/jbc.272.21.13803. [DOI] [PubMed] [Google Scholar]
- 38.Tatrai A, Lee SK, Stern PH. U-73122, a phospholipase C antagonist, inhibits effects of endothelin-1 and parathyroid hormone on signal transduction in UMR-106 osteoblastic cells. Biochim Biophys Acta. 1994;1224:575–582. doi: 10.1016/0167-4889(94)90296-8. [DOI] [PubMed] [Google Scholar]
- 39.Nishizuka Y. Intracellular signaling by hydrolysis of phospholipids and activation of protein kinase C. Science (Wash DC) 1992;258:607–614. doi: 10.1126/science.1411571. [DOI] [PubMed] [Google Scholar]
- 40.Demaurex N, Lew DP, Krause KH. Cyclopiazonic acid depletes intracellular Ca2+stores and activates an influx pathway for divalent cations in HL-60 cells. J Biol Chem. 1992;267:2318–2324. [PubMed] [Google Scholar]
- 41.Glennon MC, Bird GS, Takemura H, Thastrup O, Leslie BA, Putney JW., Jr In situ imaging of agonist-sensitive calcium pools in AR4-2J pancreatoma cells. Evidence for an agonist and inositol 1,4,5-trisphosphate–sensitive calcium pool in or closely associated with the nuclear envelope. J Biol Chem. 1992;267:25568–25575. [PubMed] [Google Scholar]
- 42.Premack BA, Schall TJ. Chemokine receptors: gateways to inflammation and infection. Nature Med. 1996;2:1174–1178. doi: 10.1038/nm1196-1174. [DOI] [PubMed] [Google Scholar]
- 43.Weiss RA, Clapham PR. Hot fusion of HIV. Nature (Lond) 1996;381:647–648. doi: 10.1038/381647a0. [DOI] [PubMed] [Google Scholar]
- 44.Lane SJ, Sousa AR, Lee TH. The role of the macrophage in asthma. Allergy. 1994;49:201–209. doi: 10.1111/j.1398-9995.1994.tb02650.x. [DOI] [PubMed] [Google Scholar]
- 45.Fuller, R.W. 1992. Asthma. Macrophages. Brit. Med. Bulletin. 65–71. [DOI] [PubMed]
- 46.Hance AJ. Pulmonary immune cells in health and disease: dendritic cells and Langerhans' cells. Eur Respir J. 1993;6:1213–1220. [PubMed] [Google Scholar]
- 47.Holt PG. Regulation of antigen-presenting cell function(s) in lung and airway tissues. Eur Respir J. 1993;6:120–129. [PubMed] [Google Scholar]
- 48.Blackshaw AJ, Levison DA. Eosinophilic infiltrates of the gastrointestinal tract. J Clin Pathol. 1986;39:1–7. doi: 10.1136/jcp.39.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu LL, Warren MK, Rose WL, Gong W, Wang JM. Human recombinant monocyte chemotactic protein and other CC chemokines bind and induce directional migration of dendritic cells in vitro. J Leukocyte Biol. 1996;60:365–371. doi: 10.1002/jlb.60.3.365. [DOI] [PubMed] [Google Scholar]
- 50.Van Snick J, Houssiau F, Proost P, Van Damme J, Renauld JC. I-309/T cell activation gene-3 chemokine protects murine T cell lymphomas against dexamethasone-induced apoptosis. J Immunol. 1996;157:2570–2576. [PubMed] [Google Scholar]
- 51.Proudfoot AE, Power CA, Hoogewerf AJ, Montjovent MO, Borlat F, Offord RE, Wells TNC. Extension of recombinant human RANTES by the retention of the initiating methionine produces a potent antagonist. J Biol Chem. 1996;271:2599–2603. doi: 10.1074/jbc.271.5.2599. [DOI] [PubMed] [Google Scholar]
- 52.Samson M, Soularue P, Vassart G, Parmentier M. The genes encoding the human CC chemokine receptors CC-CKR1 to CC-CKR5 (cmkbr1–cmkbr5) are clustered in the p21.3–p24 region of chromosome 3. Genomics. 1996;36:522–526. doi: 10.1006/geno.1996.0498. [DOI] [PubMed] [Google Scholar]
- 53.McWilliam AS, Nelson DJ, Holt PG. The biology of airway dendritic cells. Immunol Cell Biol. 1995;73:405–413. doi: 10.1038/icb.1995.63. [DOI] [PubMed] [Google Scholar]