Abstract
Interferon (IFN) exhibits a potent antiviral activity in vitro and plays a major role in the early defense against viruses. Like IFN, the proinflammatory chemokine, interleukin (IL)-8, is induced by viruses and appears in circulation during viral infections. In an in vitro cytopathic effect assay for IFN, we found that IL-8 can inhibit IFN-α activity in a dose-dependent manner. This action was reversed by specific monoclonal antibodies to IL-8. The chemokine was able to attenuate the IFN-mediated inhibition of viral replication as determined by measuring infectious virus yield. IL-8 also diminished the ability of IFN to inhibit an early stage of viral replication since IL-8 attenuated the inhibition of the formation of viral proteins. It appeared that IL-8 interfered with a late rather than an early step of IFN-mediated pathway such as early gene expression. The IL-8 inhibitory action on IFN-α antiviral activity was associated with reduced 2′,5′-A oligoadenylate synthetase activity, a pathway well correlative with the anti– encephalomyocarditis virus action of IFN-α. Understanding pathways that antagonize IFN action may lead to novel approaches to potentiate endogenous and therapeutic IFN.
IFNs are induced in many cell types in response to viruses and possess their known antiviral activity against a variety of DNA and RNA viruses (1, 2). Because of its unique antiproliferative and antiviral properties, IFN-α is being used as a therapeutic agent in a number of infectious and noninfectious diseases, and in several clinical trials. Thus, the elucidation of mechanisms that may either synergize or antagonize IFN-α antiviral action may lead to ways to maximize endogenous and exogenous IFN benefits.
We previously reported that IL-8, a chemotactic protein, is induced by cytomegalovirus (CMV)1 in the monocytic THP-1 cell line (3) and that IL-8 enhances the replication of several viruses including CMV (4), encephalomyocarditis virus (EMCV), and poliovirus (5). Others showed that viruses such as respiratory influenza virus, syncytial virus, and rotavirus induced IL-8 production (6–8). These observations prompted us to investigate the possibility that IL-8 may inhibit the antiviral action of IFN-α.
IL-8 is the best characterized member of the family of chemokines: proinflammatory cytokines that chemoattract and activate blood cells (9, 10). This protein belongs to the subfamily of α chemokines (CXC family), which are distinguished from β chemokines (CC family) by few structural and functional dissimilarities. Besides its central role in inflammation, other biological functions of IL-8 include T cell chemotaxis (11), angiogenesis (12), and hematopoiesis (13). In this paper, we report a novel function for the α chemokine, IL-8, which is inhibition of the antiviral action of IFN-α.
MATERIALS AND METHODS
Cells and Viruses.
The human epithelial amnion WISH cell line (HeLa markers) was obtained from Dr. J.A. Armstrong (University of Pittsburgh, Pittsburgh, PA). Normal human fibroblasts were prepared from foreskin. VERO (African green monkey kidney), L929 (mouse fibrosarcoma), THP-1 (human monocytic), and MRC-5 (human fibroblastic lung) cell lines were obtained from the American Type Culture Collection (Rockville, MD). All cell lines were grown at 37°C and 5% CO2 in MEM (GIBCO BRL, Gaithersburg, MD) supplemented with 10% newborn calf serum (GIBCO BRL), except for THP-1 cells which were grown in RPMI 1640 with 10% fetal bovine serum (FBS).
EMCV, also obtained from Dr. J.A. Armstrong (University of Pittsburgh, Pittsburgh, PA), was propagated in L929 cells. Poliovirus type 1, vesicular stomatitis virus (VSV; Indiana strain), and HSV-1 strain F were obtained from the American Type Culture Collection and propagated in the VERO cell line. Virus preparations were clarified by low speed centrifugation, filtered through 0.22 μm paper, and concentrated by ultracentrifugation. All viruses were titrated in VERO with resultant titers of 2 × 107, 2 × 106, 8 × 108 , and 1 × 109 PFU/ml for EMCV, poliovirus, VSV, and HSV-1, respectively. Plaque assays are described later. Viruses were aliquoted and stored at −70°C until use.
IFNs and IL-8.
Human rIL-8 was expressed in Escherichia coli as previously described (14). Biological activity was assessed using neutrophil chemotaxis multiwell Boyden chamber assay as described (15); maximum activity was observed at 10 ng/ml (15). Also, rIL-8 (Lot No. BA-044041; R&D Systems, Minneapolis, MN) was occasionally used and similar results were obtained. When both rIL-8 were calibrated against the reference preparation 89/ 520 (National Institute for Biologicals Standardization and Calibration, Hertfordshire, UK), the maximum activity was in the vicinity of 100 IU/ml. Human rIFN-α2a, obtained from Hoffman-LaRoche (Basel, Switzerland), had a specific activity of 2 × 108 IU/mg, as reported by the manufacturer. A starting solution was made and calibrated with NIH Gxa01-901-535 IFN-α reference preparation; the titer was 109 IU/ml. rIFN-γ was obtained from Genzyme (Cambridge, MA) and had a specific activity of 107 U/mg, according to the manufacturer. A starting solution was made and calibrated with NIH Gxg01-901-535 reference preparation; the titer was 106 IU/ml.
IFN Bioassay.
The tetrazolium salt (MTS) IFN microtiter assay was used to assess potency of IFN-α by measuring end point titers. The assay has been previously described in detail (16). In some experiments, the crystal violet stain assay was used (17). When using either method, the OD was correlated with the degree of protection from virus-induced cytopathic effect (18). Percent cell protection was calculated as follows: 1 − ([dilution OD − virus control OD]/[cell control OD − virus control OD]) × 100%, where OD is optical density and dilution OD refers to an average OD in triplicate wells at the dilution specified. Percent cell protections were plotted against serial dilutions of the IFN preparation. End-point titers expressed as laboratory units per milliliter (LU/ml) were taken as reciprocals of dilutions that gave 50% cell protection. IFN doses (LU/ml, reciprocal of dilutions) were corrected to IU/ml by calibration with the international reference standard described above.
Antibodies.
A mouse anti–human IL-8 mAb of an IgG1 isotype, WS-4, was generated as previously described (18). The isotype-matched normal IgG was obtained from R&D Systems. Antibodies were incubated with IL-8 for 4 h at room temperature. Different molarity ratios were first assessed to determine optimum ratio; a ratio of 3:1 (antibody/IL-8) showed maximal reversal. For immunoprecipiration experiments, antiserum to poliovirus was raised in guinea pigs; the virus was previously purified on sucrose gradient by ultracentrifugation and washed with Tris-HCl buffer (pH 7.4). The antiserum was adsorbed to VERO cells to eliminate antibodies cross-reactive to cellular proteins.
Infectious Virus Yield Titration.
For experiments that required virus yield titration, virus was added to cells for 1 h before supernatants were aspirated to remove unadsorbed virus particles. Culture plates were incubated for 24 h at 37°C. The cultures were subjected to freezing and thawing to lyse the cells. Cell lysates that contained both intracellular and extracellular particles were clarified from cell debris by centrifugation. PFUs in the resultant supernatants were quantitated according to standard methods (19) involving VERO cells. The overlay MEM contained either agar (Sigma Chemical Co., St. Louis, MO) in case of EMCV or methylcellulose (Sigma Chemical Co.) for other viruses. The plates were stained with crystal violet to quantitate PFUs.
Poliovirus Polypeptides: Metabolic Labeling, Radioimmunoprecipitation, and SDS-PAGE.
After cytokine and IFN treatments, VERO monolayers in 24-well plates were incubated with poliovirus for 1 h. The unadsorbed virus was removed by washing with PBS buffer. The cultures were incubated for 24 h with methionine-free MEM (GIBCO BRL) supplemented with 2% dialyzed FBS and 10 μCi of [35S]methionine (Amersham, Buckingham, UK). The radioactive medium was removed, and monolayers were washed in PBS. Cells were lysed in gel lysis/radioimmunoprecipitation (RIPA) buffer (10 mM Tris-HCl, 15 mM NaCl, 1.5 MgCl2, 1% Triton X-100, 0.25% deoxycholate, 1 mM PMSF, and 15 U/ml aprotinin) on ice. Lysates were mixed with anti–poliovirus 1 antiserum for overnight at 4°C. The immune complexes were precipitated using 10% suspension of protein A–Sepharose beads (Pharmacia, Uppsala, Sweden) in blocking buffer (1% BSA–PBS) for 1 h at 4°C. The beads were collected by centrifugation, washed five times, and resuspended in SDS sample buffer. The immune complexes were released into the supernatants by boiling and then electrophoresed in 12% SDS-PAGE. The gels were fixed, washed, dried, and visualized by autoradiography (Kodak XAR film; Kodak, Rochester, NY) at −70°C. Before immunoprecipitation, lysates were examined by 12% SDS-PAGE to verify quality and loading of total proteins. 14C-methylated protein molecular weight markers (14–220 kD) were used to verify the size of viral proteins.
RNA Preparation and Northern Blot Analysis.
Total RNA was extracted by guanidine isothiocyanate method (20) using Tri Reagent (Molecular Research Center, Cincinnati, OH). 20 μg of total RNA was electrophoresed through a 1.2% agarose/2.2 M formaldehyde gel. Northern transfer was performed overnight using Zeta Probe nylon membrane (Bio Rad, Hercules, CA). Membranes were baked and prehybridized in Express Hyb solution (Clonetech, Palo Alto, CA) in a Hybaid Mini Hybridization Oven (Labnet, Woodbridge, NJ). cDNA probes specific for the 0.7-kb 6–16 messenger RNA (mRNA) probe (provided by Dr. Sandra Pellegrini, Pasteur Institute, Paris, France) and 28S ribosomal RNA (American Type Culture Collection) were labeled with [32P]dCTP (Amersham) using nick translation kit (GIBCO BRL). The labeled probes were purified on Sephadex G-50 columns, denatured, added to Express Hyb solution, and hybridized for 1 h at 68°C. The blots were washed and exposed to Kodak X-Omat AR film (Sigma Chemical Co.), and the autoradiograms were subsequently developed.
Reverse Transcriptase PCR and Southern Blotting.
In brief, the reverse transcriptase (RT) reaction was performed using 5 μg total RNA, 500 ng random hexamer (Random Primers; Promega, Madison, WI), 500 μM dNTP mixture, 20 U RNAsin (Pharmacia), and 200 U of Moloney murine leukemia virus reverse transcriptase (GIBCO BRL). The samples were heated to inactivate RT. A pair of primers that amplify both IL-8R A and B (CXCR1 and CXCR2; Maxim Biotech, Inc., San Francisco, CA) was used to amplify a 680-bp fragment of the CXCR gene (21): sense: 5′CTGAACCTAGCCTTGGCCGACCT3′; antisense: 5′TAGATGAGGGGGTTGAGGCAGC3′. Hot start PCR amplification was performed using Taq DNA polymerase (Promega). cDNA was amplified for 35 cycles at 94°C for 60 s, 60°C for 60 s, and 72°C for 60 s (Gene Amp PCR System 9600; Perkin Elmer, Foster City, CA). PCR products were electrophoresed through a 2% agarose gel and visualized with ethidium bromide. Size markers (ϕX 174 DNA/HaeIII fragments) obtained from GIBCO BRL were used to verify the size of 680-bp fragments. Southern blotting using Zetaprobe nylon membrane (Bio Rad) was performed, and the membrane was hybridized with a 40-mer oligonucleotide probe that recognizes CXCR1 and CXCR2 PCR bands. Its sequence was as follows: 5′TGCTATGAGGACATGGGCAACAATACAGCA3′. The probes was labeled at 5′ end with [γ-32P]ATP (Amersham) using T4 polynucleotide kinase (New England BioLabs, Beverly, MA). Labeled probes were purified on a Sephadex G–50 column, denatured, and added to the membranes in Express Hyb solution and hybridized for 1 h at 37°C. The blots were washed and exposed to Kodak X-Omat AR film (Sigma Chemical Co.), and the autoradiograms were subsequently developed.
The pair of primers that amplify 282-bp fragment for 2′,5′-oligoadenylate synthetase (OAS) and 838-bp fragment for β actin, and the oligonucleotide probe used to confirm OAS PCR fragments were previously described in detail (5). RT-PCR for OAS and β actin gene expression was performed essentially the same as above. The PCR conditions for OAS amplification allowed at least qualitative comparisons, e.g., significant stimulation or inhibition, of signal strength on agarose gels. These comparisons were validated by generating PCR products using different cycles in which the signal strength was within linearity up to 44 cycles.
Competition Receptor Binding Assay.
The binding assay was performed in 96-well U-bottom flexible plates (Falcon, Becton Dickinson; Bedford, MA). The wells were first coated overnight with FBS to reduce nonspecific binding (22). WISH or the CXCR-positive THP-1 cells (23) were seeded at 0.5 × 106/well and incubated in a binding buffer (PBS, 1% BSA, and sodium azide) with 106 cpm of 125I–IL-8 specific activity 2,000 Ci/mmol; Amersham) in the presence or absence of 750-fold molar excess of cold IL-8 in each microwell. Plates were then incubated at 4°C for 90 min, centrifuged in an Eppendorf refrigerated centrifuge, and washed three times with PBS/BSA buffer. Cell-bound radioactivity was harvested from the wells in polystyrene tubes and quantified in gamma counter.
OAS Assay.
The assay was described in detail elsewhere (5). In brief, cells were lysed by 0.5% NP-40–containing buffer. Equal amounts (1:50 dilution) of lysates were added to tubes containing polyI polyC–agarose at bottom to bind OAS, and unbound materials were washed off by centrifugation. ATP was added to assay tubes and incubated for 1 h at 37°C. OAS converts ATP to 2-5A in which the latter was measured by competitive radioimmunoassay. The amounts of 2-5A (pmol/dl) generated reflects OAS activity. All materials in OAS assay were obtained from Eiken Chemical Co. (Tokyo, Japan).
Statistical Analysis.
All comparisons were performed with the Student's paired t test with the aid of GraphPad Prism software (San Diego, CA). Significance was reported with two-tailed P <0.005 unless otherwise described.
RESULTS
Inhibitory Effect of IL-8 on IFN-α Antiviral Action.
We found that inclusion of recombinant IL-8 in an in vitro assay for IFN activity (16, 17) significantly reduced the potency of IFN-α. The assay measures the ability of IFN to protect human WISH epithelial cells from the cytopathic, (e.g., cytotoxic) effects (CPE) induced by encephalomyocarditis virus. When target cells were pretreated with IL-8, 90% reduction in IFN-α potency (P <0.005, paired Student's t test) was consistently observed as assessed by measuring end-point titers (Fig. 1 A). When the dosage was expressed in terms of international unitage, the ED50 of IFN-α activity increased from 1 to 10 IU/ml as a result of IL-8 treatment (Fig. 1 B). IL-8 did not enhance EMCV-induced CPE at the virus challenge dose, multiplicity of infection equal to 0.1, used in the assay which gave near maximum (90– 100%) CPE. Virus-induced CPE was 90 ± 4% and 95 ± 2% in the absence or presence of IL-8 (33 ng/ml), respectively. Also, IL-8 did not affect cell viability (MTS tetrazolium assay) which was 98 ± 4.5% of cell control in the presence of IL-8.
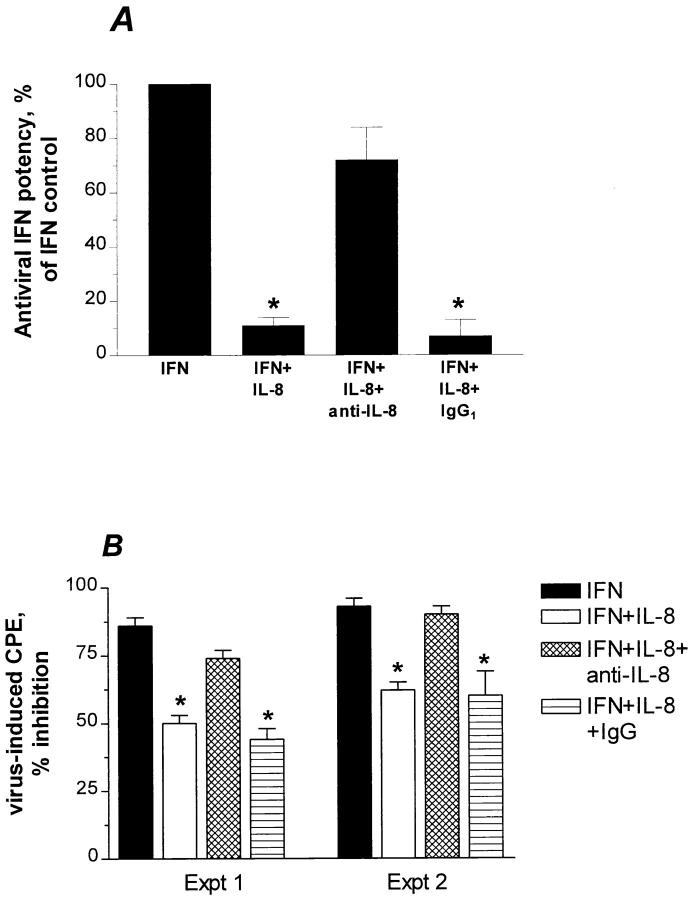
Figure 1.
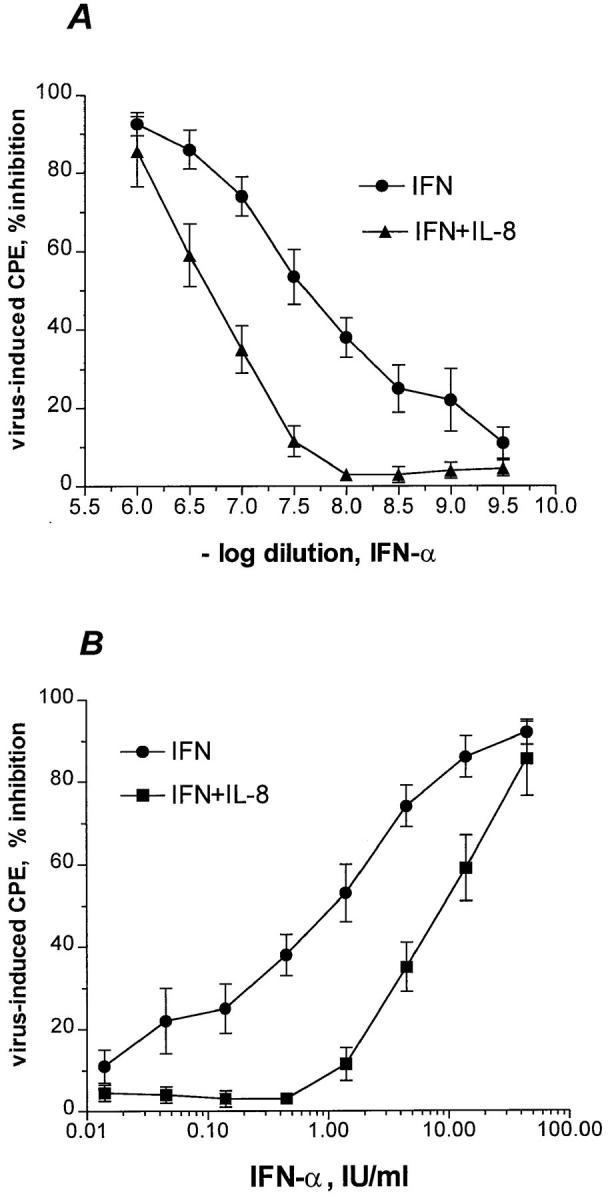
Effect of IL-8 on IFN-α antiviral potency. (A) Confluent WISH cells were treated with medium or 33 ng/ ml of rIL-8 for 20 h. The cells were then subjected to the IFN bioassay as described in Materials and Methods. (B) The reciprocals of dilutions, i.e., IFN doses in LU/ml, were corrected to IU/ml by calibration with the reference standard National Institutes of Health Gxa01-901-535. CPE refers to cytopathic effect in terms of cell destruction. Data are from mean ± SEM of four experiments.
Dependence of IL-8 Inhibitory Potency on IFN Dose, IFN Type, and IL-8 Dose.
The IL-8 inhibitory potency towards anti-EMCV IFN-α activity was more pronounced at lower IFN doses (Fig. 1). For example, at 20 IU/ml, the IFN-mediated inhibition of CPE was 90 and 70% in the absence and presence of IL-8 treatment, respectively. In contrast, at 1 IU/ml , the IFN-mediated inhibition of viral CPE decreased from 50 to 8% due to IL-8 treatment (Fig. 1 B).
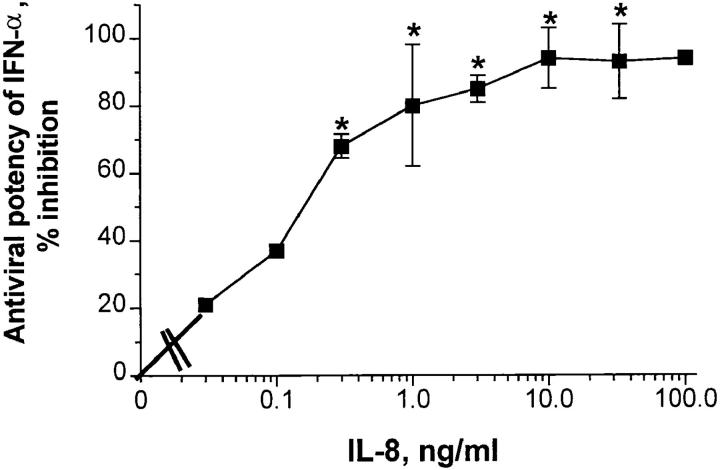
The inhibitory effect of IL-8 on IFN-α antiviral activity was dose dependent, yielding an ED50 of ∼150 pg/ml (Fig. 2). Maximum inhibition was observed at 10 ng/ml (Fig. 2). No IL-8 toxicity was seen at the doses tested as judged by MTS tetrazolium dye; for example, at 100 ng/ml, cell control OD was 1.35 ± 0.02 (n = 4), whereas in the presence of 100 ng/ml of IL-8, the OD was 1.34 ± 0.04 (n = 4). Likewise, there was no toxicity observed using either trypan blue exclusion dye or crystal violet staining; in both instances, the IL-8 controls were always 97 ± 3% of cell control.
Figure 2.
Dose-response relationships for IL-8 inhibition of IFN-α antiviral activity. Confluent WISH cells were pretreated with indicated concentrations of IL-8; cells were then used for the IFN bioassay as described in Materials and Methods. Percent inhibition refers to percent reduction in original IFN-α potency (107 IU/ml) in the absence of IL-8 treatment (IFN control, 0% inhibition). Average number of experiments (those with SEM) is four. *Statistical significance at P <0.005 when compared to no IL-8 treatment. Other points could not be statistically analyzed because n = 2 is small.
rIFN-γ appeared to be less susceptible to the inhibitory effect of IL-8 than IFN-α; in four experiments, rIFN-γ titers (106 IU/ml) were reduced by 60–80%, whereas IFN-α titers in parallel experiments were inhibited by 85–93%.
Specificity of IL-8 Inhibition of IFN-α Activity.
mAb to IL-8 (WS-4; 18), but not isotype-matched IgG reversed most of IL-8 inhibitory effect on anti-EMCV IFN-α activity as assessed by reduction in IFN titers (Fig. 3 A). Also, anti–IL-8 was able to reverse the IL-8 inhibitory action on IFN-α activity as assessed by the percentage of IFN-mediated cell protection from virally-induced CPE (Fig. 3 B).
Figure 3.
Specificity of IL-8 inhibition of IFN-α antiviral activity. (A) mAb to IL-8 (WS-4) and control isotype-matched IgG, both at a molar ratio of 3:1, were incubated before addition of IFN-α and followed by EMCV. Percent of IFN-α control (100%) refers to the original antiviral IFN-α potency in terms of IFN titers. Data are mean ± SEM of three experiments. (B) Experiments 1 and 2 denote percent inhibition of EMCV-induced CPE due to IFN-α (10 IU/ml). Cell protection was calculated as described in Materials and Methods. Results in each experiment represent the means ± SEM of triplicate cultures. *Statistical significance at P < 0.005 when compared to IFN control.
Expression of CXCR (IL-8R) in WISH Cells.
The IL-8 receptors, CXCR1 and CXCR2, are not only expressed on cells of hematopoietic origin such as neutrophils and monocytes, but also on cells of nonhematopoietic type (23–26). Using THP-1 cell line as a positive control for IL-8 receptor (23), we found that WISH cells also contained CXCR transcripts as assessed by RT-PCR (Fig. 4, top). To confirm the PCR products, a probe for highly conserved region of CXCR1 and CXCR2 was used in Southern analysis as revealed in Fig. 4 (bottom). The CXCR transcript was expressed at lower levels in WISH than in THP-1. This may be attributed to the presence of other homologous chemokine receptor sequences. The lower expression of CXCR in WISH than in THP-1 cells was also observed in radiobinding competitive experiments (Table 1). Specific binding of 125I–IL-8 due to 750-fold molar excess of unlabeled IL-8 was threefold lower in WISH than in THP-1. Also, unlabeled IL-8 competed less efficiently in THP-1 than in WISH cells (Table 1).
Figure 4.
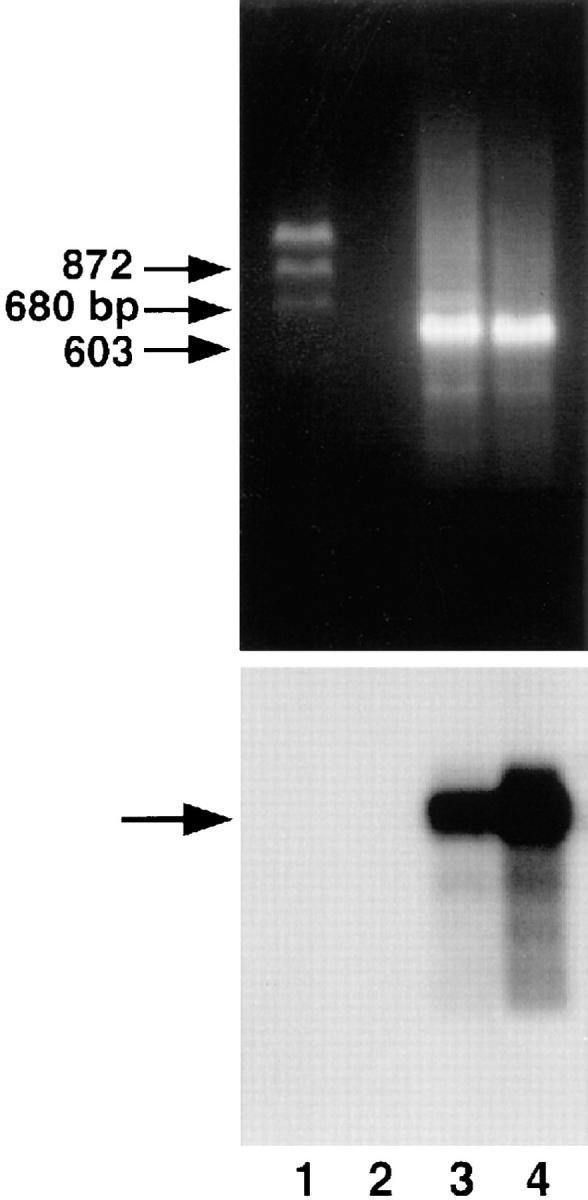
Expression of CXC (IL-8) receptor mRNA in THP-1 and WISH cell lines. Total RNA was extracted from WISH cells (lane 3) and THP-1 monocytic cells (positive control; reference 23, lane 4) and subjected to RT-PCR. PCR products generated from the primers that recognize both CXCR1 and CXCR2 corresponded to the expected size of 680 bp (middle arrow). Lane 1 denotes size markers and lane 2 represents PCR reaction including primers but no template. The lower panel shows specific signal (arrow) due to Southern hybridization with oligonucleotide directed to internal region of the PCR products (CXCR1 and CXCR2).
Table 1.
Competition of Binding by Radiolabeled and Unlabeled IL-8 to WISH and THP-1 Cell Lines*
| Total binding | ||||||||
|---|---|---|---|---|---|---|---|---|
| Cells | 125I–IL-8 | 125I–IL-8 + excess unlabeled IL-8 | Specific binding | (%)‡ | ||||
| cpm | ||||||||
| WISH | 4,350 ± 1,076 | 2,898 ± 462 | 1,451 ± 649 | (33) | ||||
| THP-1 | 8,863 ± 1,730 | 3,923 ± 816 | 4,431 ± 918 | (50) | ||||
5 × 105 cells were used with 106 cpm of 125I–IL-8 in a radiobinding competition assay in the absence or presence of 750-fold molar excess of unlabeled IL-8.
Percent of total binding. Results are the average of three independent experiments with SEM.
Effect of IL-8 on Infectious Virus Yield in IFN-treated Cells.
Not only did IL-8 attenuate IFN-α antiviral action against virus-induced CPE, but also against the total (intracellular and extracellular) virus yield from infected cells (see Fig. 5). This may indicate that IL-8 antagonized IFN-α inhibitory action on viral replication. There was antagonism by IL-8 (four- to sixfold enhancement of virus yield) in EMCV-infected WISH cells treated with IFN-α doses at 10 IU/ml and lower (Fig. 5 A). The inhibitory effect of IL-8 on IFN-α–mediated suppression of virus yield was also seen with poliovirus, as EMCV it is positive-stranded RNA virus of the family picornaviruses. In this case, there was three- to sixfold increase in virus yield as a result of IL-8 treatment in IFN-treated poliovirus-infected WISH cells (Fig. 5 B). Although there was no direct IL-8 enhancement of CPE, which was maximum at the virus doses used, there was a slight enhancement (twofold) in virus yield in IL-8–treated cells used with the picornaviruses (Fig. 5, A and B). The inhibitory effect of IL-8 on IFN-α action was demonstrated with HSV-1 in MRC-5 (Fig. 5 C). With such DNA virus, higher doses (e.g., >30 IU/ml; Fig.5 C) were required to suppress HSV-1 replication. Thus, in case of HSV-1, these high IFN-α concentrations were still subject to IL-8 effect. The inhibitory effect of IL-8 on IFN action could not be demonstrated with the negative-stranded VSV using either the virus-induced CPE (data not shown) or virus yield (Fig. 5 D) assays.
Figure 5.
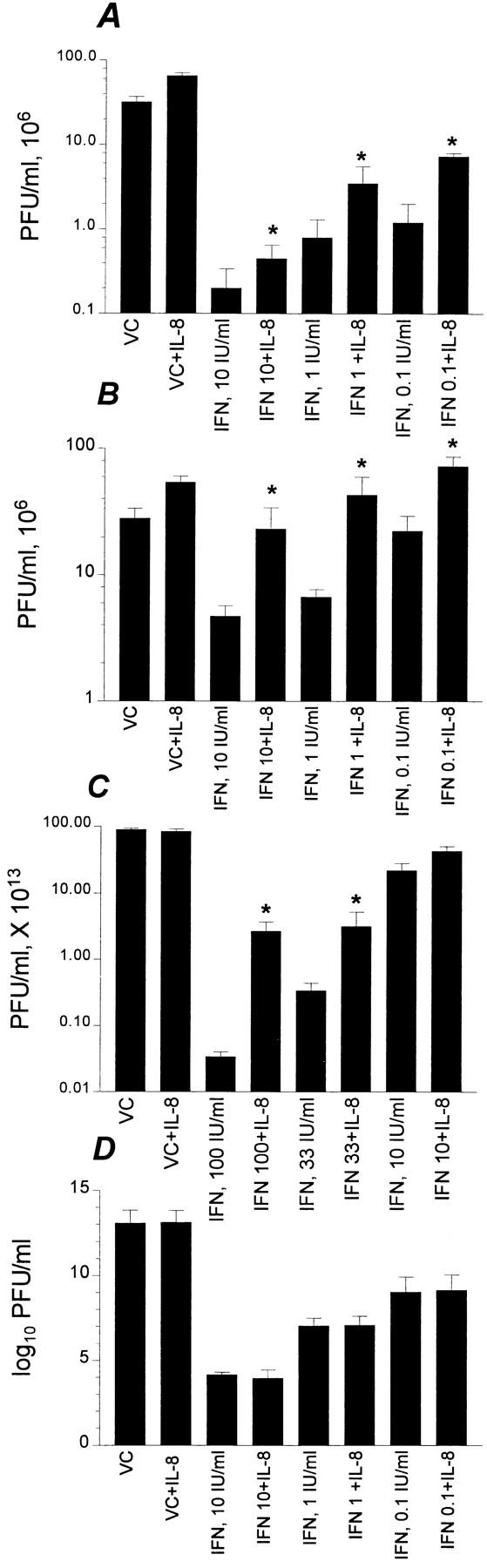
Effect of IL-8 on virus yield in IFN-treated cells. Confluent cells in microtiter plates were pretreated overnight with IL-8 (10 ng/ml) followed by IFN-α at indicated concentrations for 20 h. Viruses were added at doses that were previously determined to cause maximum CPE within 24 h. The following virus/cell systems were used: EMCV/WISH (A), poliovirus/WISH (B), HSV-1/MRC-5 (C), and VSV/normal fibroblasts (D). Infectious virus yield was quantitated in terms of PFU/ml as described in Materials and Methods. IL-8 itself had no effect on the CPE. Data are from an average of three experiments. *Statistical significance at P <0.005 when compared to IFN alone at the indicated concentrations. VC, virus control; VC+IL-8, virus control that was treated with IL-8. All other treatments, as indicated, are in the presence of virus.
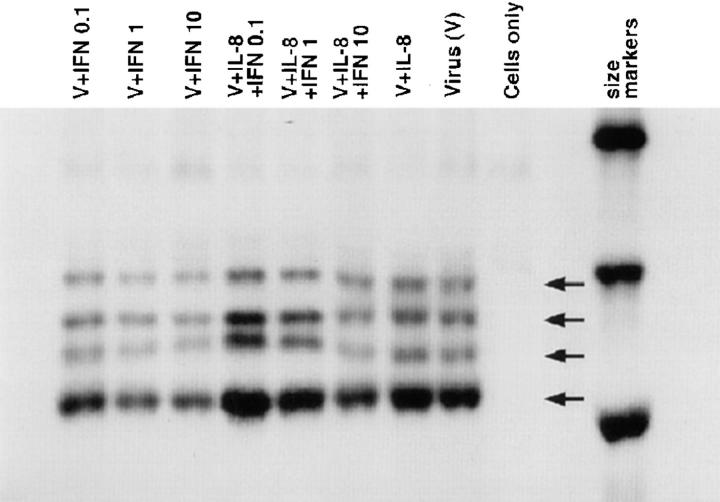
Effect of IL-8 on Poliovirus Protein Synthesis.
IFNs are known to intervene with an early stage of picornaviral replication resulting in inhibition of the formation of viral proteins (1, 2). We had neutralizing antiserum of high titer to poliovirus type 1; poliovirus as EMCV is IFN-sensitive picornavirus and has limited number of distinct proteins. Fig. 6 illustrates that the antiserum recognized at least three of the four major capsid proteins of poliovirus: VP1 (34 kD), VP2 (28 kD), and VP3 (24 kD), in addition to another band that may represent one of the precursor or intracellular proteins. The inhibition of the formation of 35S-labeled poliovirus proteins showed a clear dose response by IFN-α (Fig. 6). IL-8 was able to attenuate the IFN-mediated inhibition of the formation of poliovirus proteins (Fig. 6). IL-8 reversed the IFN-mediated inhibition of viral proteins synthesis particularly at doses 0.1 and 1 IU/ml of IFN-α. IL-8 itself did not significantly upregulate the protein bands (Fig. 6) with the cytopathic challenge used in the experiments.
Figure 6.
Attenuation of IFN-induced inhibition of metabolic formation of the major picornaviral polypeptides. Confluent VERO cells were pretreated with IL-8 (10 ng/ml) or mock for 20 h before IFN-α (20 h) treatment, and then poliovirus. Metabolic labeling, RIPA, and gel electrophoresis were described in Materials and Methods. Cells only, viral immunoprecipitates did not contain cross-reactive bands with cellular proteins as shown in the region of size markers of 21.5–30 kD. Virus control (V) shows (from top) at least three of the large polypeptides (VP1, VP2, and VP3) with an additional band of viral origin possibly corresponding to a precursor protein.
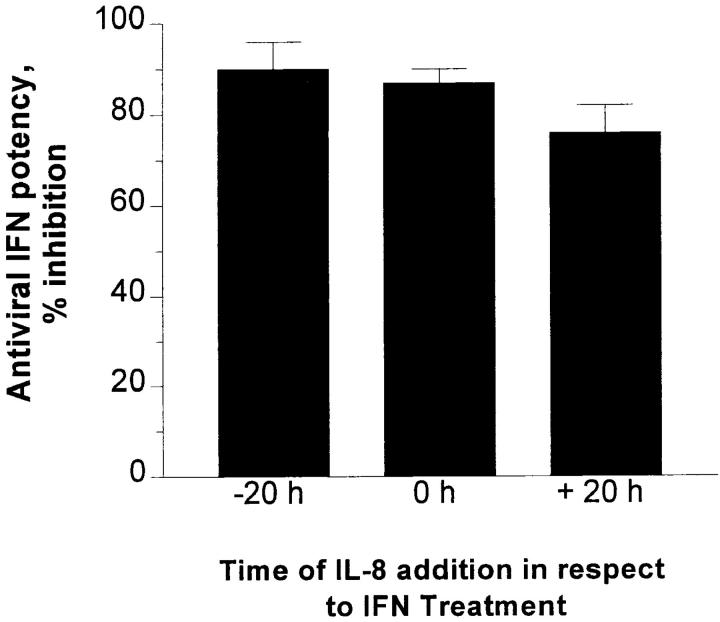
Kinetics of IL-8 Inhibitory Action on IFN-α Antiviral Activity and Lack of IL-8 Interference with Early IFN Response.
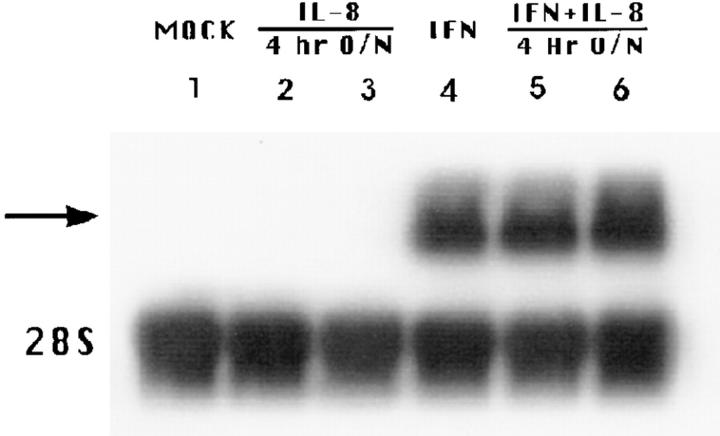
The IL-8 inhibitory effect on IFN activity was similarly demonstrated whether IL-8 was added before, simultaneously with, or as late as 20 h after IFN treatment (Fig. 7). This suggests that IL-8 exerted its inhibitory action at a late rather than early step in IFN-mediated pathway. As shown in Fig. 8, this was also supported by the lack of changes in mRNA expression of the 6-16 gene (27) that contains the IFN-α–stimulated response element that is conserved in most IFN-α responsive genes (28).
Figure 7.
Effect of IL-8 addition on IFN-α antiviral response. Confluent WISH cells were subjected to the IFN bioassay as described in Materials and Methods. IL-8 was added at three different time points in relation to addition of IFN as indicated. Percent reduction in original IFN-α potency (assayed in absence of IL-8 treatment) is shown as a function of time of IL-8 addition. Data represents an average of three experiments with SEM. There were no significant changes among the three columns.
Figure 8.
Effect of IL-8 on IFN-induced 6-16 mRNA expression. WISH cells were pretreated with medium or 100 ng/ ml IL-8 followed by medium or 10 IU/ml IFN-α for 4 h. Northern blotting was performed as described in Materials and Methods. Lane designations are as indicated. (Lane 1) Signal in cell control. (Lanes 2 and 3) Signals due to IL-8 treatment for 4 h and overnight (O/N), respectively. (Lane 4) Signal due to IFN treatment only for 4 h. (Lanes 5 and 6) Signals due to IFN with 4 h and overnight treatment of IL-8, respectively. Arrow indicates expected size of the 0.7-kb 6-16 mRNA and lower panel shows constitutive expression of 28S ribosomal RNA. Data are from one of three experiments performed.
Effect of IL-8 on the IFN-regulated OAS Activity.
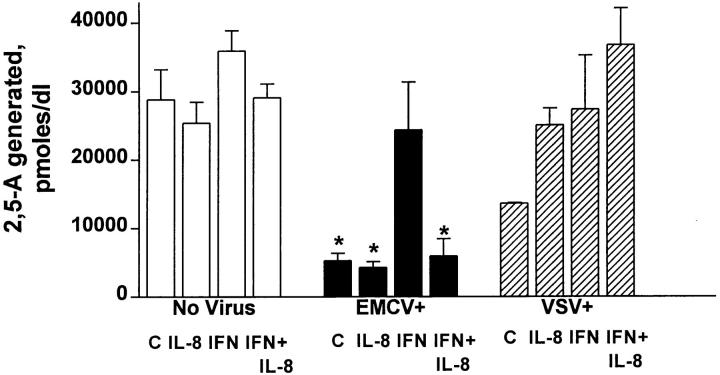
We looked at the constitutive OAS pathway, a pathway that was reported to correlate with IFN-α action against EMCV, but not VSV (1, 2). Both EMCV, substantially, and VSV, moderately, were associated with lower OAS activity, probably due to their general cytopathic perturbation of the cells (Fig. 9). However, IL-8 action was associated with further reduction in OAS activity in the EMCV-infected IFN-treated cells (Fig. 9). The IL-8 suppressive action on OAS activity in the presence of IFN-α appears to be linked to the type of the virus since this was not seen with anti-VSV IFN-α activity (Fig. 9). IL-8 (10 ng/ml) had minimum effect on the cellular constitutive OAS activity in the presence or absence of IFN-α (Fig. 9). However, at high doses, e.g., 100 ng/ml, there was 38% inhibition of OAS activity in WISH cells without affecting cell viability; in three independent experiments, the OAS activity was 25,800 ± 6,025 and 17,442 ± 3,990 in the absence or presence of 100 ng/ml, respectively.
Figure 9.
Effect of IL-8 on the IFN-regulated OAS activity. WISH monolayers in 24-well plates were treated with IL-8 (10 ng/ml), treated with IFN-α (10 IU/ml), and infected with EMCV or VSV; control treatments received no virus. Each treatment (in duplicate wells for each experiment) continued for 16–18 h. IL-8 had no effect on cell viability. WISH cells produce high constitutive levels of OAS and IFN-α at 10 IU/ ml induced further moderate OAS activity. OAS biochemical activity was assessed by measuring 2′,5′-oligoadenylate generated from ATP by OAS action (see Materials and Methods). Average number of experiments are three. *Statistical significance at P <0.01 when compared to values of any column in the group designated No Virus. C, denote cells.
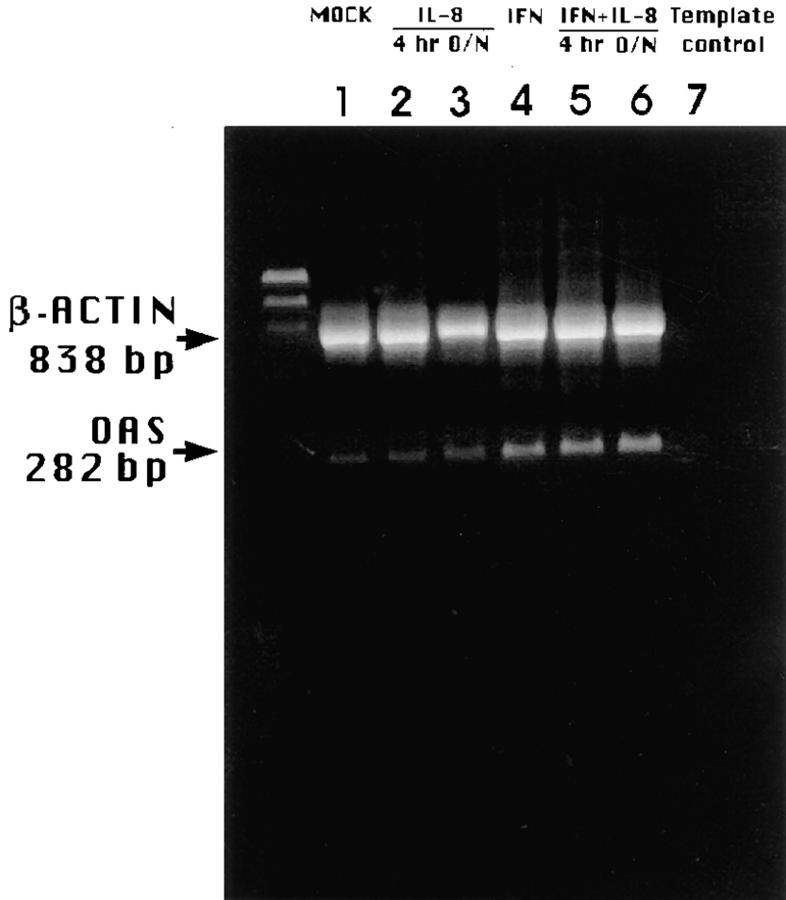
It appeared that IL-8 had no dramatic effect, e.g., significant inhibition, on IFN-α–induced mRNA expression of OAS as assessed by RT-PCR (Fig. 10), supporting the hypothesis that IL-8 apparently blocked the antiviral action of IFN-α at late, e.g., OAS activity, rather than early stage.
Figure 10.
Effect of IL-8 on IFN-regulated OAS mRNA gene expression. WISH cells were pretreated with medium or IL-8, 4 or 18 h overnight (O/ N), followed by medium or 10 IU/ml IFN-α for 4 h. RNA was reverse transcribed, amplified with primers for OAS and β actin, and gel electrophoresed. Lane designations are as indicated on figure. RT-PCR conditions for OAS gene expression allowed, at least, qualitative comparisons, e.g., significant stimulation or inhibition of signal strength. Identity of OAS fragment was confirmed with oligoprobe specific to OAS (not shown). Experiment is one of three performed.
DISCUSSION
Recently, chemokines have attracted the attention of the biomedical community because of their protective role in HIV infections and the reports of an ever increasing number of chemokines with novel functions (29–32). In this study, we have also demonstrated a novel role for one member of the chemokine family that, to the best of our knowledge, has not been previously described. In short, we have provided evidence for the IL-8 inhibition of the antiviral action of IFN-α in several virus–cell systems with emphasis on EMCV and HeLa line (WISH) as a well-studied model for IFN action.
The potency of the IL-8 inhibitory effect on IFN-α antiviral action observed here was dependent on IFN-α and IL-8 doses. The action of IL-8 was more potent at lower IFN doses (<30 IU/ml). These are still within the physiological concentrations in plasma of healthy individuals, patients with viral diseases, and even patients undergoing some IFN therapy regimens (33–35). The IL-8 doses shown in this report to be effective in inhibition of IFN-α antiviral action were also observed in plasma of subjects with various inflammatory and viral infections (36–38).
We also demonstrated here that the inhibitory action of IL-8 on IFN-α antiviral activity was observed at different stages of virus life cycle: viral protein formation, total virus yield, and CPE. It is likely that IL-8 action interferes with IFN-mediated inhibition of viral replication since both the biosynthesis of viral protein and total virus yield were compromised by IL-8. Of these three, the earliest stage is the formation of viral proteins. There are two well-characterized pathways for IFN action that results in inhibition of viral proteins, namely, OAS/RNAse L and double-stranded dependent protein kinase (PKR) pathways. OAS/RNAse L is thought to control picornaviral viruses such as EMCV and Mengo virus, but not VSV replication (39–41). Our observations have demonstrated that IL-8 inhibits IFN-α antiviral action against the picornaviruses, EMCV, and poliovirus, but not against VSV. Also, reduction in OAS activity was seen with EMCV, but not VSV in IFN-treated cells. Taking these observations together with our previous observations that showed that IL-8 action in EMCV-infected, unlike VSV-infected, cells were associated with decreased OAS activity (5), suggest that OAS is the late stage that may be subject to IL-8 action. Thus, the IL-8 selective action in certain virus–cell systems may be related to which of the multiple mechanisms that is regulated by IFN to control the replication of the virus (1, 2, 42). For example, IFN inhibition of HSV-1 replication may not be attributed to OAS pathway, and the mechanism of IL-8 inhibitory action on IFN-α activity against HSV-1 may well be different from those against the picornaviruses.
The observations regarding the timing of IL-8 addition with respect to IFN-α treatment may indicate that IL-8 induced a nearly IFN-resistant state in the cells before virus challenge. Also, IL-8 appeared to work at a stage later than IFN-induced gene expression and protein synthesis, both of which are normally completed within several hours after IFN treatment (43). This hypothesis is supported by the observation that no suppressive influence of IL-8 on IFN-induced expression of 6-16 and OAS genes was qualitatively seen. Thus, it is more likely that the mechanism of IL-8 action in inhibiting IFN-α antiviral action occurs on OAS pathway in a manner that is probably independent of gene expression.
The capacity of viruses to induce IL-8 in vitro and in vivo (3, 6–8, 36–38), the enhancement of viral replication by IL-8 (4, 5), and the interference with IFN-α antiviral action against viruses may constitute a common strategy by which viruses take advantage of the host proteins for their own survival. This is opposite to the IFN system, which is to protect cells from viruses, and also shown to inhibit IL-8 synthesis (44).
In the present investigation, we used picornaviruses, such as EMCV, that are common in the study of IFN system (1, 40–42). Aside from EMCV and poliovirus, IFN-α– mediated inhibition of HSV-1 replication seems to be also compromised by IL-8. However, it is not known whether IL-8 inhibits IFN action against other viruses such as HIV. Recently, it was reported that β chemokines (RANTES, macrophage inhibitory protein–1α, and –1β) and the α chemokine stromal-derived factor 1 can inhibit HIV binding to the coreceptor CC-CKR-5 and CXC-CKR-3, respectively (for review see reference 29). This is not necessarily in conflict with the notion of a potential antagonizing effect of IL-8 on IFN action in HIV infections. IL-8 binds to different receptors, CXCR1 and CXCR2, that are expressed by several types of leukocytes and cell types of nonhematopoietic origin including WISH epithelial cells (our results), fibroblasts, endothelial cells, and keratinocytes (23– 26).
The presence of IL-8 induced by viruses or other stimuli may contribute, at least partly, to the low potency of IFN in vivo during therapy and disease. Careful assessment of these hypothesis in animal models is required. Interference with IL-8 production or action may suppress viral activity and/or augment endogenous IFN-α antiviral action against selected viruses. Also, intervention of IL-8 action or production may be useful as a mean of supplementing IFN-α therapy and hence, enhancing of IFN-α antiviral potency or reduction of its toxicity.
Acknowledgments
We would like to thank Dr. John A. Armstrong (University of Pittsburgh, Pittsburgh, PA), and Dr. Malcolm Paterson (King Faisal Specialist Hospital and Research Center, Riyadh, Saudi Arabia) for critical review of the manuscript. We also thank Dr. Sandra Pellegrini (Pasteur Institute, Paris, France) not only for supplying the 6-16 cDNA probe, but also for her helpful comments. The technical assistance of Maud Dzimiri is acknowledged. The authors thank Royspec Purchasing Services of King Faisal Hospital and Research Center in Maryland (Hanover, MD) for expeditious shipping of biological and other materials.
Footnotes
Abbreviations used in this paper: CMV, cytomegalovirus; CPE, cytopathic effects; EMCV, encephalomyocarditis virus; FBS, fetal bovine serum; LU, laboratory units; mRNA, messenger RNA; MTS, tetrazolium salt; OAS, 2′,5′-oligoadenylate synthetase; RIPA, radioimmunoprecipitation; RT, reverse transcriptase; VSV, vesicular stomatitis virus.
Part of this work was presented at the first joint meeting of the International Cytokine Society (ICS) and International Society for Interferon and Cytokine Research (ISICR), Geneva, Switzerland, October 1996.
REFERENCES
- 1.Pestka S, Langer JA, Zoon KC, Samuel CE. Interferons and their actions. Annu Rev Immunol. 1987;56:727–777. doi: 10.1146/annurev.bi.56.070187.003455. [DOI] [PubMed] [Google Scholar]
- 2.Sen GC, Lengyel P. The interferon system. A bird's view of its biochemistry. J Biol Chem. 1982;267:5017–5020. [PubMed] [Google Scholar]
- 3.Murayama T, Ohara Y, Obuchi M, Khabar KSA, Higashi H, Mukaida N, Matsushima K. Human cytomegalovirus induces IL-8 production by a human monocytic cell line, THP-1, through acting concurrently on AP-1 and NF-kB binding sites of the interleukin-8 gene. J Virol. 1997;71:5692–5695. doi: 10.1128/jvi.71.7.5692-5695.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murayama T, Kuno K, Jisaki F, Obuchi M, Sakamuro D, Furukawa T, Mukaida N, Matsushima K. Enhancement of human cytomegalovirus replication in a human lung fibroblast cell line by interleukin-8. J Virol. 1994;68:7582–7585. doi: 10.1128/jvi.68.11.7582-7585.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khabar KSA, Al-Zoghaibi F, Murayama T, Matsushima K, Mukaida N, Siddiqui Y, Dhalla M, Al-Ahdal MN. Interleukin-8 selectively enhances cytopathic effect (CPE) induced by positive-strand RNA viruses in the human WISH cell line. Biochem Biophys Res Commun. 1997;235:774–778. doi: 10.1006/bbrc.1997.6872. [DOI] [PubMed] [Google Scholar]
- 6.Choi AM, Jacoby DB. Influenza virus A infection induces interleukin-8 gene expression in human airway epithelial cells. FEBS Lett. 1992;309:327–329. doi: 10.1016/0014-5793(92)80799-m. [DOI] [PubMed] [Google Scholar]
- 7.Fiedler MA, Wereke-Dollries K, Stark JM. Respiratory syncytial virus increases IL-8 gene expression and protein release in A549 cells. Am J Physiol. 1996;269:865–872. doi: 10.1152/ajplung.1995.269.6.L865. [DOI] [PubMed] [Google Scholar]
- 8.Sheth R, Anderson J, Sato T, Oh B, Hempson SJ, Rollo E, Mackow ER, Shaw RD. Rotavirus stimulate IL-8 secretion from cultured epithelial cells. Virology. 1996;221:251–259. doi: 10.1006/viro.1996.0374. [DOI] [PubMed] [Google Scholar]
- 9.Baruch-Ben A, Michiel DF, Oppenheim JJ. Signals and receptors involved in recruitment of inflammatory cells. J Biol Chem. 1995;270:11703–11706. doi: 10.1074/jbc.270.20.11703. [DOI] [PubMed] [Google Scholar]
- 10.Oppenheim JJ, Zachariae COC, Mukaida N, Matsushima K. Properties of the novel proinflammatory supergene “intercrine” cytokine family. Annu Rev Immunol. 1991;9:617–618. doi: 10.1146/annurev.iy.09.040191.003153. [DOI] [PubMed] [Google Scholar]
- 11.Taub DD, Anver M, Oppenheim JJ, Longo DL, Murphy WJ. T lymphocyte recruitment by interleukin-8. IL-8 induced degranulation of neutrophils releases potent chemoattractants for human T lymphocytes both in vitro and in vivo. J Clin Invest. 1996;97:1931–1941. doi: 10.1172/JCI118625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koch AE, Polverini PJ, Kunkel SL, Harlow LA, DiPietro LA, Elner VM, Elner SG, Strieter RM. Interleukin-8 as a macrophage-derived mediator of angiogenesis. Science (Wash DC) 1992;258:1798–1801. doi: 10.1126/science.1281554. [DOI] [PubMed] [Google Scholar]
- 13.Cacalano G, Lee J, Kikly K, Ryan AM, Pitts-Meek S, Hultgren B, Wood WI, Moore MW. Neutrophil and B cell expansion in mice that lack the murine IL-8 receptor homolog. Science (Wash DC) 1994;265:682–684. doi: 10.1126/science.8036519. [DOI] [PubMed] [Google Scholar]
- 14.Matsushima K, Morishita K, Yoshima T, Lavu S, Kobayashi Y, Lew W, Appella E, Kung HF, Leonard EJ, Oppenheim JJ. Molecular cloning of a human monocyte-derived neutrophils chemotactic factor (MDNCF) and the induction of MDNCF mRNA by interleukin-1 and tumor necrosis factor. J Exp Med. 1988;167:1883–1893. doi: 10.1084/jem.167.6.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furuta R, Yamagishi J, Kotani H, Sakamoto F, Fukui T, Matsui Y, Sohmura Y, Yamada M, Yoshimura T, Larsen CG, Oppenheim JJ, Matsushima K. Production and characterization of recombinant human neutrophil chemotactic factor. J Biochem (Tokyo) 1989;106:436–441. doi: 10.1093/oxfordjournals.jbchem.a122870. [DOI] [PubMed] [Google Scholar]
- 16.Khabar KSA, Alzoghaibi F, Dzimiri M, Taha M, Al-Tuwaijri A, Al-Ahdal MN. MTS interferon assay: a simplified cellular dehydrogenase assay for interferon activity using a water-soluble tetrazolium salt. J Interferon Cytokine Res. 1996;16:31–33. doi: 10.1089/jir.1996.16.31. [DOI] [PubMed] [Google Scholar]
- 17.Armstrong JA. Cytopathic effect inhibition assay for interferon: microculture plate assay. Methods Enzymol. 1980;78:381–387. doi: 10.1016/0076-6879(81)78145-x. [DOI] [PubMed] [Google Scholar]
- 18.Ko YN, Mukaida N, Pamyutich A, Voitenk NN, Matsushima K, Kawai T, Kasahara T. Establishment of a sensitive enzyme-linked immunosorbent assay for human interleukin-8. J Immunol Methods. 1992;149:227–235. doi: 10.1016/0022-1759(92)90254-q. [DOI] [PubMed] [Google Scholar]
- 19.Lewis, J.A. 1991. Antiviral activity of cytokines. In Cytokines: A Practical Approach. F.R. Balkwill, editor. Oxford University Press, Oxford. 109–120.
- 20.Chomczynski P, Sacchi N. Single step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 21.Murphy PM, Tiffany HL. Cloning of complementary DNA encoding a functional human interleukin-8 receptor. Science (Wash DC) 1991;253:1280–1283. doi: 10.1126/science.1891716. [DOI] [PubMed] [Google Scholar]
- 22.Higuchi M, Aggarwal BB. Microtiter plate radioreceptor assay for tumor necrosis factor and its receptor in large numbers of samples. Anal Biochem. 1992;204:53–58. doi: 10.1016/0003-2697(92)90138-w. [DOI] [PubMed] [Google Scholar]
- 23.Grob PM, David E, Warren TC, DeLeon RP, Farina PR, Homon CA. Characterization of a receptor for human monocyte-derived neutrophil chemotactic factor/ interelukin-8. J Biol Chem. 1990;265:8311–8316. [PubMed] [Google Scholar]
- 24.Kemeny L, Kenderssy AS, Ocsovszky I, Michel G, Ruzicka T, Dobozy A. Interleukin-8 induces HLA-DR expression on cultured human keratinocytes via specific receptors. Int Arch Allergy Immunol. 1995;106:251–256. doi: 10.1159/000236866. [DOI] [PubMed] [Google Scholar]
- 25.Mueller SG, Schraw WP, Richmond A. Melanoma growth stimulatory activity enhances the phosphorylation of the class II interleukin-8 receptor in non-hematopoietic cells. J Biol Chem. 1994;269:1973–1980. [PubMed] [Google Scholar]
- 26.Schönbeck U, Brandt E, Petersen F, Dieter-Han F, Loppnow H. IL-8 specifically binds to endothelial but not smooth muscle cells. J Immunol. 1995;154:2375–2383. [PubMed] [Google Scholar]
- 27.Pellegrini S, John J, Shearer M, Kerr IM, Stark GR. Use of a selectable marker regulated by alpha interferon to obtain mutations in the signaling pathway. Mol Cell Biol. 1989;9:4605–4612. doi: 10.1128/mcb.9.11.4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darnell JE, Kerr IM, Stark GR. JAK-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science (Wash DC) 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 29.Balter M. A second coreceptor for HIV in early stages of infection. Science (Wash DC) 1996;272:1740. doi: 10.1126/science.272.5269.1740. [DOI] [PubMed] [Google Scholar]
- 30.Godiska R, Chantry D, Raport CJ, Sozzani S, Allavena P, Leviten D, Mantovani A, Gray PW. Human macrophage-derived chemokine (MDC), a novel chemoattractant for monocytes, monocyte-derived dendritic cells, and natural killer cells. J Exp Med. 1997;185:1595–1604. doi: 10.1084/jem.185.9.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mackay CR. Chemokine receptors and T cell chemotaxis. J Exp Med. 1996;184:799–802. doi: 10.1084/jem.184.3.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patel, V.P., B.L. Kreider, Y. Li, H. Li, K. Leung, T. Salcedo, B. Nardelli, V. Pippalla, S. Gentz, R. Thotakura, et al. Molecular and functional characterization of two novel human C–C chemokines as inhibitors of two distinct classes of myeloid progenitors. J. Exp. Med. 185:1163–1172. [DOI] [PMC free article] [PubMed]
- 33.Bernier J, Reuter A, Vrindts-Gevaert Y, Franchimont P. Radioimmunoassay of leukocyte (alpha) interferon and its application to some clinical conditions. J Nucl Med. 1984;25:765–772. [PubMed] [Google Scholar]
- 34.Bornemann LD, Spiegel HE, Dziewanowska ZE, Krown SE, Colburn WA. Intravenous and intramuscular pharmokinetics of recombinant leukocyte A interferon. Eur J Clin Pharmacol. 1985;28:469–471. doi: 10.1007/BF00544369. [DOI] [PubMed] [Google Scholar]
- 35.Shiozawa S, Yoshikawa N, Iijima K, Negishi K. A sensitive radioimmunoassay for circulating alpha-interferon in the plasma of healthy children and patients with measles virus infection. Clin Exp Immunol. 1988;73:366–369. [PMC free article] [PubMed] [Google Scholar]
- 36.Matsumoto T, Mike T, Nelson RP, Trudeau WL, Lockey RF, Yodoi J. Elevated serum levels of IL-8 patients with HIV infections. Clin Exp Immunol. 1993;93:149–151. doi: 10.1111/j.1365-2249.1993.tb07957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnston SL. Natural and experimental rhinovirus infections of the lower respiratory tract. Am J Respir Crit Care Med. 1995;152:S46–52. doi: 10.1164/ajrccm/152.4_Pt_2.S46. [DOI] [PubMed] [Google Scholar]
- 38.al-Wabel A, al-Knawy B, Raziuddin S. Interleukin-8 and granulocyte–macrophage colony-stimulating factor secretion in hepatocellular carcinoma and viral chronic active hepatitis. Clin Immunol Immunopathol. 1995;74:231–235. doi: 10.1006/clin.1995.1034. [DOI] [PubMed] [Google Scholar]
- 39.Chebath J, Benech P, Revel M, Vegneron M. Constitutive expression of (2′–5′) oligo A synthetase confers resistance to picornavirus infection. Nature (Lond) 1987;330:587–588. doi: 10.1038/330587a0. [DOI] [PubMed] [Google Scholar]
- 40.Hassel BA, Zhou A, Stomayor C, Maran A, Silverman RH. A dominant negative mutant of 2-5A– dependent RNase suppresses antiproliferative and antiviral effects of interferon. EMBO (Eur Mol Biol Organ) J. 1993;12:3297–3304. doi: 10.1002/j.1460-2075.1993.tb05999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rysiecki G, Gewart D, Williams B. Constitutive expression of (2′–5′)-oligo A synthetase cDNA results in increased antiviral activity and growth suppression. J Interferon Res. 1990;9:649–657. doi: 10.1089/jir.1989.9.649. [DOI] [PubMed] [Google Scholar]
- 42.Samuel CE. Antiviral action of interferon: interferon-regulated cellular proteins and surprisingly selective antiviral activities. Virology. 1991;183:1–11. doi: 10.1016/0042-6822(91)90112-o. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka N, Taniguchi T. Cytokine gene expression: regulatory cis-elements and DNA binding factors involved in the interferon system. Adv Immunol. 1992;52:263–281. doi: 10.1016/s0065-2776(08)60877-9. [DOI] [PubMed] [Google Scholar]
- 44.Oliveira IC, Mukaida N, Matsushima K, Vilcek J. Transcriptional inhibition of the interleukin-8 gene by interferon is mediated by the NF-kB site. Mol Cell Biol. 1994;14:5300–5308. doi: 10.1128/mcb.14.8.5300. [DOI] [PMC free article] [PubMed] [Google Scholar]