Abstract
TRAIL-R3, a new member of the TRAIL receptor family, has been cloned and characterized. TRAIL-R3 encodes a 299 amino acid protein with 58 and 54% overall identity to TRAIL-R1 and -R2, respectively. Transient expression and quantitative binding studies show TRAIL-R3 to be a plasma membrane–bound protein capable of high affinity interaction with the TRAIL ligand. The TRAIL-R3 gene maps to human chromosome 8p22-21, clustered with the genes encoding two other TRAIL receptors. In contrast to TRAIL-R1 and -R2, this receptor shows restricted expression, with transcripts detectable only in peripheral blood lymphocytes and spleen. The structure of TRAIL-R3 is unique when compared to the other TRAIL receptors in that it lacks a cytoplasmic domain and appears to be glycosyl-phosphatidylinositol–linked. Moreover, unlike TRAIL-R1 and -R2, in a transient overexpression system TRAIL-R3 does not induce apoptosis.
The TNF family of cytokines and receptors has been shown to play a pivotal role in the maintenance of homeostasis in multiple biological networks, including the immune system (1, 2). Particular interest has been generated by the finding that certain members of the TNF family are capable of inducing programmed cell death or apoptosis. In addition to TNF and Fas ligand, TRAIL, a recently identified member of the TNF family, has been shown to induce apoptosis in a wide variety of transformed cell lines of diverse origin (3). Unlike Fas ligand, the expression of which is predominantly restricted to activated T cells (4) and sites of immune privilege (5), TRAIL message is widely expressed (3). This suggests that either the receptor for TRAIL is restricted in distribution, or that TRAIL is capable of transducing different signals via one or multiple receptors, as is the case for TNF. Two receptors for TRAIL, TRAIL-R1 or DR4 (6) and TRAIL-R2 (7), have been recently characterized. Interestingly, both receptors show widespread distribution and are capable of mediating apoptosis.
In an attempt to identify novel proteins related to the above-mentioned inducers of apoptosis, we searched for sequences with homology to known members of the TNF family of cytokines and receptors. Such molecules may provide additional means to regulate the process of programmed cell death, and may lead not only to further understanding of immune regulation, but also to better intervention strategies in the battle against defects of the immune system. Here we describe the identification and characterization of a new TRAIL receptor which, unlike the previously characterized TRAIL-R1 and -R2, does not signal apoptosis and appears to be glycosyl-phosphatidylinositol (GPI)–linked to the plasma membrane.
MATERIALS AND METHODS
Isolation of TRAIL-R3 cDNA.
A cDNA sequence (data available from EMBL/GenBank/DDBJ under accession number T71406) with homology to TRAIL-R1 (6) and -R2 (7), was identified using the full length TRAIL-R1 sequence to perform a Blast search of the NCBI (National Center for Biotechnology Information, National Institutes of Health, Bethesda, MD) EST (expressed sequence tag) database. A putative third TRAIL receptor was identified by sequencing the I.M.A.G.E. clone containing this EST (I.M.A.G.E. Consortium Homo sapiens cDNA clone 110226; reference 8). Additional cDNAs encoding TRAIL-R3 were subsequently identified from a human foreskin fibroblast cDNA library using a 32P-dCTP random prime–labeled PCR product encompassing the cysteine-rich extracellular domain of the putative TRAIL-R3.
Plasmid Construction.
A full length TRAIL-R3 transcript, using Met 41 as the initiator methionine, was cloned into the pDC409 mammalian expression vector (9) by PCR. The TRAIL-R3–Fc fusion chimera was constructed as described (9), by fusing the extracellular domain, encoded between Met 41 and Ala 216, to the Fc portion of a mutein human IgG1 sequence.
Transient Transfection and Measurement of X-gal Expression.
CVI/EBNA cells (CRL 10478; American Type Culture Collection, Rockville, MD) (1.65 × 105 cells) were transfected with 1.5 μg (1.0 μg of test plasmid and 0.5 μg of pDC409-Escherichia coli lacZ) of DNA by the DEAE-dextran method (10). After 48 h cells were washed with PBS, fixed with glutaraldehyde, and stained with X-gal (5-bromo-4-chloro-3-indoxyl-β-d-galactopyranoside) for β-galactosidase activity. A reduction in cells stained indicates loss of β-galactosidase expression and correlates with death of cells that express the protein(s) cotransfected with the lacZ gene (11). Each experiment was repeated three times.
Scatchard Analysis.
Equilibrium-binding isotherms between the TRAIL ligand and three TRAIL receptors were determined by Scatchard analysis using either purified Fc proteins (TRAIL-R1–Fc and -R2–Fc) bound to plates previously coated with goat anti–human Fc, or transfected CVI/EBNA cells transiently expressing TRAIL-R3. Transfected cells were replated after 24 h in 24 well plates at a density of 50,000 cells/well and left to recover overnight. The cells were then incubated (30 min at room temperature) in binding medium (3% BSA, 20 mM Hepes, pH 7.4, 0.15 M NaCl, 0.04% NaN3) with serial dilutions (2×) of soluble Leucine-Zipper (LZ) human TRAIL (7) starting at a concentration of 5 μg/ml. The cells were washed with binding medium to remove unbound LZ-TRAIL, and incubated (30 min at room temperature) in binding medium with 125I-labeled M15 anti-LZ mAb (125 ng/ml). The cells were then washed, removed from the plates with trypsin, and the radioactivity in the cell suspension was measured. A similar procedure was used for plates carrying bound Fc proteins, except that specifically bound ligand was released with 0.1 M glycine HCl, pH 3. In both experiments, nonspecific binding was determined by inclusion of a 500-fold molar excess of unlabeled M15 anti-LZ antibody in two duplicate reaction mixtures containing the first two dilutions of LZ-TRAIL. Specific binding values were calculated by subtracting linearly extrapolated nonspecific binding from each data point. The data were plotted in a Scatchard coordinate system with a nonlinear least squared fit using RS1 software (BBN Software Products Corporation, Cambridge, MA).
Jurkat Killing Assay.
Concentrated supernatants (25×) containing equivalent amounts (75–125 μg/ml) of TRAIL-R1–Fc, -R2–Fc, -R3–Fc, and CD30-Fc were added to Jurkat cells (104 cells/well) simultaneously with LZ-TRAIL (150 ng/ml). Percentage viability was measured after 16–20 h by MTT dye conversion (12). The highest MTT reading, obtained in the absence of LZ-TRAIL, was used as the maximum viability value.
GPI Linkage Analysis.
CVI/EBNA cells (105) transfected with (a) TRAIL-R1, (b) -R2, (c) -R3, (d) LERK-3 (13), or (e) pDC409 vector were harvested 48 h after transfection with 50 mM EDTA (10 min at 37°C), and incubated with or without 1 U/ml recombinant phosphatidylinositol–specific phospholipase C (PI-PLC; Oxford Glyco Sciences, Bedford, MA) (1 h, 37°C). The cells were then washed and incubated (30 min at room temperature) with either LZ-TRAIL (5 μg/ml) followed by 125I-labeled M15 anti-LZ antibody (125 ng/ml) or Hek-Fc, the LERK-3 cognate (5 μg/ml) followed by 125I-labeled goat anti–human Fc (125 ng/ml). Free and bound probes were separated by microfuging duplicate 60-μl aliquots of the suspension through a pthalate oil mixture (14). The counts in the free (supernatant) and bound (cell pellet) probes were measured. Each experiment was repeated four times.
Northern Analysis.
A human multiple tissue Northern blot (Clontech, Palo Alto, CA) was probed with a 32P-dCTP random prime–labeled PCR product encompassing the extracellular domain of TRAIL-R3 from Met 41 to Thr 201. Hybridization was performed at 63°C in Stark's buffer overnight (15). The blot was then washed three times in 2× SSC, 0.1% SDS at 68°C and exposed to film (X-OMAT; Eastman Kodak Co., Rochester, NY) 16–72 h at −70°C.
Chromosomal Mapping.
Chromosomal location was determined using two independent radiation hybrid panels, the Stanford G3 Radiation Hybrid Panel and the Genebridge 4 Radiation Hybrid Panel (Research Genetics, Huntsville, AL). The panels were screened with two oligonucleotide primers (5′CTTCCTTACCTGAAAGGTTCAGGTAGG3′ and 5′CTCTTGGACTTGGCTGGGAGATGTG3′) capable of reliably and specifically amplifying human TRAIL-R3 from genomic DNA. The results were electronically submitted to the appropriate servers for linkage analysis.
RESULTS AND DISCUSSION
Isolation of TRAIL-R3 cDNA.
The NCBI EST database was screened with the TRAIL-R1 sequence to determine the potential existence of additional TRAIL receptors. Three EST sequences (data available from EMBL/GenBank/ DDBJ under accession numbers T71406, AA150849, and AA150541) were identified showing partial identity to the nucleotide sequence of TRAIL-R1 (6) and -R2 (7). An 897-nucleotide open reading frame (ORF) was obtained by direct sequencing of T71406 (I.M.A.G.E. Consortium Clone 110226) (8) (Fig. 1 A). This ORF, referred to as TRAIL-R3, has 54 and 52% nucleotide and 58 and 54% amino acid (aa) identity to TRAIL-R1 and -R2, respectively. The authenticity of this cDNA was confirmed by analysis of a second full length cDNA clone isolated by screening the human foreskin fibroblast library with a probe encompassing the cysteine-rich extracellular domain of the putative TRAIL-R3.
Figure 1.
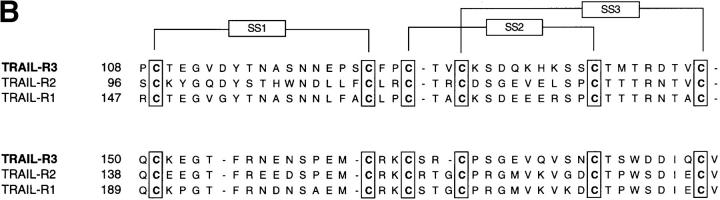
TRAIL-R3 is a novel member of the TRAIL receptor family. (A) The nucleotide and aa sequence of TRAIL-R3 is shown. The aa sequence is started at the first Met; the two potential initiator codons are boxed. The predicted NH2-terminal leader cleavage site is indicated by the triangle. The COOH-terminal hydrophobic domain is marked by a dashed line. Five pseudo-repeats in the linker region, separating the extracellular domain from the hydrophobic COOH terminus, are shown in boxes. (B) Alignment of the extracellular domains of TRAIL-R3, -R2, and -R1 shows conservation of two of the cysteine-rich pseudo-repeats characteristic of the TNF receptor family. Conserved cysteine residues are boxed. Predicted disulfide bonds are shown. These sequence data are available from EMBL/GenBank/DDBJ under accession number AF014794.
The TRAIL-R3 transcript encodes a 299-aa protein with a predicted signal sequence cleavage site after aa 69, a 121-aa extracellular cysteine-rich domain, an 88-aa extracellular linker sequence, and a 21-aa hydrophobic COOH-terminal sequence. Like TRAIL-R1, this transcript carries two methionines within the first 60 aa of the ORF. Analysis of signal peptide cleavage sites predicts the mature protein to start at Arg 70, suggesting that Met 41 is probably the start codon. Unlike the previously characterized TRAIL receptors, the predicted TRAIL-R3 protein does not contain a cytoplasmic domain (Fig. 1 A). Like TRAIL-R1 and -R2, the extracellular domain of TRAIL-R3 contains only two of the four cysteine-rich pseudo-repeats characteristic of the extracellular domain of most members of the TNF receptor family (Fig. 1 B). The TRAIL-R3 cysteine-rich extracellular domain is separated from a COOH-terminal hydrophobic region by an 88-aa linker sequence. This linker contains five copies of a 15-aa pseudo-repeat (Fig. 1 A); a single copy of this repeat is found in the 31-aa linker region of TRAIL-R2, but is absent in TRAIL-R1.
TRAIL-R3 Binds TRAIL.
Given the observed aa identity in the extracellular ligand binding domains of TRAIL-R3, -R2, and -R1, we predicted that TRAIL-R3 would bind TRAIL. To test this hypothesis, TRAIL-R3 was transiently expressed in CVI/EBNA cells. Transfected cells were then tested for their ability to bind TRAIL using the very sensitive autoradiographic analysis previously described (16). Binding of TRAIL was observed to CVI/ EBNA cells transfected with TRAIL-R3, but not to vector transfected cells (data not shown), demonstrating that this receptor is expressed on the cell surface and that it is a cognate for the TRAIL ligand.
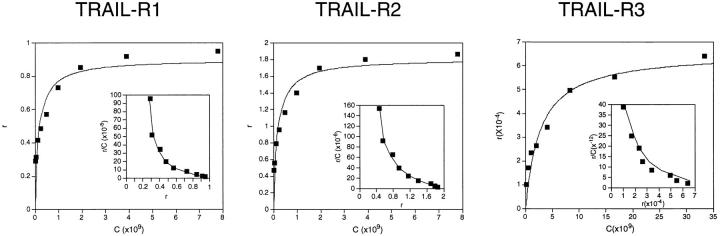
The equilibrium-binding isotherm between soluble TRAIL ligand and TRAIL-R3 was determined using a recombinant receptor transiently expressed on CVI/EBNA cells. For comparison, the isotherms of TRAIL-R1 and -R2 were determined using purified Fc proteins as the substrate for the binding studies (see Materials and Methods). Binding of the receptors to TRAIL was achieved using LZ-TRAIL and 125I-labeled M15 anti-LZ antibody. All three receptors bound LZ-TRAIL in a specific, saturable fashion, and Scatchard analysis using nonlinear least squared regression revealed binding sites with comparable affinities (K d(HIGH) = 0.04–0.36 nM; K d(LOW) = 0.38–9.0 nM), indicating that TRAIL binds equally well to all three receptors (Fig. 2).
Figure 2.
Equilibrium binding isotherms of TRAIL-R3, -R2, and -R1. Full length TRAIL-R3 transiently expressed on CVI/EBNA cells and purified TRAIL-R1 and -R2 Fc proteins were used in equilibrium binding assays with LZ-TRAIL + 125I-labeled M15 anti-LZ antibody, as described above (see Materials and Methods). The binding data plotted in the Scatchard coordinate system is shown. The membrane-bound TRAIL-R3 protein binds TRAIL with similar affinity as TRAIL-R1 and -R2 soluble Fc proteins.
TRAIL-induced Apoptosis is Inhibited by Soluble TRAIL-R3–Fc.
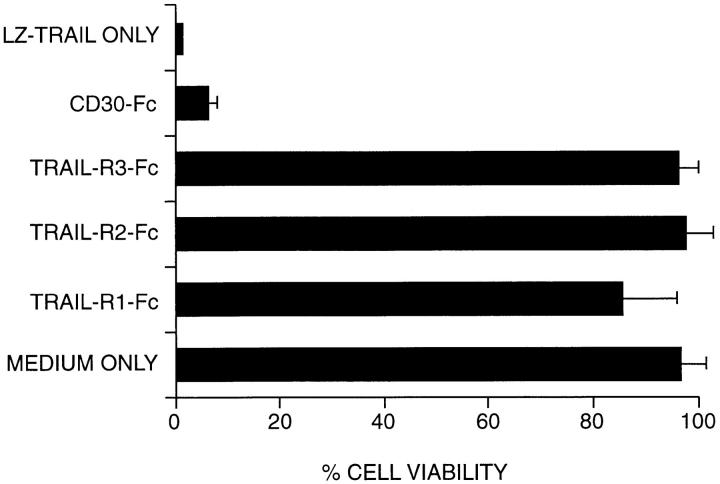
Soluble fusion proteins comprising the ligand binding domain of receptors fused to the Fc domain of human IgG1 have proven to be potent inhibitors of ligand-mediated activity (17, 18). Thus, a soluble TRAIL-R3–Fc was constructed to determine its ability to impede TRAIL-mediated activities. The Jurkat T-cell line, previously shown to die in response to TRAIL, was cultured with soluble LZ-TRAIL in the presence of concentrated supernatants from CVI/EBNA cells transiently expressing soluble (a) TRAIL-R3–Fc, (b) -R2–Fc, (c) -R1–Fc, or (d) CD30-Fc. Specific and complete inhibition of LZ-TRAIL–mediated apoptosis was obtained with TRAIL-R3–Fc, -R1–Fc, and -R2–Fc, but not with CD30-Fc (Fig. 3).
Figure 3.
TRAIL-R3–Fc inhibits TRAIL-mediated killing. Jurkat cells treated with LZ-TRAIL (150 ng/ml) were cultured for 16 h with concentrated supernatants containing soluble TRAIL-R3–Fc, -R2–Fc, -R1– Fc or CD30-Fc proteins. All three TRAIL-R–Fc proteins block TRAIL-mediated apoptosis of Jurkat cells as measured by MTT conversion (12). The maximum viability value corresponds to the MTT reading in the absence of LZ-TRAIL (MEDIUM ONLY). The minimum viability value corresponds to the MTT reading in the presence of LZ-TRAIL (LZ-TRAIL ONLY).
TRAIL-R3 Does Not Induce Apoptosis by Overexpression.
Fas and TNF-R1 are prototypic triggers of apoptosis. In addition, four new members of the TNFR family are able to induce apoptosis. A common feature of these receptors, which include DR-3 (19), CAR-1 (20), and the two receptors for TRAIL (6, 7), is that they share an ∼80-aa region of homology, referred to as the “death domain,” within their cytoplasmic domains. This region appears to be critical for the induction of apoptosis by Fas, TNFR-1 and DR-3 (19, 21, 22).
As for Fas, TNFR-1, and DR3, overexpression of TRAIL receptors 1 and 2 in a transient transfection system results in ligand-independent apoptosis (6, 7). Unlike TRAIL-R1 and -R2, TRAIL-R3 lacks a cytoplasmic domain, the region that normally encodes the death domain. Therefore, we expected TRAIL-R3 to be unable to transduce an apoptotic signal. Indeed, transient overexpression of TRAIL-R3 did not lead to cell death (data not shown).
TRAIL-R3 Is GPI Linked.
The sequence of TRAIL-R3 is unusual in that the COOH-terminal hydrophobic region is not followed by a cytoplasmic domain. However, despite the lack of a typical type I transmembrane protein structure, we have shown that a recombinant form of TRAIL-R3 is expressed on the cell surface of transfected cells and is capable of binding TRAIL with high affinity.
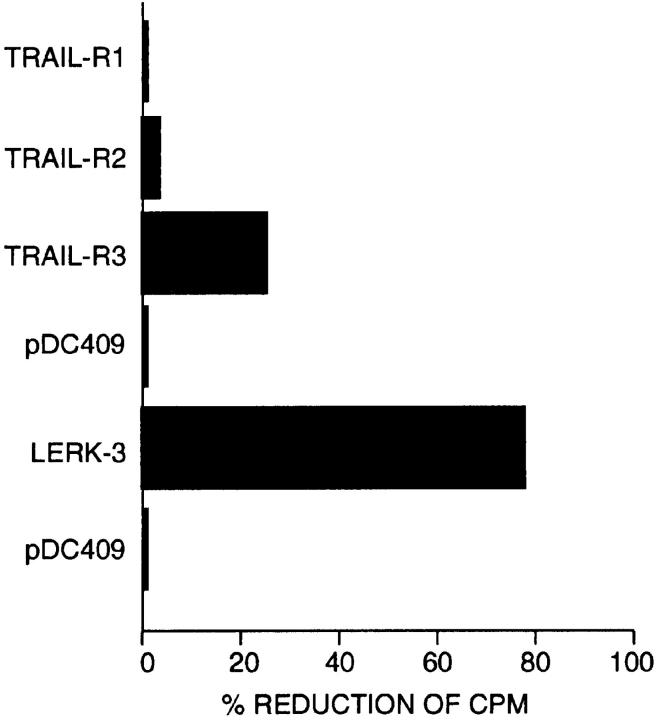
Several proteins can stably associate with the external surface of the cell membrane by covalent linkage to glycolipids. These include several members of the immunoglobulin superfamily (23), as well as some leucocyte surface proteins such as Ly-6 (24). The distinguishing features of such proteins include (a) the presence of a hydrophobic NH2-terminal signal peptide, which typically directs the protein to the endoplasmic reticulum, (b) a second hydrophobic region at the COOH terminus, and (c) the lack of a cytoplasmic domain. Since the structure of TRAIL-R3 fulfills all of the above requirements, we tested the possibility that this receptor is membrane bound through a GPI anchor. Given that most, but not all, GPI-anchored proteins can be released from cell surfaces by PI-PLC, we treated CVI/EBNA cells transfected with TRAIL-R3 with this enzyme. TRAIL-R1 and -R2 were used as negative controls and the GPI-linked LERK-3 protein (13) as a positive control. Analysis of TRAIL binding to TRAIL-R3–expressing cells after treatment with PI-PLC indicates that ∼30% of the membrane-bound TRAIL-R3 protein is displaced from the cell surface by PI-PLC (Fig. 4). Approximately 80% of the GPI-linked LERK-3 was displaced by PI-PLC treatment (Fig. 4). As expected, PI-PLC treatment did not affect the TRAIL-R1 and -R2 transmembrane proteins. The poor sensitivity of TRAIL-R3 to PI-PLC–mediated release suggests that anchoring of TRAIL-R3 to the cell membrane is mediated by phospholipid bonds only partially hydrolyzed by phospholipase C (25). The functional significance of GPI-anchoring TRAIL-R3 to the cell surface, as indeed is the case for GPI-linked proteins in general, remains to be determined. It has been suggested that GPI anchors allow differential protein release. Therefore, it is plausible that the structure of TRAIL-R3 developed as a mechanism for expeditious release of this receptor, thus providing a soluble inhibitor of TRAIL-mediated activities. Alternatively, rapid downregulation of TRAIL-R3 may be required for TRAIL to signal through TRAIL-R1 and -R2. In addition, it is possible that, like other GPI-anchored molecules, TRAIL-R3 may be capable of direct signaling (26).
Figure 4.
TRAIL-R3 can be partially displaced from the cell membrane by phospholipase C. The effect of PI-PLC treatment of CVI/EBNA cells transiently transfected with TRAIL-R1, -R2, -R3, LERK-3, or empty pDC409 vector is shown. After treatment or no treatment with PI-PLC, the cells were incubated with either LZ-TRAIL followed by 125I-labeled M15 anti-LZ antibody or Hek-Fc followed by 125I-labeled goat anti–human–Fc. The radioactivity in the free and bound probes, separated by microfuging through a pthalate oil mixture, was counted and the results plotted. PI-PLC treatment resulting in lowered bound probe counts (cpm) indicates displacement of cell surface proteins that bind the labeled probe.
TRAIL-R3 Shows Restricted Tissue Distribution.
The tissue distribution of TRAIL-R3 mRNA was determined by Northern blot analysis (Fig. 5). TRAIL-R3 message was clearly detected only in PBLs. A weak signal (not visible in Fig. 5) was observed, after prolonged exposure, in spleen. Five transcripts of ∼1.3, 2.5, 3.0, 4.0, and 7.0 kb were detected. The restricted distribution of TRAIL-R3 markedly contrasts with the wide-spread distribution of -R1 (6) and -R2 (7).
Figure 5.
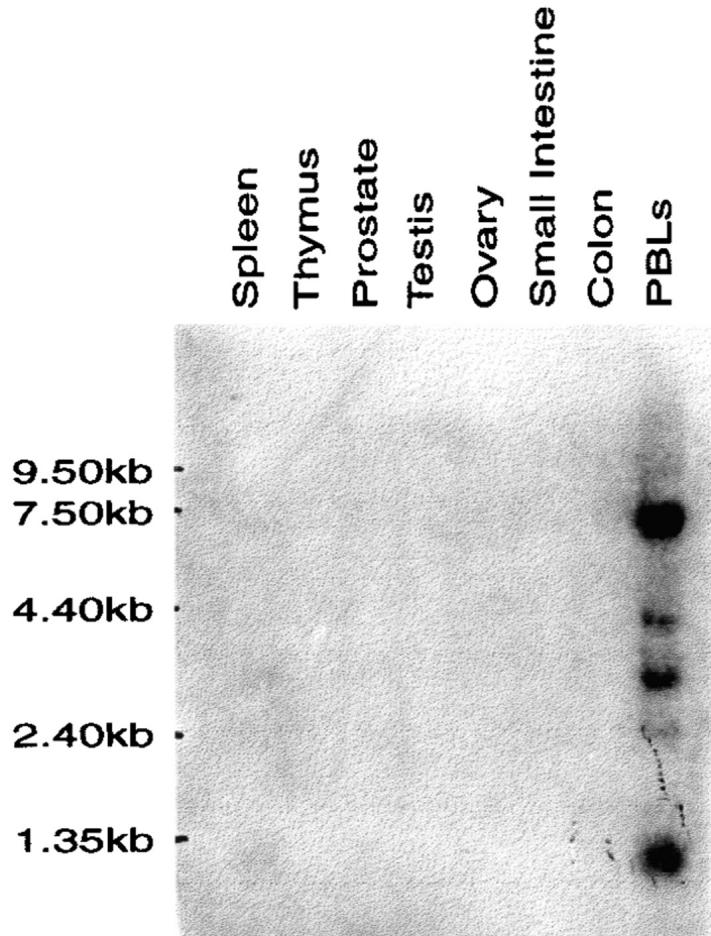
TRAIL-R3 shows restricted tissue expression. Northern analysis showing the distribution of TRAIL-R3 mRNA in whole human tissues. The source of the mRNA is shown above each lane. The position of RNA size markers is shown on the left. Multiple transcripts are detected in PBLs.
The TRAIL-R3 Gene Is Located on Human Chromosome 8p.
The chromosomal location of TRAIL-R3 was determined by PCR analysis of two independent radiation hybrid panels. The TRAIL-R3 gene has been mapped to human chromosome 8p22-21, ∼49 cM from the telomere, in close proximity to a cluster which also encodes the genes for TRAIL-R1 and -R2. This cluster is reminiscent of the TNFR gene clusters on chromosome 1p (TNFR-2, CD30, OX40) and on chromosome 12p (CD27, LTβR, TNFR-1). However, the high degree of nucleotide identity shared by the three TRAIL receptors, combined with their close chromosomal proximity, suggests that these loci have arisen recently as duplications of a precursor sequence.
Conclusions.
In conclusion, we have cloned and characterized TRAIL-R3, a new member of the TRAIL receptor family. We have clearly demonstrated that TRAIL-R3 exists as a cell surface molecule capable of binding TRAIL. However, the structure of this protein is unusual. Unlike TRAIL-R1 and -R2, -R3 appears to be GPI-linked and lacks a cytoplasmic region, including the death domain. Thus, as expected, TRAIL-R3 is unable to induce apoptosis. Of further interest is the restricted distribution of the TRAIL-R3 message. Unlike the other two TRAIL receptors, which are widely expressed, TRAIL-R3 transcripts are only present in PBLs and spleen. The significance of this finding remains to be determined; it is tantalizing to speculate that in these tissues TRAIL-R3 acts as an inhibitor of TRAIL-mediated apoptosis by competing with TRAIL-R1 and -R2 for binding to the ligand.
The identification of TRAIL-R3 adds new complexity to the emerging TRAIL receptor subfamily and is reminiscent of the intricacy surrounding the dual receptors for TNF. By analogy to the latter system, it is possible that novel TRAIL ligands are yet to be characterized. Continued evaluation of the biological activities of TRAIL-R3 and detailed characterization of this receptor family will provide important insights into the in vivo functions of these proteins.
Acknowledgments
The authors are grateful to C. Rauch for purification of LZ-TRAIL; K. Maggiora and A. Learned for transfections; and M. Petersen for purification of soluble Fc proteins.
REFERENCES
- 1.Cosman D. A family of ligands for the TNF receptor superfamily. Stem Cells. 1994;12:440–455. doi: 10.1002/stem.5530120501. [DOI] [PubMed] [Google Scholar]
- 2.Smith CA, Farrah T, Goodwin RG. The TNF receptor superfamily of cellular and viral proteins: activation, costimulation, and death. Cell. 1994;76:959–962. doi: 10.1016/0092-8674(94)90372-7. [DOI] [PubMed] [Google Scholar]
- 3.Wiley SR, Schooley K, Smolak PJ, Din WS, Huang C-P, Nicholl JK, Sutherland GR, Davis-Smith T, Rauch C, Smith CA, Goodwin RG. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity. 1995;3:673–682. doi: 10.1016/1074-7613(95)90057-8. [DOI] [PubMed] [Google Scholar]
- 4.Suda T, Okazaki T, Naito Y, Yokota T, Arai N, Ozaki S, Nakao K, Nagata S. Expression of the Fas ligand in cells of T cell lineage. J Immunol. 1995;154:3806–3813. [PubMed] [Google Scholar]
- 5.Griffith TS, Brunner T, Fletcher S, Green DR, Ferguson TA. Fas ligand–induced apoptosis as a mechanism of immune privilege. Science (Wash DC) 1995;270:1189–1192. doi: 10.1126/science.270.5239.1189. [DOI] [PubMed] [Google Scholar]
- 6.Pan G, O'Rourke K, Chinnaiyan AM, Gentz R, Ebner R, Ni J, Dixit VM. The receptor for the cytotoxic ligand TRAIL. Science (Wash DC) 1997;276:111–113. doi: 10.1126/science.276.5309.111. [DOI] [PubMed] [Google Scholar]
- 7.Walczak H, Degli-Esposti MA, Johnson RS, Smolak PJ, Waugh JY, Boiani N, Timour MS, Gerhart MJ, Smith CA, Goodwin RG, Rauch CT. TRAIL-R2: a novel apoptosis-mediating receptor for TRAIL. EMBO (Eur Mol Biol Organ) J. 1997;16:5386–5397. doi: 10.1093/emboj/16.17.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lennon G, Auffray C, Polymeropoulos M, Soares MB. The I.M.A.G.E. Consortium: an integrated molecular analysis of genomes and their expression. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- 9.Smith CA, Gruss HJ, Davis T, Anderson D, Farrah T, Baker E, Sutherland GR, Brannan C, Copeland NG, Jenkins NA, et al. CD30 antigen, a marker for Hodgkin's lymphoma, is a receptor whose ligand defines an emerging family of cytokines with homology to TNF. Cell. 1993;73:1349–1360. doi: 10.1016/0092-8674(93)90361-s. [DOI] [PubMed] [Google Scholar]
- 10.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 545 pp.
- 11.Hsu H, Xiong J, Goeddel DV. The TNF receptor 1–associated protein TRADD signals cell death and NF-κ B activation. Cell. 1995;81:495–504. doi: 10.1016/0092-8674(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 12.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 13.Kozlosky CJ, Maraskovsky E, McGrew JT, VandenBos T, Teepe M, Lyman SD, Srinivasan S, Fletcher FA, Gayle RB, III, Cerretti DP, Beckmann MP. Ligands for the receptor tyrosine kinases hek and elk: isolation of cDNAs encoding a family of proteins. Oncogene. 1995;10:299–306. [PubMed] [Google Scholar]
- 14.Urdal DL, Call SM, Jackson JJ, Dower SK. Affinity purification and chemical analysis of the interleukin-1 receptor. J Biol Chem. 1988;263:2870–2877. [PubMed] [Google Scholar]
- 15.Wahl GM, Stern M, Stark GR. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci USA. 1979;76:3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gearing DP, King JA, Gough NM, Nicola NA. Expression cloning of a receptor for human granulocyte-macrophage colony-stimulating factor. EMBO (Eur Mol Biol Organ) J. 1989;8:3667–3676. doi: 10.1002/j.1460-2075.1989.tb08541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohler KM, Torrance DS, Smith CA, Goodwin RG, Stremler KE, Fung VP, Madani H, Widmer MB. Soluble tumor necrosis factor (TNF) receptors are effective therapeutic agents in lethal endotoxemia and function simultaneously as both TNF carriers and TNF antagonists. J Immunol. 1993;151:1548–1561. [PubMed] [Google Scholar]
- 18.Alderson MR, Tough TW, Davis-Smith T, Braddy S, Falk B, Schooley KA, Goodwin RG, Smith CA, Ramsdell F, Lynch DH. Fas ligand mediates activation-induced cell death in human T lymphocytes. J Exp Med. 1995;181:71–77. doi: 10.1084/jem.181.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chinnaiyan AM, O'Rourke K, Yu G-L, Lyons RH, Garg M, Duan DR, Xing L, Gentz R, Ni J, Dixit VM. Signal transduction by DR3, a death domain–containing receptor related to TNFR-1 and CD95. Science (Wash DC) 1996;274:990–992. doi: 10.1126/science.274.5289.990. [DOI] [PubMed] [Google Scholar]
- 20.Brojatsch J, Naughton J, Rolls MM, Zingler K, Young JAT. CAR1, a TNFR-related protein, is a cellular receptor for cytopathic avian leukosis-sarcoma viruses and mediates apoptosis. Cell. 1996;87:845–855. doi: 10.1016/s0092-8674(00)81992-3. [DOI] [PubMed] [Google Scholar]
- 21.Tartaglia LA, Ayres TM, Wong GH, Goeddel DV. A novel domain within the 55 kd TNF receptor signals cell death. Cell. 1993;74:845–853. doi: 10.1016/0092-8674(93)90464-2. [DOI] [PubMed] [Google Scholar]
- 22.Itoh N, Nagata S. A novel protein domain required for apoptosis. Mutational analysis of human Fas antigen. J Biol Chem. 1993;268:10932–10937. [PubMed] [Google Scholar]
- 23.Williams AF, Barclay AN. The immunoglobulin superfamily—domains for cell surface recognition. Annu Rev Immunol. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]
- 24.Hammelburger JW, Palfree RG, Sirlin S, Hammerling U. Demonstration of phosphatidylinositol anchors on Ly-6 molecules by specific phospholipase C digestion and gel electrophoresis in octylglucoside. Biochem Biophys Res Commun. 1987;148:1304–1311. doi: 10.1016/s0006-291x(87)80275-9. [DOI] [PubMed] [Google Scholar]
- 25.Cross GAM. Glycolipid anchoring of plasma membrane proteins. Annu Rev Cell Biol. 1990;6:1–39. doi: 10.1146/annurev.cb.06.110190.000245. [DOI] [PubMed] [Google Scholar]
- 26.McGrew JT, Rock KL. Stimulation of human Jurkat cells by monoclonal antibody crosslinking of transfected–Ly-6A.2 (TAP) molecules. Cell Immunol. 1991;137:118–126. doi: 10.1016/0008-8749(91)90062-g. [DOI] [PubMed] [Google Scholar]