Abstract
In normal mice, major histocompatibility complex (MHC) proteins are bound to many different peptides, derived from the proteins of their host. In the thymus, the diversity of this collection of MHC + peptide ligands allows thymocytes bearing many different T cell receptors (TCRs) to mature by low avidity reactions between the MHC + peptide ligands and the thymocyte TCRs. To investigate this problem, the selection of T cells specific for a well-studied combination of MHC + peptide, IEk + moth cytochrome c 88–103 (MCC), was investigated. Mice were created that expressed IEk bound to a single peptide, either a variant of MCC in which a critical TCR contact residue, 99K, was changed to A, or a variant of a mouse hemoglobin 64–76 (Hb) peptide, 72A. IEk bound to the MCC variant caused the clonal deletion of some T cells specific for the IEk + MCC ligand; nevertheless, it also positively selected many T cells that could react with this ligand. Some of the TCRs on the selected T cells were related to those on cells from normal mice and some were not. IEk bound to the Hb variant, on the other hand, did not select any T cells which could react with IEk + MCC. These results demonstrate that although positive selection is a partially degenerate event, the sequence of the peptide involved in positive selection controls the selected repertoire.
Many mysteries about the processes that drive thymocyte positive and negative selection have recently been solved. It is now known that thymocytes are selected to mature based on low avidity reactions between the α/β TCRs they bear and combinations of MHC protein and peptide in the thymus cortex (1–6). Reactions that are of too high avidity cause thymocytes to die by clonal deletion (7). Likewise, reactions that are of too low avidity also cause thymocytes to die, in this case of neglect.
Thymocytes bearing a particular TCR can be positively selected by a particular MHC protein bound to a number of different peptides (2–4, 8). Also, a particular MHC + peptide combination can positively select thymocytes bearing many different TCRs (9–13). Perhaps these degeneracies are due to the fact that many different TCR and MHC + peptide combinations can achieve the affinity and concentrations necessary to reach the avidity of reaction required for positive selection.
Positive selection is not a completely degenerate phenomenon, however. Some peptides bound to a certain MHC protein cannot select thymocytes bearing a particular TCR, even though another peptide bound to the same MHC can manage this (2–4, 14, 15). Likewise, a single MHC + peptide combination probably does not select thymocytes bearing as diverse a collection of TCRs as does the same MHC protein bound to the 2,000 or more peptides with which it is engaged in normal thymuses (9–13). The first experiments done on this subject suggested that the selecting peptide might be related in structure to the peptide that would later be able to drive a productive response by a T cell bearing a particular TCR (2–4). Later experiments indicated that this view was too extreme and that the selecting and activating peptides might not have to be obviously related (8, 15, 16). Some of these experiments were limited, however, by the fact that they were done in vitro using TCR transgenic thymocytes; thus, only a few different TCRs could be tested. Also, most of the experiments involved selection on class I MHC and few involved selection on class II.
To address this problem, we created a collection of mice in which almost all class II proteins of a particular type were occupied by a single peptide (11). The current experiments were done to find out whether the TCR repertoire selected by a single MHC + peptide combination would be related to that selected by the same MHC bound to the many mouse peptides with which it is engaged in normal mice. We chose to study a T cell specificity which has been very well examined in normal mice, that of T cells specific for the moth cytochrome c peptide 88–103 (MCC)1 bound to IEk (17–21). We found that IEk bound to a variant of MCC in which a central TCR contact residue (22), 99K, had been exchanged for an A selected T cells which could react with IEk + MCC and which bore TCRs which were closely related in sequence to those selected in normal mice by IEk bound to mouse peptides.
Materials and Methods
Construction of Transgenes.
We have previously described the production of genes coding for soluble IEβk covalently linked via a flexible linker at its NH2-terminal end to several peptides that can bind to intact IEk protein (23). Two of these constructs, coding for IEβk bound to MCC with a substitution of A for K at position 99 (99A) or for mouse hemoglobin βd 64–76 (Hb) with a substitution of A for N at position 72 (72A) were modified for injection into mice. Modifications included the removal of the thrombin sites from the flexible linkers that bound the peptide to the IEβk protein and introduction of genetic material coding for wild-type IEβk transmembrane and cytoplasmic domains (gift of Dr. H. Fischer). These genes were cloned into a plasmid (DOI) containing a promoter from IEα (gift of Drs. D. Mathis and C. Benoist, I.G.B.M.C. Strasbourg France; reference 24). cDNA coding for IEαd (gift of Dr. R. Germain, NIAID, NIH) was also introduced separately into DOI as was cDNA coding for the wild-type IEβk protein without covalently linked peptides. The α chain construct was prepared for injection by digestion with HhaI; the β chain constructs were cut with XbaI and NruI, and the fragments were purified by electrophoresis and on glass beads before injection.
Mice.
B10.BR and B10.M mice were purchased from the Jackson Laboratory (Bar Harbor, ME). All other animals were bred in the Animal Care Facility at the National Jewish Medical and Research Center (Denver, CO).
Transgenic mice were produced by injection of the IEα gene together with one of the IEkβ genes into fertilized B10.M eggs. Mice were screened for expression of the transgenes by the ability of their peripheral blood cells to stimulate T cell hybridomas specific for IEk bound to the covalently linked peptides, KC99A-4.18 for the 99A transgene and KH-8.3 for the 72A transgene or, for the wild type IEk transgene, by stimulation of KH-8.3 after addition of Hb peptide.
One founder mouse of each strain was crossed with animals lacking invariant chain, IiKO, gift of Drs. E. Bikoff and R. Germain (25). The F1 mice were crossed and F2 animals selected for further breeding based on their expression of the IE transgene, lack of expression of Ii, expression of H2f, and lack of expression of H2b. Progeny for breeding were also selected, by measuring the percentage of CD4+ Vβ3+ T cells they contained, for absence of either of the 129/J-derived mouse mammary tumor viruses that produce superantigens known to delete mouse T cells bearing Vβ3. 129/Js are, of course, the originators of the Ii deficiency. This last selection was very important for subsequent experiments because the TCR repertoire to be examined was that for IEk bound to MCC, a TCR repertoire that is rich in T cells bearing Vβ3 (17–21). Therefore, unless otherwise stated, all the transgenic mice used in this study had genotypes that were transgene+, H2f+, H2b−, IiKO and negative for superantigens which react with Vβ3.
Antibodies and Flow Cytometry.
Single cell suspensions from thymus or lymph nodes or spleen were filtered through nylon mesh (Falcon, Becton Dickinson, Franklin Lakes, NJ), washed in balanced salts solution (BSS) and stained with antibodies at 1–3 × 107 cells/ml in staining buffer (BSS, 0.1% sodium azide, 2% fetal bovine serum). Before analysis for class II MHC expression, thymus cells were enriched for large cells with a low speed spin, 500 g for 2 min, after which supernatant cells were discarded.
Cells were stained and analyzed for expression of TCR Vα, Vβ, CD4, and CD8 as previously described (26). Expression of the IEk transgenes was evaluated using biotinylated 14-4-4. The biotinylated, fluorosceinated, or phycoerythrin labeled antibodies were prepared in our laboratory or purchased from PharMingen Corp. (San Diego, CA). Streptavidin conjugated to fluoroscein, phycoerythrin, or cychrome was also purchased from PharMingen Corp. Cells were incubated for 30 min at room temperature with the primary antibody in the presence of 10% normal mouse serum and a 10% culture supernatant of the anti-Fc receptor antibody 2.4G2 (27) to block nonspecific binding. Cells were then washed three times before the addition of the secondary reagents and then incubated with these materials at 4°C for 30 min. Cells were then washed and staining profiles analyzed on a FACScan® instrument (Beckton Dickinson Inc., Mountain View, CA). At least 104 cells were analyzed for each assay. Analysis of anti-IEk staining of thymus stromal cells was accomplished by gating on the cells in the thymus with the largest forward and side scatter values as previously described (11).
Production of Chimeric Mice.
8–12-wk-old mice were irradiated with 950 rads from a 137Cs source and immediately afterwards reconstituted with fetal liver cells from 7–8 pooled day 14 fetuses, the progeny of the cross of two IEk wild-type (wt) Ii+/− mice. Therefore, the majority of donors expressed Ii and the IEkwt transgene without covalently bound peptides. Mice were maintained on acidified chlorinated water and immunized with antigen 8 wk later.
Production and Analysis of Antigen-specific T Cell Hybridomas.
Chimeric and normal mice were primed with MCC in complete Freund's adjuvant in the base of the tail. T cell hybridomas were prepared from these animals as previously described (28). In brief, lymph node cells were harvested from the draining nodes of the immunization 7 d later. The cells were incubated for 4 d with MCC and then live cells were purified and cultured for 3 more d with saturating amounts of IL-2. The live cells were then fused to an α−β− variant of BW5147, BWα−β− (28). Hybridomas were assayed for their ability to react with IEk in the presence or absence of added MCC. Presenting cells were B10.BR spleen cells which express IEk at high levels, or cells from IEwt transgenic mice, which express IEk at low levels (Fig. 1). All hybridomas used for further analysis expressed high levels of TCR and CD4.
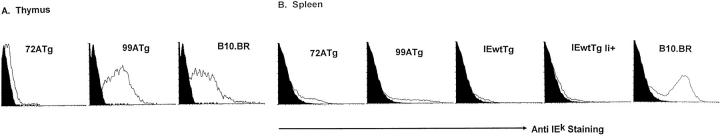
Figure 1.
(A) IEk expression on transgenic thymus cells. Large cells from the thymuses of 72ATg and 99ATg and control B10.BR and Tg− mice were prepared and stained with anti-IEk as described in Materials and Methods. Here, the staining profile of thymus cells from the various mice overlaid on the stained background control of Tg− thymus cells. (B) IEk expression on transgenic spleen cells. Spleen cells from 72ATg, 99ATg, IEwtTg, and IEwtTg Ii+ mice and from control B10.BR and Tg− animals were prepared and stained with anti-IEk as described in Materials and Methods. Here, the staining profile of the spleen cells from the various types of mice overlaid on the stained background control of Tg− spleen cells is shown. For both A and B, the results are representative of at least three independent experiments.
Results
Characterization of Transgenic Mice.
In the past, T cell TCR repertoires for several peptides, including MCC and Hb, presented by IEk, have been very well studied (17–21, 29). The T cells bearing these TCRs were all selected in the thymus by reaction between their TCRs and IEk bound to one or more of the mouse peptides with which it is normally engaged in animals. We wished to compare these well-known TCR repertoires with those that might be selected by IEk bound to a single peptide. To this end we created transgenic mice expressing IEk bound covalently to single peptides and no other IE proteins. The peptides chosen were related to those used in the previous repertoire studies and were MCC with a substitution of A for K at position 99 or Hb with a substitution of A for N at position 72. These altered peptides will be called 99A and 72A, respectively, in this paper. Their sequences are shown in Table 1.
Table 1.
Peptides Involved in these Experiments
| Name | Origin | Sequence | ||
|---|---|---|---|---|
| 72A | Mouse Hbβd 64–76 72A | GKKVITAF A EGLK | ||
| 99A | MCC 88–103 99A | ANERADLIAYL A QATK | ||
| MCC | MCC 88–103 | ANERADLIAYL K QATK |
Amino acids known to point into the IEk-binding cleft are underlined. The amino acids which have been changed from the wild-type peptide and the amino acid from which they were changed are boldfaced.
At the moment, mice that express IE transgenes and no other class II proteins cannot be constructed. It is, however, possible to breed mice that express transgenic IE proteins and no other IE molecule. This is best done by introduction of IEα and IEβ transgenes into H2f or H2q animals since these two haplotypes do not have functional IEα or IEβ genes of their own (30). Therefore, the IE transgenes described in this paper were injected into the fertilized eggs of H2f B10.M mice. As described in Materials and Methods, mice transgenic for IEk bound covalently to 99A, or for IEk bound covalently to 72A, were identified by the ability of their peripheral blood lymphocytes to stimulate T cell hybridomas specific for the two IEk + peptide combinations. Mice transgenic for IEk with no covalently attached peptide were identified by the ability of their peripheral blood cells to stimulate a T cell hybridoma, KH-8.3, after addition of its target peptide, Hb.
We have previously shown that the presence of Ii caused the removal of the covalently bound peptide from class II in cells (11). To prevent this, each of the transgenic lines was crossed with invariant chain deficient mice (IiKO) and the progeny were intercrossed to produce animals which were H2f homozygous IiKO and lacked the 129/J-derived mouse mammary tumor viruses that code for superantigens which delete thymocytes bearing Vβ3.
These crosses produced three lines of animals, all H2f and IiKO, and expressing IEwt or IEk covalently bound to 99A or 72A. The mouse lines were called IEwtTg, 99ATg, and 72ATg, respectively. H2f IiKO animals lacking any transgenes were called Tg−. A list of the mice used in this paper, and their characteristics, is shown in Table 2.
Table 2.
Mice Used in these Experiments
| Mouse | Transgenes | Expression of | ||||
|---|---|---|---|---|---|---|
| Class II | Ii | |||||
| B10.M | None | IAf | Yes | |||
| Tg− | None | IAf | No | |||
| 72ATg | IEα + IEβk-72A | IAf IEk-72A | No | |||
| 99ATg | IEα + IEβk-99A | IAf IEk-99A | No | |||
| IEwtTg | IEα + IEβk | IAf IEk | No | |||
| IEwtTg Ii+ | IEα + IEβk | IAf IEk | Yes | |||
| B10.BR | None | IAf IEk | Yes | |||
Thymus and spleen cells from these animals were stained to determine expression of the transgenes. As shown in Fig. 1, 99ATg mice expressed the product of the transgenes at high levels on the large cells in its thymus. They also expressed high levels of the product on their spleen cells, although not as many spleen cells in 99ATg mice were IEk+ as in control B10.BR animals. Levels approximated those on the equivalent B10.BR H2k cells. 72ATg spleen cells stained less well, and expression of this transgene was very low in the thymus. Expression of the IEwt transgene was very low on spleen cells, regardless of whether or not the mice expressed Ii. The promoter used in these experiments, although derived from the IEα gene itself, is known to drive variable levels of expression of class II proteins in transgenics (24). These results are probably manifestations of this phenomenon.
To establish the distribution of IEk-bearing cells in the thymuses of these mice, frozen sections of their thymuses were stained with biotinylated anti-IEk and streptavidin-coupled horseradish peroxidase. Biotinylated anti-IAb or anti-Kk served as controls. As expected, both the cortex and medulla of B10.BR thymuses stained brightly with the anti-IEk reagent. No staining above background was observed with the Tg− or 72ATg thymuses, indicating that expression of the transgene was very low on both cortical and medullary epithelium in the latter mouse. Anti-IEk stained the cortical epithelium of 99ATg animals almost as brightly as it stained that of B10.BRs, with intense reticular patterning. By contrast there was no staining above background in the medulla of the 99ATg animals. These results demonstrated that the 99A transgene was well expressed in the cells responsible for positive selection of class II– restricted T cells, thymus cortical epithelium, but poorly, if at all, expressed on medullary thymus epithelium. T cell tolerance to the transgene in 99ATg mice must be established by contact with thymus cortical epithelium or bone marrow–derived cells.
Staining experiments of this type did not reveal whether or not the IE proteins in the 99ATg and 72ATg mice were still bound to the covalent peptides with which they had been engaged genetically. To find out whether displacement with other peptides had occurred, cells from the transgenic mice were tested for their ability to present exogenously added peptides to T cells specific for IEk bound to the exogenous peptide. Thymus and spleen cells from the 99ATg mice presented exogenous peptides very poorly (Fig. 2). By contrast, thymus and spleen cells from the IEwtTg animals presented peptide fairly well, and thymus and spleen cells from B10.BR animals had very good activity.
Figure 2.
Thymus and spleen cells from 72ATg and 99ATg mice present exogenous peptides poorly. Thymus and spleen cells were isolated from various types of mice as described in Materials and Methods. They were cultured with different concentrations of Hb and a T cell hybridoma, KH-8.3, specific for IEk + Hb. The ability of the cells to present the exogenously added peptide was assayed by measurement of production of IL-2 by the T cell hybridoma 24 h later. The results are representative of at least three independent experiments.
Spleen and thymus cells from 72ATg mice presented exogenous antigen even less well than those of 99ATg animals (data not shown), indicating that the 72A peptide, like the 99A peptide, was not displaced from the IEk molecule in Ii− animals.
These experiments demonstrated that virtually all of the IEk molecules on the surfaces of 99ATg and 72ATg cells were occupied by the 99A and 72A peptides, respectively. Even though the IEwtTg cells bore only small amounts of IEk on their surfaces as detected by staining, they still had more IE molecules on their surfaces, which could present exogenous peptides, than 99ATg cells did.
Cells Bearing Either of Two TCRs Specific for IEk + MCC Are Deleted in 99ATgs.
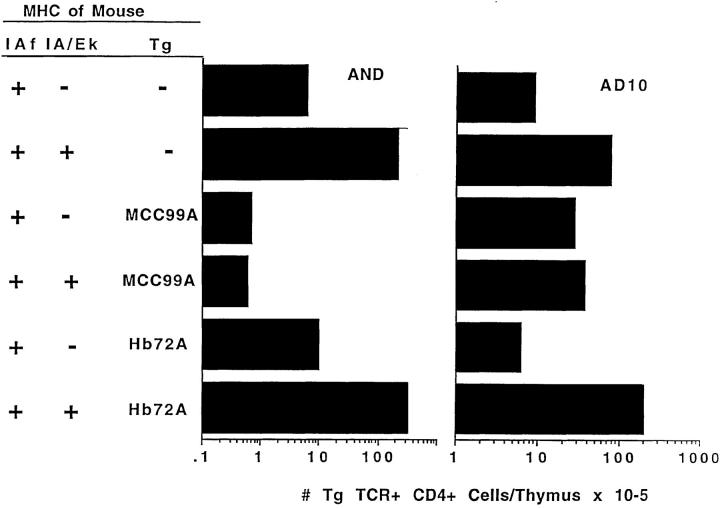
A number of mice have been produced that express TCR transgenes specific for IEk + MCC. To find out whether the 99ATg or 72ATg mice could positively select thymocytes bearing these TCRs, mice expressing two of these TCRs, AD10 and AND (31), were crossed with the 99ATg and 72ATg animals. Progeny (which were still Ii+) were tested for expression of the TCR and IE transgenes and for expression of H2k and H2f. They were then tested for selection of the thymocytes bearing the transgenic TCRs by estimation of the numbers of CD4+ mature thymocytes bearing high levels of the relevant Vα and Vβ regions, Vα11 and Vβ3.
The results in Fig. 3 show that CD4+ thymocytes bearing either of the two transgenes were selected with low efficiency in nontransgenic H2f homozygous mice. This was not because H2f proteins caused deletion of thymocytes bearing these TCRs since CD4+ thymocytes bearing either the AND or AD10 TCRs appeared in large numbers in H2fxk mice, positively selected by the wild-type IEk protein + some unknown mouse peptide.
Figure 3.
IEk-99A causes the clonal deletion of T cells bearing canonical IEk + MCC–reactive TCRs. Animals expressing the AND or AD10 TCR transgenes were crossed with 72ATg or 99ATg mice, and the animals were intercrossed such that some were H2fxk and others were H2f homozygous. Thymocytes from these animals were stained with anti-Vα11, anti-Vβ3, and CD4. The counts are those of cells/thymus-bearing CD4 and high levels of Vα11 and Vβ3. The results shown are the averages of between one and three mice of each type.
Introduction of the 99ATg did cause deletion, however, as witnessed by the fact that mature CD4+ T cells bearing the transgenic TCRs were lower in number in H2fxk mice that expressed the 99ATg than they were in animals of the same MHC type that did not express the transgene. T cells bearing the AD10 TCR have previously been reported to be antagonized and deleted by very high concentrations of MCC99A bound to IEk (32), so this result was not surprising. Thymocytes bearing the AND TCR were more efficiently deleted in 99ATg mice than were thymocytes bearing the AD10 TCR. The AND and AD10 TCRs differ by a single amino acid. The AND TCR has an alanine residue in its Vα CDR3 region, four amino acids COOH terminal to the invariant cysteine, whereas the AD10 TCR has a threonine at this position (28). Apparently this change causes the two TCRs to have slightly different affinities for IEk bound to MCC 99A, resulting in different efficiencies of deletion of thymocytes bearing the two different TCRs.
These experiments were done with mice that still expressed Ii and hence the MCC99A peptide was probably displaced from some of the 99ATg class II proteins in the mice (11). Thus the fact that expression of the 99ATg in H2f homozygous mice increased the numbers of mature CD4+ thymocytes bearing the AD10 TCR could not be properly evaluated. It is possible that this indicated that the 99ATg could both positively and negatively select thymocytes bearing this TCR, i.e., the affinity of AD10 for IEk bound to 99A was in the marginal zone which has been noted before for other TCR, MHC + peptide combinations. Alternatively, positive selection in this case might have been driven by interactions between the AD10 TCR and 99ATg proteins from which the MCC99A peptide had been displaced. Future experiments will resolve this.
Expression of the 72ATg did not affect the fate of the thymocytes in any animal, suggesting either that this protein was not expressed at high enough levels to be effective and/or that the AND and AD10 TCRs had no productive affinity for this IEk–peptide combination.
These experiments showed that some TCRs specific for IEk + MCC have a high enough affinity for IEk-99A to cause thymocytes bearing them to be deleted in 99ATg mice. Hence, the extent of the TCR repertoire for IEk + MCC in 99ATg mice is limited, not only by failure to positively select, but also by clonal deletion of the relevant cells.
The 99ATg Selects T Cells that Can React with IEk + MCC.
IiKO mice that express class II proteins covalently bound to peptides that can occupy their grooves cannot be primed with foreign proteins or peptides (data not shown). Therefore, the ability of T cells from the class II–peptide transgenic mice to react with foreign proteins or peptides could not be evaluated in intact mice. To circumvent this problem, the transgenic mice were irradiated and reconstituted with fetal liver from H2f animals transgenic for IEkwtTg genes and expressing Ii. In the chimeric animals that were thus created, T cells were positively selected on host thymus cortical epithelial cells, and could be primed with foreign peptides presented on the IEkwt expressing fetal liver derived cells. The T cells must also have been tolerant to the low levels of IEk bound to mouse peptides expressed on IEkwt Ii+ and Ii− cells.
Three types of chimeric mice were produced in which the hosts were 99ATg, 72ATg, or Tg−. After T cells had matured in the chimeras, they were primed with MCC peptide and the primed T cells were harvested and converted into T cell hybridomas as described in Materials and Methods. As additional controls, hybridomas were also produced in the same fashion from MCC primed, nonchimeric H2f mice with or without Ii expression, and from nonchimeric IEwtTg mice with or without Ii expression.
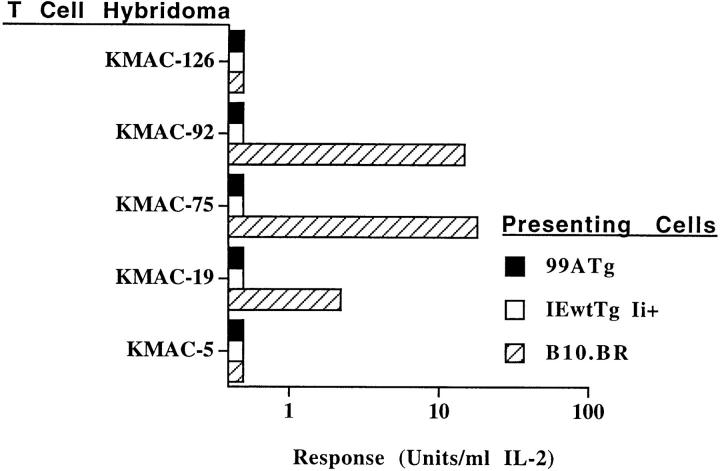
Hybridomas were tested for their ability to respond to B10.BR, IEwtTg Ii+, or 99ATg spleen cells. Examples of the reactivities of some of the hybridomas from 99ATg chimeric mice are shown in Fig. 4. None of the hybridomas responded to IEwtTg Ii+ or 99ATg cells. Thus the hybridomas were tolerant to class II proteins at the levels they were expressed in the chimeric mice. Several of the hybridomas did, however, respond to B10.BR spleen cells. Anti-IE addition to the stimulation cultures showed that this was due to reaction with IEk (data not shown). The parent T cells for these hybridomas must have been tolerant to IEk bound to the mouse peptides with which it is normally engaged at the very low levels at which it was expressed in the chimeric animals (Fig. 1), but were not tolerant to the same combination of IEk and peptides at the high levels at which it is expressed on B10.BR cells. These hybridomas were probably another example of the fact that selection on a particular MHC–peptide combination causes cells to mature, which are likely to react with that same MHC protein bound to other peptides, particularly when expressed at high levels (11–13).
Figure 4.
Some IEk + MCC–reactive T cells from 99ATg mice react with IEk + normal mouse peptides. T cell hybridomas specific for IEk + MCC were prepared from 99ATg animals as described in Materials and Methods and were assayed for their ability to respond to spleen cells from various types of mice. The results shown are typical of two independent experiments.
The hybridomas were also tested for their ability to respond to different concentrations of MCC presented by B10.BR cells (Fig. 5). Many of the hybridomas responded well to this antigen and were as sensitive to low doses of the peptide as a typical hybridoma prepared from primed T cells from a normal mouse. Some of the hybridomas, for example KMAC-92, which responded to B10.BR cells in the absence of peptide responded better when increasing doses of MCC were added to the cultures.
Figure 5.
IEk + MCC–specific T cells from 99ATg mice are as reactive with their ligand as T cells from normal mice are. The response of 99ATg T cell hybridomas to IEk + MCC was compared with that of a representative T cell hybridoma, 5KC-73.8/S1.6, from normal mice. The results shown are typical of two independent experiments.
The numbers of hybridomas obtained from two mice of each type are shown in Table 3. When T cells cannot react with the immunizing MHC + peptide combination, immunized lymph nodes give rise to few T cell blasts, and few T cell hybridomas are produced (our unpublished observations). This was manifest in this experiment by the fact few hybridomas were obtained from fusion of MCC-immunized T cells from some of the mice that did not express an IEk protein and Ii in their thymus stromal cells. Analysis of the numbers of hybridomas that could react with IEk + MCC supported this conclusion. No hybridomas with this specificity were obtained from mice which did not express IEk on their thymus stromal cells. This included the Tg− mice that contained fetal liver–derived cells expressing IEwt and Ii. Hence, as previously reported, the MHC class II proteins on the fetal liver–derived cells in these chimeras could not participate in positive selection (33, 34).
Table 3.
IEk-99A Selects T Cells that Can React with IEk + MCC
| Mouse | IE on | No. Hybridomas | ||||||
|---|---|---|---|---|---|---|---|---|
| Thymus stroma | Fetal liver– derived cells | Tested | IEk + MCC Rx | |||||
| IEwt Ii+/−→ Tg− | None | IEwt | 6 | 0 | ||||
| IEwt Ii+/−→ 72ATg | 72A | IEwt | 92 | 0 | ||||
| IEwt Ii+/−→ 99ATg | 99A | IEwt | 116 | 21* | ||||
| B10.M | None | None | 91 | 0 | ||||
| Tg− | None | None | 6 | 0 | ||||
| IEwt Ii+ | IEwt Ii+ | IEwt Ii+ | 126 | 66 | ||||
| IEwt | IEwt | IEwt | 2 | 0 | ||||
Eight of these hybridomas had identical TCR α and β chain sequences.
No hybridomas that could react with IEk + MCC were obtained from IEwtTg mice. Thus, either the peptides that occupied the grooves of IEk in IiKO animals did not allow selection of IEk + MCC–reactive T cells, or the levels of IEk in these animals were too low to allow selection of such cells (14). Also, no T cell hybridomas that could react with IEk + MCC were obtained from 72ATg chimeric mice. This might have been due either to an inability of IEk bound to the 72A peptide to select such cells, or it may have been due to the very low levels of expression of this combination in the thymuses of the transgenic animals. Some of the hybridomas that were produced in the fusion of T cells from 72ATg chimeric mice were specific for IAf bound to MCC (data not shown). It is interesting that T cells with this specificity were not produced from fusions of T cells from mice that lacked an IEk transgene. Future experiments will examine this phenomenon in more detail.
A number of hybridomas specific for IEk + MCC were obtained from 99ATg chimeric mice. These hybridomas could not have been positively selected on the fetal liver– derived cells in the chimeras since similar hybridomas did not appear in the other chimeras prepared with the same fetal liver. The results with primed B10.M or Tg− mice demonstrate that these hybridomas could not have been selected on IAf. Finally, these hybridomas were not selected on the small amounts of IEk on the 99ATg thymus epithelial cells from which the 99A peptide had been displaced since there were fewer of these proteins in 99ATg thymuses than there were in IEwtTg thymuses (see data in Fig. 2) and mice of the latter type failed to produce T cells with this specificity. These controls proved that the T cell parents of the IEk + MCC–reactive hybridomas obtained from the chimeric 99ATg mice must have been positively selected on the 99ATg.
IEk Bound to MCC99A Selects IEk + MCC–reactive T Cells which Have TCRs Similar to those Selected by IEk in Normal Mice.
The IEk + MCC–reactive T cell hybridomas listed in Table 3 were stained with anti-Vα and anti-Vβ antibodies. Table 4 lists the combinations found on these cells. We were surprised to find that only about a quarter of the hybridomas from IEwt Ii+ animals bore the pairing of Vα11 Vβ3, which is usually associated with recognition of IEk + MCC. Even though an overall analysis of this type for responses to IEk + MCC 88–103 (in contrast to IEk + pigeon cytochrome C 88–104) is not available in the literature, the impression is that almost all T cells in normal mice specific for IEk + MCC bear Vα11 and Vβ3. Perhaps the difference between the results in the literature and those reported here is due to the low levels of IEk on the selecting thymus epithelium in the IEwt Ii+ mice used in our experiments.
Table 4.
Vα and Vβ Use by IEk + MCC–reactive T Cells
| Selecting IEk | Percentage (No.) of IEk + MCC–reactive T cells using: | |||||||
|---|---|---|---|---|---|---|---|---|
| Vα11/Vβ3 | Vα11/Vβ8 | Vα11/Other Vβ | Other Vα/Vβ | |||||
| IEwt Ii+ | 26 (7) | 11 (3) | 26 (7) | 37 (9) | ||||
| 99A | 45 (9*) | 10 (2) | 40 (8) | 5 (1) | ||||
T cell hybridomas were analyzed for Vα and Vβ expression as described in Materials and Methods.
Includes eight with identical TCRs.
Clearly, T cells selected on IEk bound to 99A could bear the same Vα–Vβ combinations, including Vα11 and Vβ3, as those selected on IEk bound to many mouse peptides (17–21).
The receptors from the 99ATg chimera derived T cells bearing Vα11 and Vβ3 or Vβ8.3 were sequenced (Table 5 and 6). The TCR sequences from eight of the nine hybridomas bearing Vα11 and Vβ3 were identical. The T cell parents of these cells must have come from the same expanded clone. This result suggested that there were relatively few IEk + MCC–specific T cells in the 99ATg chimeric mice since hybrids derived from a single clone are not usually found in fusions to antigen specific T cells from normal mice (P. Marrack, personal observation).
Table 5.
TCR α Chain Sequences
| T cell | Selecting | CDR3 | Jα | |||||
|---|---|---|---|---|---|---|---|---|
| IE | Vα | |||||||
| Consensus | IEk | 11.1 | CAAEASS TNKLVFG | – | ||||
| 2B4 | IEk | 11.2 | CAALRVTGGNNKLTFG | 47 | ||||
| KMAC-92* | 99A | 11.2 | CAAGASS FNKLVFG | 42 | ||||
| KMAC-19 | 99A | 11.2 | CAAEASG SWQLIFG | 17 | ||||
| KMAC-75 | 99A | 11.1 | CAAEASN TNKVVFG | 27 | ||||
| KMAC-126 | 99A | 11.1 | CAAEAAN NNAPRFG | 27 | ||||
Sequences of IEk + MCC–reactive TCRs from 99ATg mice. The TCR α chain sequences of IEk + MCC–reactive T cells from 99ATg mice are shown together with consensus sequences derived from the literature (15, 17–21, 34).
Identical sequences were found in seven other T cell hybridomas from the same fusion.
Consensus sequences for the CDR3 regions of TCRs specific for IEk + MCC and selected in normal IEk+ mice were derived from the literature (17–21, 35; Tables 5 and 6). Most of the α chain CDR3s of these TCRs used Vα11.1, had an E 3 amino acids COOH terminus to the conserved C, and had CDR3s which were otherwise rich in compact amino acids such as A and S. A notable exception to these rules is the α chain of the well-known T cell hybridoma, 2B4, which uses Vα11.2 and has a lengthier CDR3 (18, 20). The literature suggests two different consensus sequences for the CDRs of the TCR β chains. Both are shown in Tables 5 and 6. Consensus sequence 1 was most commonly reported in the past (17–21), with consensus sequence 2 derived from that of 2B4 and a collection of sequences from mice expressing IEk and heterozygous for Ii expression (15).
Table 6.
TCR β Chain Sequences
| T cell | Selecting | CDR3 | Jα | |||||
|---|---|---|---|---|---|---|---|---|
| IE | Vβ | |||||||
| Consensus 1 | IEk | 3 | CASSLNNANSDYTFG | 1.2 | ||||
| Consensus 2 | IEk | 3 | CASSLNWGQDTQYFG | 2.5 | ||||
| KMAC-92* | 99A | 3 | CASSLNWGGDTQYFG | 2.5 | ||||
| KMAC-19 | 99A | 3 | CASSLNWGGGEQYFG | 2.6 | ||||
| KMAC-75 | 99A | 8.3 | CASSPGTGASDYTFG | 1.2 | ||||
| KMAC-126 | 99A | 8.3 | CASSDAQNTGQLYFG | 2.2 | ||||
Sequences of IEk + MCC–reactive TCRs from 99ATg mice. The TCR-β chain sequences of IEk + MCC–reactive T cells from 99ATg mice are shown together with consensus sequences derived from the literature (15, 17–21, 34).
Identical sequences were found in seven other T cell hybridomas from the same fusion.
The CDR3 sequences of the 99ATg T cells are also shown in Tables 5 and 6. Like 2B4 and unlike the norm, the two Vβ3-bearing T cell hybridomas bore Vα11.2. Vα11.1 in combination with Vβ3 may cause a higher reactivity for IEk + MCC99A than Vα11.2 does since most of the Vα11.1-bearing, IEk + MCC reactive T cells from normal mice are activated or deleted by IEk + MCC99A, but 2B4 is not (21, 32, 35). Consequently, the Vα11.1- and Vβ3-bearing T cells may have been deleted in 99ATg mice, leaving as IEk + MCC–reactive T cells only those which bore, with Vβ3, Vα11.2. The two Vα11 family members differ by only five amino acids (36). Only one of these changes, position 72 in the CDR4 loop which is an R in Vα11.1 and S in Vα11.2, maps to any region that might contact MHC and peptide (37, 38). Perhaps the shorter, uncharged side chain of the S in Vα11.2 and/or the presence of bulky amino acids in the TCR α and β chains reduced contact between the TCR and IEk + 99A, and thus allowed IEk + MCC–reactive T cells bearing Vα11.2 to be positively selected and not deleted by the 99ATg.
Other than this, the α chain CDR3 sequences of the Vβ3- and Vβ8.3-bearing IEk + MCC–reactive TCRs obtained from 99ATg mice were similar in sequence and length to the consensus with the exception that one lacked the acidic amino acid usually found three residues COOH terminus to the conserved cysteine, and was thus similar in sequence to the CDR3 of a TCR reported to cross-react between MCC and MCC99E (39), and two contained a bulky aromatic amino acid.
The CDR3 sequences of the two Vβ3-bearing TCRs obtained from 99ATg mice were very similar in sequence to that of consensus 2, the group which includes the 2B4 TCR (Tables 5 and 6). As discussed above, this similarity may have been driven by the requirement for a low reactivity with IEk bound to MCC99A.
The results in Tables 5 and 6 did suggest one other idea: most of the TCR α and β chains which could contribute to TCRs that could react with IEk + MCC might now be known. Thus, several of the sequences from the 99ATg mice were identical to sequences that had already been described. The α chain of KMAC-75 was identical to that of a previously described hybridoma, 5C.C7 (20). The α chain CDR3 sequence of KMAC-19 was identical to that previously reported for a TCR selected on wild-type IEk in Ii+/− mice (15). This previous publication did not mention whether the TCR in question bore Vα11.1 or Vα11.2.2. Likewise, the β chain sequence of KMAC-19 was identical to that previously described for a T cell selected by IEk bound to MCC (15).
Discussion
The complete repertoire of T cells selected on the 99ATg and specific for IEk + MCC was not assessed in this paper since the T cells we isolated came from mice that contained fetal liver cells bearing IEk engaged by normal mouse peptides, albeit at low levels. The T cells in the chimeras were thus positively selected by IEk bound to a single peptide, and were tolerant to IEk bound to many normal mouse peptides at low levels, but not necessarily at high levels. This was manifested by the fact that none of the T cell hybridomas obtained from the chimeras reacted with IEk on IEwtTg Ii+ cells, although some of the hybridomas did react with IEk on B10.BR cells. Nevertheless, it is likely that expression of even the low levels of IEk found in the chimeras did cause deletion of some IEk + mouse peptide– reactive T cells, and thus reduce the total diversity of TCRs found in the chimeras. Moreover, all the mice contained an additional class II protein, IAf, and it is likely that at least some selected T cells were deleted by recognition of this protein, bound to its array of mouse peptides (11).
Assessment of the ability of IEk bound to 99A to select IEk + MCC–reactive T cells was also limited by the fact that some TCRs specific for this latter ligand also react with high affinity with the 99A selecting ligand (32). This was demonstrated in this paper by the deletion in 99ATg mice of T cells bearing the well known IEk + MCC reactive TCRs, AND or AD10 (20, 31, 32).
Overall, therefore, the repertoire for IEk + MCC in the transgenic chimeras was limited to some extent by negative selection on two types of ligands, low levels of wild-type IEk or IAf bound to mouse peptides, and IEk-99A.
In spite of these two limitations, it was clear that T cells reactive with IEk + MCC could be positively selected by IEk-99A, a closely related ligand. A previous publication showed that MCC with substitutions of E for K at position 99, or E for T at position 102, and indeed even MCC itself could positively select T cells reactive with IEk + MCC (15). Assuming that the affinity/avidity hypothesis of positive selection is correct, these results show that TCRs with high affinity for IEk + MCC can have an appropriate selecting affinity for IEk + any of these MCC variants or even for IEk + MCC itself at the concentrations that these ligands were expressed in the thymus.
Selection is peptide specific. T cells specific for IEk + MCC cannot be selected by IEk bound to any peptide. Such T cells were not positively selected in animals expressing IEk but not Ii (reference 14 and this paper). Apparently none of the peptides that were bound to IEk in IiKO animals could participate in selecting T cells that can react with IEk + MCC. Also, as shown in this paper, IEk + MCC–reactive T cells were not obtained from animals expressing IEk bound to 72A. Likewise, T cells with this specificity were not selected by IEk bound to a lambda repressor peptide (15).
Our result with the 72A peptide is quite surprising since it has previously been shown that unaltered Hb 67–76 could select MCC-reactive T cells (15). The 72A peptide differed from that previously used by the conservative change of A for N at the central, T cell contact residue of the peptide, and by a longer NH2-terminal tail protruding from the IEk-binding groove. MCC itself has a basic amino acid, K, at the equivalent position. Perhaps TCRs that will react with MCC are selected better by Hb with the hydrophilic amino acid N at this position than Hb with the shorter, more hydrophobic A. 99A, however, also has an A at the equivalent position and, as shown in this paper, 99A selected MCC-reactive TCRs well.
Alternatively or as well, perhaps the difference was due to the level of expression of the IEk–peptide conjugates. The IEk-99A transgenic protein was expressed at much higher levels per cell than the IEk-72A conjugate. This may have caused positive selection of thymocytes bearing TCRs with very low affinity for the 99A peptide, whereas low affinity positive selection may not have been possible in the 72ATg mice. Recent measurement of the affinity of a TCR on one of the IEk + MCC–specific T cells described in this paper showed that it did indeed have extremely low affinity for IEk-99A.
Finally, the difference may be due to the fact that the T cells used in this paper were tolerant to IEwt and, less likely, to IAf. Tolerance to IEk plus all the peptides to which it is bound in normal mice may have deleted all the T cells selected on IEk-72A that could react with IEk + MCC. We do not think this is the explanation, however, because preliminary experiments with 72ATg T cells that are not tolerant to IEwt suggest that these T cells are also unable to react with IEk + MCC.
Some of the TCRs specific for IEk + MCC that were positively selected in the 99ATg mice were quite similar to those found in normal animals. Again, this was quite surprising since many T cells with this specificity that bore TCRs like those in wild-type animals must have been deleted in the 99ATg animals, as exemplified by the disappearance of T cells bearing the transgenic TCRs, AD10 and AND, in these mice. It is worth noting, however, that the TCR repertoire of the 99ATg animals was not completely included within that of normal animals since two different TCRs which used Vβ8.3 were obtained from the 99ATgs, and IEk + MCC–reactive T cells bearing this Vβ have not previously been reported from normal mice, although one T cell with this specificity and this Vβ was reported from mice heterozygous for Ii expression (15).
Finally, although IEk + MCC–reactive T cells bearing identical TCRs have yet to be independently isolated, these data support the notion that the complete TCR repertoire for this MHC + peptide combination is nearing saturation. Several of the chains of the TCRs isolated from our transgenic mice were identical in sequence to chains that have been described previously. Presumably the numbers of TCR α and β chains that can contribute to TCRs that can react with IEk + MCC are limited, and this fact is reflected in the fact that particular sequences for α or β are being reported with increased frequency.
Acknowledgments
The authors thank Dean Becker for his help with production and breeding of transgenic mice and Ella Kushnir, Patricia Mount, and Dr. Gail Ackermann for support in the Animal Care Facility at National Jewish Medical and Research Center, Denver, CO. They also thank Drs. Stephen Hedrick, Elizabeth Bikoff, Ron Germain, Laurie Glimcher, Diane Mathis, and Christophe Benoist for their generous gifts of transgenic and knockout mice, DNA constructs, and antibodies.
This work was supported by United States Public Health Service grants AI-17134, AI-18785, AI-22295, and AI-29544.
Footnotes
Abbreviations used in this paper: Hb, mouse hemoglobin 64-76; Ii, invariant chain; MCC, moth cytochrome c peptide 88–103; wt, wild-type.
References
- 1.Lo D, Ron Y, Sprent J. Induction of MHC- restricted specificity and tolerance in the thymus. Immunol Res. 1986;5:221–232. doi: 10.1007/BF02919203. [DOI] [PubMed] [Google Scholar]
- 2.Hogquist KA, Jameson CS, Heath WR, Howard JL, Bevan MJ, Carbone FR. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 3.Ashton-Rickardt PG, Bandiera A, Delaney JR, Van Kaer L, Pircher HP, Zinkernagel RM, Tonegawa S. Evidence for a differential avidity model of T cell selection in the thymus. Cell. 1994;76:651–663. doi: 10.1016/0092-8674(94)90505-3. [DOI] [PubMed] [Google Scholar]
- 4.Sebzda E, Wallace VA, Mayer J, Yeung RSM, Mak TW, Ohashi PS. Positive and negative thymocyte selection induced by different concentrations of a single peptide. Science (Wash DC) 1994;263:1615–1618. doi: 10.1126/science.8128249. [DOI] [PubMed] [Google Scholar]
- 5.Alam SM, Travers PJ, Wung JL, Nasholds W, Redpath S, Jameson SC, Gascoigne NR. T-cell receptor affinity and thymocyte positive selection. Nature (Lond) 1996;381:616–620. doi: 10.1038/381616a0. [DOI] [PubMed] [Google Scholar]
- 6.Hogquist KA, Tomlinson AJ, Kieper WC, McGargill MA, Hart MC, Naylor S, Jameson SC. Identification of a naturally occurring ligand for thymic positive selection. Immunity. 1997;6:389–399. doi: 10.1016/s1074-7613(00)80282-4. [DOI] [PubMed] [Google Scholar]
- 7.Kappler JW, Roehm N, Marrack P. T cell tolerance by clonal elimination in the thymus. Cell. 1987;49:273–280. doi: 10.1016/0092-8674(87)90568-x. [DOI] [PubMed] [Google Scholar]
- 8.Pawlowski TJ, Singleton MD, Loh DY, Berg R, Staerz UD. Permissive recognition during positive selection. Eur J Immunol. 1996;26:851–857. doi: 10.1002/eji.1830260419. [DOI] [PubMed] [Google Scholar]
- 9.Ashton-Rickardt PG, Van Kaer L, Schumacher TNM, Ploegh HL, Tonegawa S. Peptide contributes to the specificity of positive selection of CD8+ T cells in the thymus. Cell. 1993;73:1041–1049. doi: 10.1016/0092-8674(93)90281-t. [DOI] [PubMed] [Google Scholar]
- 10.Hogquist KA, Gavin MA, Bevan MJ. Positive selection of CD8+ T cells induced by major histocompatibility complex binding peptides in fetal thymus organ culture. J Exp Med. 1993;177:1469–1473. doi: 10.1084/jem.177.5.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ignatowicz L, Kappler J, Marrack P. The repertoire of T cells selected by a single MHC/peptide ligand. Cell. 1996;84:521–529. doi: 10.1016/s0092-8674(00)81028-4. [DOI] [PubMed] [Google Scholar]
- 12.Miyazaki T, Wolf P, Tourne S, Waltzinger C, Dierich A, Barois N, Ploegh H, Benoist C, Mathis D. Mice lacking H2-M complexes, enigmatic elements of the MHC class II peptide-loading pathway. Cell. 1996;84:531–541. doi: 10.1016/s0092-8674(00)81029-6. [DOI] [PubMed] [Google Scholar]
- 13.Martin WD, Hicks GG, Mendiratta SK, Leva HI, Ruley HE, Van Kaer L. H2-M mutant mice are defective in the peptide loading of class II molecules, antigen presentation and T cell repertoire selection. Cell. 1996;84:543–550. doi: 10.1016/s0092-8674(00)81030-2. [DOI] [PubMed] [Google Scholar]
- 14.Tourne S, Nakano N, Benoist C, Mathis D. The influence of invariant chain on the positive selection of single T cell receptor specificities. Eur J Immunol. 1995;25:1851–1856. doi: 10.1002/eji.1830250709. [DOI] [PubMed] [Google Scholar]
- 15.Nakano N, Rooke R, Benoist C, Mathis D. Positive selection of T cells induced by viral delivery of neopeptides to the thymus. Science (Wash DC) 1997;275:678–683. doi: 10.1126/science.275.5300.678. [DOI] [PubMed] [Google Scholar]
- 16.Ignatowicz L, Rees W, Pacholczyk R, Ignatowicz H, Kushnir E, Kappler J, Marrack P. Peptides involved in positive selection and activation of T cells are not necessarily related in sequence. Immunity. 1997;7:179–186. doi: 10.1016/s1074-7613(00)80521-x. [DOI] [PubMed] [Google Scholar]
- 17.Fink PJ, Matis LA, McElligott DL, Bookman M, Hedrick SM. Correlations between T-cell specificity and the structure of the antigen receptor. Nature (Lond) 1986;321:219–226. doi: 10.1038/321219a0. [DOI] [PubMed] [Google Scholar]
- 18.Winoto A, Urban JL, Lan NC, Goverman J, Hood L, Hansburg D. Predominant use of a Vα gene segment in mouse T-cell receptors for cytochrome c. Nature (Lond) 1986;324:679–682. doi: 10.1038/324679a0. [DOI] [PubMed] [Google Scholar]
- 19.Sorger SB, Hedrick SM, Fink PJ, Bookman MA, Matis LA. Generation of diversity in T cell receptor repertoire specific for pigeon cytochrome c. J Exp Med. 1987;165:279–301. doi: 10.1084/jem.165.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hedrick SM, Engel I, McElligott DL, Fink PJ, Hsu M-L, Hansburg D, Matis LA. Selection of amino acid sequences in the beta chain of the T cell receptor. Science (Wash DC) 1988;235:1541–1544. doi: 10.1126/science.2832942. [DOI] [PubMed] [Google Scholar]
- 21.Sorger SB, Paterson Y, Fink PJ, Hedrick SM. T cell receptor junctional regions and the MHC molecule affect the recognition of antigenic peptides by T cell clones. J Immunol. 1990;144:1127–1135. [PubMed] [Google Scholar]
- 22.Fremont DH, Hendrickson W, Marrack P, Kappler J. Crystal structures of an MHC class II molecule with covalently bound single peptides. Science (Wash DC) 1996;272:1001–1004. doi: 10.1126/science.272.5264.1001. [DOI] [PubMed] [Google Scholar]
- 23.Kozono H, Parker D, White J, Marrack P, Kappler J. Multiple binding sites for bacterial superantigens on soluble class II MHC molecules. Immunity. 1995;3:187–196. doi: 10.1016/1074-7613(95)90088-8. [DOI] [PubMed] [Google Scholar]
- 24.Kouskoff V, Fehling HJ, Lemeur M, Benoist C, Mathis D. A vector driving the expression of foreign cDNAs in the MHC class II positive cells of transgenic mice. J Immunol Methods. 1993;166:287–291. doi: 10.1016/0022-1759(93)90370-m. [DOI] [PubMed] [Google Scholar]
- 25.Bikoff EK, Huang LY, Episkopou V, van Meerwijk J, Germain RN, Robertson EJ. Defective major histocompatibility complex class II assembly, transport, peptide acquisition and CD4+T cell selection in mice lacking invariant chain expression. J Exp Med. 1993;177:1699–1712. doi: 10.1084/jem.177.6.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scherer MT, Ignatowicz L, Pullen A, Kappler J, Marrack P. The use of mammary tumor virus (Mtv)-negative and single-Mtvmice to evaluate the effects of endogenous viral superantigens on the T cell repertoire. J Exp Med. 1995;182:1493–1504. doi: 10.1084/jem.182.5.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Unkeless JC. Characterization of a monoclonal antibody directed against mouse macrophage and lymphocyte Fc receptors. J Exp Med. 1979;150:580–596. doi: 10.1084/jem.150.3.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White J, Blackman M, Bill J, Kappler J, Marrack P, Gold DP, Born W. Two better cell lines for making hybridomas expressing specific T cell receptors. J Immunol. 1989;143:1822–1825. [PubMed] [Google Scholar]
- 29.Hsu BL, Donermeyer DL, Allen PM. TCR recognition of the Hb (64-76)/IEk determinant: single conservative amino acid changes in the complementarity-determining region 3 dramatically alter antigen fine specificity. J Immunol. 1996;157:2291–2298. [PubMed] [Google Scholar]
- 30.Mathis D, Benoist C, Williams V, Kantor M, McDevitt H. Several mechanisms can account for defective Eα gene expression in different mouse haplotypes. Proc Natl Acad Sci USA. 1983;80:273–277. doi: 10.1073/pnas.80.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaye J, Vasquez NJ, Hedrick SM. Involvement of the same region of the T cell antigen receptor in thymic selection and foreign peptide recognition. J Immunol. 1992;148:3342–3353. [PubMed] [Google Scholar]
- 32.Page DM, Alexander J, Snoke K, Appella E, Sette A, Hedrick SM, Grey HM. Negative selection of CD4+ CD8+ thymocytes by T-cell receptor peptide antagonists. Proc Natl Acad Sci USA. 1994;91:4057–4061. doi: 10.1073/pnas.91.9.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sprent J, Gao EK, Kanagawa O, Webb SR. T-cell selection in the thymus. Princess Takamatsu Symp. 1988;19:127–136. [PubMed] [Google Scholar]
- 34.Markowitz JS, Auchinscloss H, Jr, Grusby MJ, Glimcher LH. Class II–positive hematopoetic cells cannot mediate positive selection of CD4+ lymphocytes in class II– deficient mice. Proc Natl Acad Sci USA. 1993;90:2779–2783. doi: 10.1073/pnas.90.7.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spain LM, Jorgensen JL, Davis MM, Berg LJ. A peptide antagonist prevents the differentiation of T cell receptor transgenic thymocytes. J Immunol. 1994;152:1709–1717. [PubMed] [Google Scholar]
- 36.Arden B, Clark SP, Mak TW. Mouse T-cell receptor variable gene segment families. Immunogenetics. 1995;42:501–530. doi: 10.1007/BF00172177. [DOI] [PubMed] [Google Scholar]
- 37.Garcia KC, Degano M, Stanfield RL, Brunmark A, Jackson MR, Peterson PA, Teyton L, Wilson IA. An αβ T cell receptor structure at 2.5A and its orientation in the TCR–MHC complex. Science (Wash DC) 1996;274:209–219. doi: 10.1126/science.274.5285.209. [DOI] [PubMed] [Google Scholar]
- 38.Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE, Wiley DC. Structure of the complex between human T-cell receptor viral peptide and HLA-A2. Nature (Lond) 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 39.Jorgensen JL, Esser U, Fazekas de St B, Groth, Reay PA, Davis MM. Mapping T-cell receptor-peptide contacts by variant peptide immunization of single-chain transgenics. Nature (Lond) 1992;355:224–230. doi: 10.1038/355224a0. [DOI] [PubMed] [Google Scholar]