Abstract
Intramuscular and intracutaneous immunization with naked DNA can vaccinate animals to the encoded proteins, but the underlying mechanisms of antigen presentation are unclear. We used DNA that encodes an A/PR/8/34 influenza peptide for CD4 T cells and that elicits protective antiviral immunity. DNA-transfected, cultured muscle cells released the influenza polypeptide, which then could be presented on the major histocompatibility complex class II molecules of dendritic cells. When DNA was injected into muscles or skin, and antigen-presenting cells were isolated from either the draining lymph nodes or the skin, dendritic, but not B, cells presented antigen to T cells and carried plasmid DNA. We suggest that the uptake of DNA and/or the protein expressed by dendritic cells triggers immune responses to DNA vaccines.
Independent pioneering studies by Ito (1) and Atanasiu (2) demonstrated the induction of papillomas in rabbits by injection of DNA extracted from the Shope papilloma virus, and the induction of antiviral antibodies by injection of newborn hamsters with polyoma virus DNA, respectively. Wolff et al. (3) then showed that direct gene transfer into muscles could lead to the expression of a reporter gene. After the consideration of genetic or DNA immunization as a realistic option for vaccine development (4), a large number of reports have described long-lasting humoral and cellular immune responses after in vivo administration of plasmids encoding foreign genes, including the nucleoprotein and hemagglutinin (HA)1 genes of the influenza virus (5, 6). These immune responses were obtained using intramuscular, intracutaneous, or intradermal injection of naked DNA. The intracellular plasmids can persist as episomes for long periods in transfected cells (7), where transcription of the foreign genes leads to de novo protein synthesis (1).
In a previous study, we had shown that immunization of BALB/c mice with a genetically engineered Ig–TB chimeric protein induced anti–HA150-159 antibodies and primed HA110-120–specific T cells (8). The Ig–TB chimera was engineered in such a way that the CDR2 and CDR3 loops of the VH region of a self-Ig contained the major B cell epitope HA150-159 and the immunodominant CD4 T cell epitope HA110-120 of the HA of the A/PR/8/34 influenza virus. We also found (9) that intramuscular immunization with a pVH–TB plasmid, encoding the VH region of Ig–TB under the cytomegalovirus promoter, was able to induce (a) CD4 proliferation and antibody responses against HA110-120 and HA150-159 epitopes, respectively, and (b) protective immunity in mice challenged with a lethal dose of A/PR/8/34 influenza virus. The pVH–TB plasmid persisted in the muscles of immunized mice for several months. In this study, we use this system to study the cellular mechanisms responsible for the activation of CD4 T cells after immunization with naked DNA.
It is well known that the induction of cellular immune responses requires processing of proteins by APCs and the recognition by T cells of the generated peptides in association with MHC molecules (10). Therefore, one may envisage several mechanisms responsible for the T cell activation induced by DNA immunization: (a) the protein encoded by the foreign gene is synthesized by the host cells, secreted, and then taken up by APCs, which process and present the peptides; (b) APCs take up the plasmid, synthesize and process the product of the foreign gene, and then present the peptides to T cells; or (c) transfected muscle or skin cells directly present peptides encoded by the foreign gene. In this study, we present evidence that mechanisms 1 and 2 can charge MHC class II molecules of dendritic cells with DNA-encoded peptides.
Materials and Methods
Mice.
BALB/c mice, 6–8 wk old, were purchased from The Jackson Laboratory (Bar Harbor, ME). Transgenic BALB/c mice for a TCR recognizing the HA110-120 epitope of the HA of the A/PR/8/34 influenza virus in association with I-Ed molecules were provided by Dr. Harald von Boehmer (Institut Necker, Paris, France).
Antigens.
The synthetic HA110-120 (SFERFEIFPKE) and HA150-159 (WLTEKEGSYP) peptides correspond to a CD4 T and B cell epitope, respectively, of HA of the A/PR/8/34 influenza virus. The peptides, with or without the NH2-terminal cysteine residue, were prepared by Fmoc Technology, purified by reverse phase–HPLC on a C2/C18 column (Pharmacia Biotech, Inc., Piscataway, NJ) and analyzed by amino acid sequencing on a gas phase sequencer (Porton Instruments, San Diego, CA).
APCs.
2PK3 B lymphoma cells (H-2d haplotype; American Type Culture Collection, Rockville, MD), purified B cells, dendritic cells, and Langerhans cells were used as APCs in T cell activation assays. The B and dendritic cells were obtained from BALB/c mice immunized with 30 μg of pVH–TB plasmid, or plasmid control, in the biceps and scapular muscles on days 0 and 2 and the mice were killed on days 4, 5, or 9. Brachial and axillary nodes were cut into small pieces in medium containing 100 U/ml collagenase D (Boehringer Mannheim, Indianapolis, IN), and the free cells were collected after vigorous pipetting. The tissues were then digested with 400 U/ml collagenase at 37°C for 30–40 min and passed through fine stainless steel mesh to remove undigested connective tissues. The cells were suspended in dense BSA and centrifuged at 1,000 g for 30 min. The low density cells were collected, washed with Ca2+, Mg2+-free HBSS twice, and stained with PE-B220 and FITC-CD11c (PharMingen, San Diego, CA). The populations were sorted into highly purified B220+CD11c− B cells and B220−CD11c+ dendritic cells (see Results). Langerhans cells were purified from mice injected subcutaneously in the dorsal side of each ear with 30 μg VH–TB plasmid or plasmid control in saline and the mice were killed 6 h after injection. Langerhans cells were allowed to migrate from sheets of skin for 4 d as previously described (11).
T Cell Hybridoma (TcH).
14-3-1 TcH expressing the 14.3.d TCR specific for HA110-120 peptide in association with I-Ed MHC class II molecules was obtained from Dr. Klauss Karjalainen (Basel Institute for Immunology, Basel, Switzerland). This TcH contains a chimeric LacZ gene under the control of the IL-2 promoter that can be used as an early indicator of activation (12). The hybridoma was grown in a selection medium containing IMDM supplemented with 10% FCS, sodium pyruvate 1 mM, gentamycin, 50 μM 2-ME, and 0.1 mg/ml hygromycin B (Sigma Chemical Co., St. Louis, MO).
Myoblast Cell Lines.
G7 myoblasts (H-2K) were transfected with pVH–TB plasmid or empty plasmid (pcDNA3; Invitrogen, San Diego, CA) as control, using the calcium phosphate transfection method (Invitrogen). Transfected cells were selected in collagen-coated dishes (150 mg/100 ml) in DMEM supplemented with 10% FCS, 10% horse serum, and 800 μg/ml G418.
RIA.
VH–TB polypeptide in the supernatants of the G7/pVH– TB myoblast cultures was determined by capture RIA. In brief, 96-well plates coated with 50 μg/ml anti–HA150-159 B2H1 mAb were blocked with 3% BSA/PBS. Cell culture supernatants (100 μl) were added overnight at 4°C. Plates were washed, incubated for 2 h at 37°C with 10 μg/ml affinity-purified rabbit anti–HA110-120 Abs (13), washed again, incubated for 2 h at room temperature with affinity-purified 125I-goat anti–rabbit IgG Abs (50,000 cpm/well), and then bound radiolabeled Abs were measured in a γ counter. To estimate the amount of VH–TB polypeptide secreted by the G7/VH–TB transfectants in culture, the cpm values obtained in RIA were integrated on a calibration curve constructed with 2BH1 mAb or PY102 mAb as previously described (9).
Enrichment of VH–TB Polypeptide from the Cell Culture Supernatants.
50 ml of cell culture supernatant collected over 24 h from cultured 106 G7/pVH–TB or G7/pC myoblasts was precipitated with 33% saturated ammonium sulfate for 2 h at room temperature. Precipitates were centrifuged and resuspended in 10 ml PBS, dialyzed extensively against PBS in Spectrapor bags (mol wt CO: 1,000; Sigma Chemical Co.), and then concentrated on Carbowax (Sigma Chemical Co.) 20,000 up to 1 ml. 100 μl of the 33% ammonium sulfate–precipitated fraction was used for the activation assays.
T Cell Activation Assay.
Various numbers of APCs suspended in 200 μl IMDM complete medium were incubated in polystyrene tubes together with a constant number of 14-3-1 TcHs (2 × 105) and graded amounts of antigens. After various intervals of incubation, 200 μl of 2 mM fluorescein di-β-d-galactopyranoside in distilled water (FDG; Sigma Chemical Co.) was added to the cell suspension for 1 min at 37°C. IMDM (3.8 ml) was then added and the tubes were placed on ice for 60 min. Cells were pelleted at 3,000 rpm, fixed with 1% paraformaldehyde in PBS, and the percentage of β-galactosidase+–activated TcH was scored among 5,000 cells by cytofluorometry as previously described (14).
Lymphokine Assay.
Either 2 × 105 splenocytes or 2 × 104 skin cells, including positively selected MHC class II Langerhans cells, were incubated for 24 h with 2 × 105 TcH in the absence or presence of 25 μg/ml HA110-120 peptide. The amount of IL-4 and IFN-γ secreted in the cell culture supernatants was determined by an ELISA kit according to the manufacturer's instructions (Biosource International, Camarillo, CA).
PCR Analysis.
Cells were digested for 3 h at 65°C with 0.1 mg/ml proteinase K in 0.1 M Tris-acetate buffer, pH 7.5, 0.2% SDS, 5 mM EDTA, and 200 mM NaCl. DNA was phenol-chloroform extracted, precipitated with 0.1 vol of isopropanol, and resuspended in TE buffer, pH 8.0. DNA from 2 × 105 cells was used for PCR analysis. Detection of pVH–TB plasmid in various APCs was done by PCR using specific primers for the 5′ and 3′ ends of pVH–TB chimeric gene (VH–TB-F and VH–TB-R; reference 9) or primers annealing in the 5′ and 3′ flanking regions of the multiple cloning site of pcDNA3 where the VH–TB gene was inserted (T7 and Sp6). As a control of integrity of DNA, the genomic IgG2a–CH1 exon (codons 135-223) was amplified using primers GGCTCCTCGGTGACTCTAGGATGC (forward) and CATGAATTCTGGGCTCAATTTTCTTGTCC (reverse). PCR conditions were: 30 s at 95°C, 30 s at 55°C, and 1 min at 72°C for 38 cycles. The PCR products were then analyzed by electrophoresis in 1% agarose gels.
Results
Transfected Myoblasts Secrete VH–TB Polypeptide for Processing by APCs.
We analyzed the ability of myocytes to secrete the protein encoded by the pVH–TB plasmid and its presentation by different APCs. We used a myoblast cell line (H-2k) transfected either with pVH–TB plasmid (G7/pVH– TB) or with empty plasmid as control, or pC (G7/pC). After 24 h of culture, 106 G7/pVH–TB cells in 10 ml of culture secreted 14.9 ng/ml of VH–TB polypeptide (mol wt: 10,000), as measured by a capture RIA using plates coated with 2BH1 mAb (anti–HA150-159 peptide) and revealed with 125I-rabbit anti–HA110-120 antibodies.
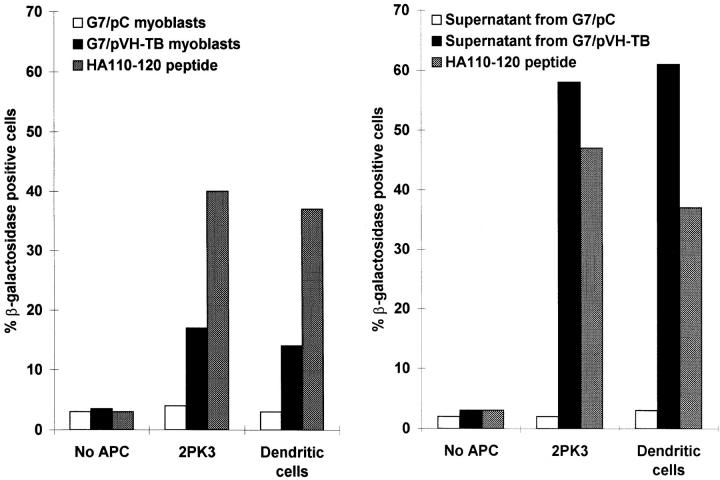
We then investigated the ability of various APCs to activate the 14.3.1 TcH when cocultured with G7/pVH–TB and G7/pC myoblasts. 14.3.1 TcH recognizes the HA110-120 in association with IEd molecules (15). Data presented in Fig. 1 (left) show that a substantial activation was obtained when the TcHs were cocultured with G7/pVH–TB myoblasts and either 2PK3 B lymphoma cells or bone marrow– derived dendritic cells. Similar results were obtained when purified CD4+ transgenic T cells expressing the HA110-120–specific TCR were used in the proliferation assay (data not shown). The ammonium sulfate–precipitated fraction of the cell culture supernatants from G7/pVH–TB cells, but not from G7/pC cells, also activated the specific TcH in the presence of either 2PK3 cells or bone marrow–derived dendritic cells (Fig. 1, right). The stronger TcH activation obtained with the ammonium sulfate fraction is due to the higher concentration of VH–TB polypeptide from cell culture supernatant. The observed activation of TcH by ammonium sulfate fraction of myoblast culture supernatant ruled out the extracellular processing of the polypeptide.
Figure 1.
Presentation of the HA110-120 peptide to the specific TcH. (Right) 2PK3 B lymphoma cells (2 × 105) or bone marrow–derived dendritic cells (5 × 104) were cultured with G7/pVH–TB , G7/pC myoblasts (2 × 105 cells), or HA110-120 peptide (30 μg/ml). After 48 h, 2 × 105 14.3.1 TcH cells were added for 12 h. (Left) 2PK3 B lymphoma cells (105) or bone marrow–derived dendritic cells (105) were cultured for 12 h with 2 × 105 14.3.1 TcH cells in the presence of HA110-120 synthetic peptide (30 μg/ml), or with the saturated ammonium sulfate fraction of the cell culture supernatants from either G7 myoblasts or G7/pVH–TB myoblasts. The percentage of TcH activated cells was determined by FACS® analysis.
Presentation of the T Helper Epitope of VH–TB Polypeptide by Dendritic Cells after Intramuscular Injection with pVH–TB Plasmid.
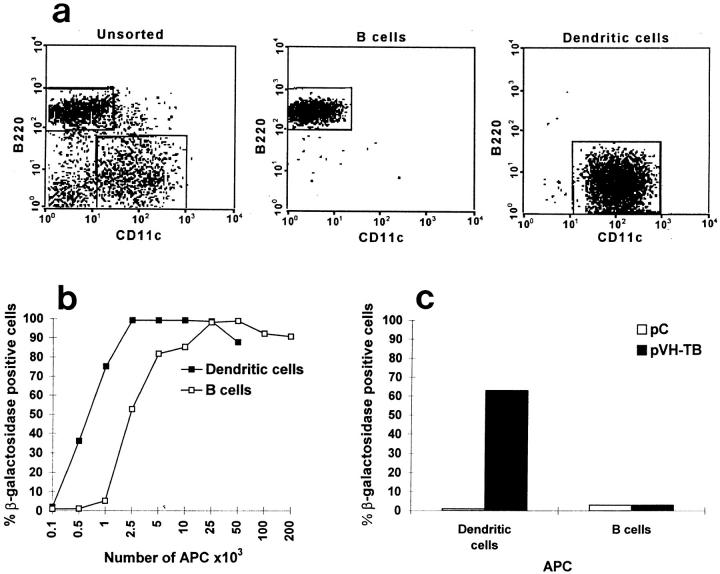
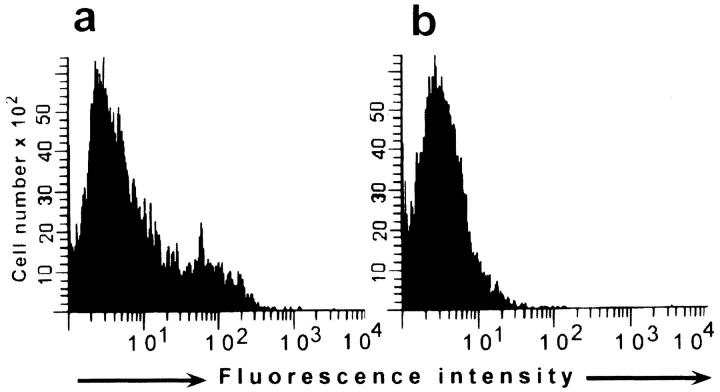
To assess the role of professional APCs in priming specific T cells after the intramuscular immunization with naked DNA, we studied the ability of sorted B cells (CD11c−B220+) and dendritic cells (CD11c+B220−) from mice immunized with pVH–TB to activate the HA110-120–specific TcH. Mice were immunized in the biceps and scapular muscles with 30 μg of pVH–TB or pC on days 0 and 2, and were killed 2 d after the last immunization. The high purity of the sorted APCs is shown in Fig. 2 a.
Figure 2.
Activation of the HA110-120–specific TcH by various numbers of dendritic and B cells. Various numbers of brachial and axillary lymph node, dendritic cells, or B cells were purified by cell sorting (a) from BALB/c mice immunized in the biceps and scapular muscles with pC and cultured (b) with TcH (2 × 105) for 24 h in the presence of HA110-120 peptide (25 μg/ ml). Dendritic or B cells isolated from mice immunized with pVH–TB or pC were cultured for 24 h with TcH (2 × 105) in the absence of exogenous antigen (c). The percentage of TcH-activated cells was determined by FACS® analysis.
First, we evaluated the efficiency of presentation of the HA110-120 T cell epitope by the B and dendritic cells from mice immunized with plasmid control. Various numbers of B or dendritic cells from the brachial and axillary lymph nodes were cocultured for 24 h with the specific TcH and HA110-120 peptide (Fig. 2 b). A lower number of dendritic than of B cells was required to achieve similar degrees of TcH activation, but both populations of APCs could stimulate the TcH.
We then studied the antigen presentation by sorted B and dendritic cells from animals immunized with pVH–TB plasmid, but without further addition of HA110-120 peptide to the cultures. The dendritic cells isolated from regional lymph nodes induced a significant activation of the TcH. In contradistinction, B cells isolated from the same lymph nodes were unable to activate the TcH (Fig. 2 c).
We estimated that 2 × 105 lymph node–derived dendritic cells in the absence of peptide induced an equivalent degree of activation to that induced by 750 dendritic cells cultured in the presence of peptide. This suggests that ∼0.4% dendritic cells from mice immunized intramuscularly with pVH–TB plasmid were able to stimulate the HA110-120–specific T cells. In the functional assay, we could not detect the presence of T cell epitope on APCs unless we enriched the dendritic cells by cell sorting; partially enriched preparations of dendritic cells (10–20% purity) from mice vaccinated with pVH–TB plasmid did not produce reliable stimulation of the TcH.
In a repeat experiment, dendritic and B cells were purified at 3 and 7 d after DNA immunization. The dendritic cells at each time point stimulated the HA110-120–specific T cells comparably. Again, B cells failed to stimulate T cells.
Presence of Plasmid DNA in Lymph Node Dendritic Cells.
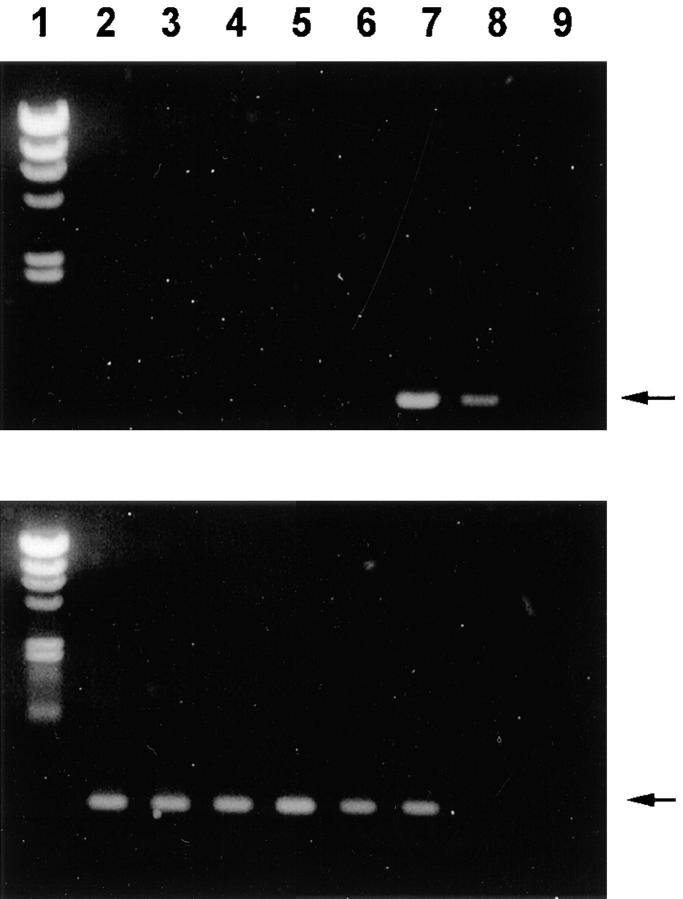
The above results (Fig. 2) demonstrated that, after intramuscular immunization, dendritic cells expressed T cell epitopes that were encoded by the naked DNA. It is possible that the dendritic cells had either taken up the plasmid itself or were expressing epitopes acquired from proteins contained in transfected myoblasts (shown in Fig. 1). To assess the former possibility, we used PCR to look for DNA in the dendritic and B cells from mice given the pC plasmid control or the pVH–TB vaccine. We found that dendritic cells, which had been purified by sorting as shown in Fig. 2 a, selectively expressed pVH–TB sequences (Fig. 3, lane 7). B cells and preparations that were partially enriched in dendritic cells did not contain detectable vaccine sequences (Fig. 3, lanes 6 and 5, respectively). As a control for the integrity of the DNA in each cell preparation, we amplified the genomic IgG2a-CH1 exon (Fig. 3, bottom). Using primers annealing in plasmid sequences (T7 and SP6), we also detected transfected cells only in the sorted dendritic cell fraction of mice immunized with pVH–TB and pC plasmid.
Figure 3.
Detection of VH– TB gene in dendritic cells from mice immunized intramuscularly with pVH–TB plasmid. DNA extracted from 2 × 105 dendritic or B cells from brachial and axillary lymph nodes was amplified by PCR using VH–TB-F and VH– TB-R primers annealing in the 5′ and 3′ end regions of the VH–TB gene, respectively (top) or, as control of integrity of DNA, primers annealing in the flanking regions of the genomic IgG2a-CH1 exon (bottom). Lane 1, DNA markers (λ/HindIII); lane 2, nonfractionated cells from mice immunized with pC; lane 3, B cells from mice immunized with pC; lane 4, dendritic cells from mice immunized with pC; lane 5, nonfractionated cells from mice immunized with pVH–TB; lane 6, B cells from mice immunized with pVH–TB; lane 7, dendritic cells from mice immunized with pVH–TB; lane 8, purified pVH–TB plasmid (5 ng); lane 9, negative control. Specific bands are shown by arrows.
Presentation of the T Helper Epitope in the pVH–TB Plasmid by Langerhans Cells after Intracutaneous Immunization with Naked DNA.
Both intracutaneous and intradermal routes of administration are currently used for the immunization with naked DNA (16). The major APCs in skin are the epidermal dendritic, or Langerhans, cells (17). To investigate the cellular mechanism of subcutaneous DNA immunization, Langerhans cells were isolated from the ears of B6 × D2 F1 (H-2b × H-2d) mice. 6 h after the injection of 30 μg pVH– TB or pC, the ear skin was explanted and cultured for 4 d, during which time cells emigrated from the skin. The emigrated cells were tested for the ability to activate HA110-120–specific TcH. A substantial activation was obtained when Langerhans cells from mice immunized with pVH–TB were cultured with TcH in the absence of exogenous antigen. This activation was measured by the production of IL-2 (Fig. 4 a), IFN-γ, and IL-4 (Table 1, group 7). Langerhans cells from mice immunized with pC failed to activate the TcH in absence of exogenous antigen (Fig. 4 b; Table 1, group 5).
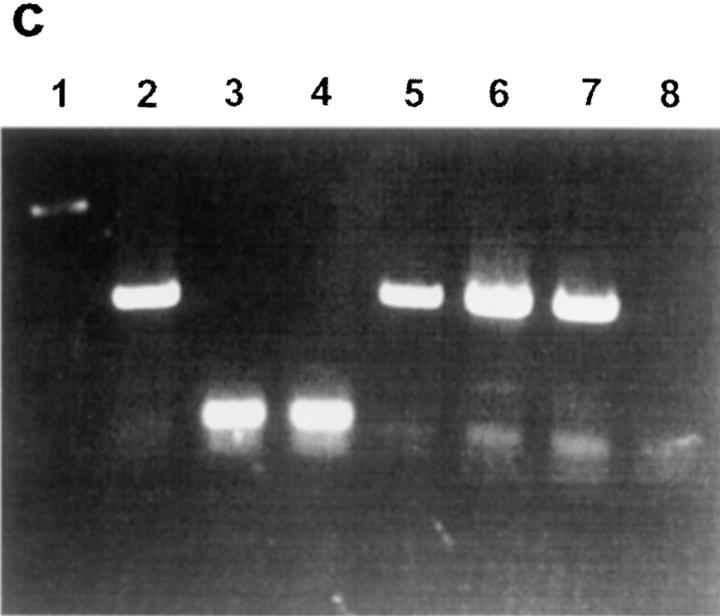
Figure 4.
Activation of a HA110-120–specific TcH by enriched Langerhans cells transfected in vivo with pVH– TB plasmid. Langerhans cells (2 × 105) were obtained from mice immunized intracutaneously with pVH–TB plasmid and cultured for 24 h with 14.3.1 TcH (2 × 105). FACS® analysis shows that Langerhans cells from animals immunized with pVH–TB activated 26% of the HA110-120–specific TcH (a), but Langerhans cells from animals immunized with pC did not activate any of the TcH (b). (c), PCR analysis of pVH– TB and pC plasmids using T7 and Sp6 primers in skin cells from mice immunized intracutaneously. Lane 1, molecular markers; lane 2, purified pVH–TB plasmid; lane 3, purified pC plasmid; lane 4, skin cells from mice immunized with pC; lane 5, skin cells from mice immunized with pVH–TB; lane 6, MHC class II–positive Langerhans cells from mice immunized with pVH–TB; lane 7, MHC class II–negative skin cells from mice immunized with pVH– TB; lane 8, negative control.
Table 1.
Cytokine Production of 14.3.1 TcH upon Activation with Langerhans Cells from BALB/c Mice Immunized with pVH–TB Plasmid
| Group | APC | Ag | IFN-γ | IL-4 | ||||
|---|---|---|---|---|---|---|---|---|
| 30 μg | pg/ml | pg/ml | ||||||
| 1 | nil | nil | 0 | 0 | ||||
| 2 | BALB/c splenocytes | nil | 0 | 5 ± 5 | ||||
| 3 | BALB/c splenocytes | HA110-120 | 58 ± 6 | 27 ± 3 | ||||
| 4 | BALB/c splenocytes | HA150-159 | 3 ± 1 | 0 | ||||
| 5 | LC (pC) nonfractionated | nil | 0 | 0 | ||||
| 6 | LC (pC) nonfractionated | HA110-120 | 161 ± 11 | 512 ± 18 | ||||
| 7 | LC (pVH–TB) nonfractionated | nil | 87 ± 3 | 58 ± 5 | ||||
| 8 | LC (pVH–TB) nonfractionated | HA110-120 | 110 ± 9 | 364 ± 24 | ||||
| 9 | LC (pVH–TB) MHC class II+ | nil | 143 ± 7 | 88 ± 8 | ||||
| 10 | LC (pVH–TB) MHC class II− | nil | 2 ± 1 | 0 |
Because the Langerhans cell population was contaminated with other cells, primarily keratinocytes, we positively selected MHC class II–positive cells using iron beads coated with anti-IAd mAbs. In the absence of exogenous antigen, only MHC class II–positive cells from DNA-vaccinated mice presented the HA110-120 peptide to the specific TcH, which secreted a significant amount of IFN-γ and IL-4 (Table 1, group 9). Negatively selected MHC class II–negative skin cells failed to activate the TcH (Table 1, group 10).
The Presence of Plasmid DNA in Dendritic Cells from the Skin of Mice Vaccinated by the Intracutaneous Route.
The data depicted in Fig. 4 c show specific PCR bands amplified from the VH–TB gene sequences in both MHC class II–positive and MHC class II–negative skin cells from animals immunized with pVH–TB. Sequences of the pC were also detected in skin cells. This suggests that MHC class II–positive cells (Langerhans cells) were transfected in vivo with naked DNA. Taken together, the results in Fig. 4 indicate that different skin cells can be transfected in vivo with naked DNA, but only Langerhans cells are able to present the HA110-120 epitope to the T helper cells.
Discussion
Our observations provide mechanisms underlying two recent observations. Ulmer et al. (18) showed that F1(H-2d × H-2k) mice injected with a myoblast cell line (H-2k) transfected with a foreign gene developed H-2d– and H-2k– restricted immunity. Corr et al. (19) used chimeric mice, in which the haplotype of bone marrow was mismatched with the haplotype of somatic cells at the site of injection, to demonstrate that the priming of specific CTLs was driven by the MHC from the bone marrow–derived APCs.
First, we demonstrated that a VH–TB polypeptide, encoded by a DNA vaccine, is secreted by G7/pVH–TB myoblasts and can be processed and presented by other APCs to the HA110-120–specific T cells. Second, our in vivo results showed that the dendritic cells from mice immunized with pVH–TB were able to activate T cells, whereas B cells were not. These data indicate that dendritic cells can play a crucial role in triggering immune responses subsequent to DNA immunization. Third, we found that, in spite of the paucity of APCs in muscles, dendritic but not B cells were transfected in vivo with pVH–TB plasmid. Condon et al. (20) have obtained cytological evidence for the in vivo expression of a protein encoded by a foreign gene in dendritic cells. Our data demonstrate for the first time that purified dendritic cells carry plasmid DNA and present the corresponding CD4 T helper epitope to antigen-specific T cells.
Likewise, after intradermal injection of DNA, the pVH– TB plasmid was detected in both MHC class II–positive and MHC class II–negative skin cells. In vivo transfection of skin cells has been reported (21), but the ability of these cells to activate specific T cells has not been investigated. Our results demonstrated that only MHC class II–positive dendritic cells were able to activate the specific CD4 T cells.
We attempted to estimate the frequency of dendritic cells that could express genes encoded by the DNA vaccine. We injected mice with DNA for a green fluorescence protein that produced bright fluorescence upon transfection of cultured cell lines, using either a CMV or HIV-1 promoter. 0–5 h after injection, the skin was removed and the emigrated dendritic cells were collected over a 5-d culture period. However, at no time could we detect fluorescent dendritic cells by FACS® or by fluorescence microscopy, probably because the frequency of DNA-bearing dendritic cells was very low (<1/200 cells).
In conclusion, we have found that dendritic cells can play a crucial role in the initiation of T helper immune responses by DNA immunization. Subsequent to intramuscular or subcutaneous immunization with DNA, myocytes or MHC class II–negative dermal cells can be transfected and can secrete the protein encoded by the foreign gene, which is then presented by dendritic but not by most B cells. In addition, the dendritic cells carry plasmid DNA and thereby may be able to express vaccine epitopes directly.
Acknowledgments
This work was supported by grants from the National Institute of Allergy and Infectious Diseases (Bethesda, MD) to C.A. Bona (AI-37115) and R.M. Steinman (AI-13013, AI-40874), Alliance Pharmaceutical Corp. (San Diego, CA), and the Japanese Ministry of Education, Science and Culture (grants 08282104, 08044271).
Footnotes
Abbreviations used in this paper: HA, hemagglutinin; pC, plasmid control; TcH, T cell hybridoma.
References
- 1.Ito Y. A tumor-producing factor extracted by phenol from papillomatous tissue (Shope) of cottontail rabbits. Virology. 1960;12:596–601. doi: 10.1016/0042-6822(60)90182-3. [DOI] [PubMed] [Google Scholar]
- 2.Atanasiu P. Production of tumors in hamsters by inoculation of DNA extracted from cultures of tissues infected with polyoma virus. CR Acad Sci. 1962;254:4228–4230. [PubMed] [Google Scholar]
- 3.Wolff JA, Malone RW, Williams P, Chong W, Acsadi G, Jani A, Felgner PL. Direct gene transfer into mouse muscle in vivo. Science (Wash DC) 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 4.Tang D, Devit M, Johnson SA. Genetic immunization is a simple method for eliciting an immune response. Nature (Lond) 1992;356:152–155. doi: 10.1038/356152a0. [DOI] [PubMed] [Google Scholar]
- 5.Ulmer JB, Donnelly JJ, Parker SE, Rhodes GH, Felgner PL, Dwarki VJ, Gromkowski SH, Deck RR, Dewitt CM, Friedman A, et al. Heterologous protection against influenza by injection of DNA encoding a viral protein. Science (Wash DC) 1993;259:1745–1749. doi: 10.1126/science.8456302. [DOI] [PubMed] [Google Scholar]
- 6.Fynan EF, Robinson HL, Webster RG. Use of DNA encoding influenza hemagglutinin as an avian influenza vaccine. DNA Cell Biol. 1993;12:785–789. doi: 10.1089/dna.1993.12.785. [DOI] [PubMed] [Google Scholar]
- 7.Pardoll DM, Beckerleg AM. Exposing the immunology of naked DNA vaccines. Immunity. 1995;3:165–169. doi: 10.1016/1074-7613(95)90085-3. [DOI] [PubMed] [Google Scholar]
- 8.Brumeanu T-D, Zaghouani H, Dehazya P, Wolf I, Bot A, Bona CA. Engineering of doubly antigenized immunoglobulins expressing T and B viral epitopes. Immunotechnology (Amst) 1996;2:85–95. doi: 10.1016/1380-2933(96)85196-7. [DOI] [PubMed] [Google Scholar]
- 9.Casares S, Brumeanu T-D, Bot A, Bona CA. Protective immunity elicited by vaccination with DNA encoding for a B epitope and a T helper epitope of A/PR/8/34 influenza virus. Viral Immunol. 1997;10:129–136. doi: 10.1089/vim.1997.10.129. [DOI] [PubMed] [Google Scholar]
- 10.Braciale TL, Braciale VL. Antigen presentation: structural themes and functional variations. Immunol Today. 1992;12:124–129. doi: 10.1016/0167-5699(91)90096-C. [DOI] [PubMed] [Google Scholar]
- 11.Ortner U, Inaba K, Koch F, Heine M, Miwa M, Schuler G, Romani N. An improved isolation method for murine migratory cutaneous dendritic cells. J Immunol Methods. 1996;193:71–79. doi: 10.1016/0022-1759(96)00058-0. [DOI] [PubMed] [Google Scholar]
- 12.Weber S, Trannecker A, Olivery F, Gerhard WV, Karjalainen K. Specific low-affinity recognition of MHC complex plus peptide by soluble T-cell receptor. Nature (Lond) 1992;356:793–796. doi: 10.1038/356793a0. [DOI] [PubMed] [Google Scholar]
- 13.Brumeanu T-D, Zaghouani H, Kohanski R, Bona CA. A sensitive method to detect defined peptide among those eluted from murine MHC class II molecules. J Immunol Methods. 1993;160:65–71. doi: 10.1016/0022-1759(93)90009-v. [DOI] [PubMed] [Google Scholar]
- 14.Nolan GP, Fiering S, Nicolas JF, Herzanberg LA. Fluorescence-activated cell analysis and sorting of viable mammalian cells based on beta-galactosidase activity after transduction of Escherichia coliLacZ. Proc Natl Acad Sci USA. 1988;85:2603–2608. doi: 10.1073/pnas.85.8.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirberg J, Baron A, Jacob S, Rolink A, Karjalainen K, von Boehmer H. Thymic selection of CD8 single positive cells with a major histocompatibility complex–restricted receptor. J Exp Med. 1994;180:25–34. doi: 10.1084/jem.180.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ertl ECJ, Xiang ZQ. Genetic immunization. Viral Immunol. 1996;9:1–9. doi: 10.1089/vim.1996.9.1. [DOI] [PubMed] [Google Scholar]
- 17.Bos JD, Kapsenberg ML. The skin immune system: progress in cutaneous biology. Immunol Today. 1993;14:75–78. doi: 10.1016/0167-5699(93)90062-P. [DOI] [PubMed] [Google Scholar]
- 18.Ulmer JB, Deck RR, Dewitt CM, Donnelly JJ, Liu MA. Generation of MHC class-I restricted T lymphocytes by expression of a viral protein in muscle cells: antigen presentation by non-muscle cells. Immunology. 1996;89:59–67. doi: 10.1046/j.1365-2567.1996.d01-718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corr M, Lee JL, Carson DA, Tighe H. Gene vaccination with naked plasmid DNA: mechanism of CTL priming. J Exp Med. 1996;184:1555–1560. doi: 10.1084/jem.184.4.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Condon C, Watkins SC, Celluzzi CM, Thompson K, Falo LD. DNA-based immunization by in vivo transfection of dendritic cells. Nat Med. 1996;10:1122–1128. doi: 10.1038/nm1096-1122. [DOI] [PubMed] [Google Scholar]
- 21.Hengge UR, Walker PS, Vogel JC. Expression of naked DNA in human, pig, and mouse skin. J Clin Invest. 1996;97:2911–2916. doi: 10.1172/JCI118750. [DOI] [PMC free article] [PubMed] [Google Scholar]