Abstract
Natural killer (NK) cells exhibit cytotoxicity against variety of tumor cells and virus-infected cells without prior sensitization and represent unique lymphocytes involved in primary host defense. NKR-P1 is thought to be one of NK receptors mediating activation signals because cross-linking of NKR-P1 activates NK cells to exhibit cytotoxicity and IFN-γ production. However, molecular mechanism of NK cell activation via NKR-P1 is not well elucidated. In this study, we analyzed the cell surface complex associated with NKR-P1 on NK cells and found that NKR-P1 associates with the FcRγ chain which is an essential component of Fc receptors for IgG and IgE. The association between FcRγ and NKR-P1 is independent of Fc receptor complexes. Furthermore, NK cells from FcRγ-deficient mice did not show cytotoxicity or IFN-γ production upon NKR-P1 cross-linking. Similarly, NK1.1+ T cells from FcRγ-deficient mice did not produce IFN-γ upon NKR-P1 crosslinking. These findings demonstrate that the FcRγ chain plays an important role in activation of NK cells via the NKR-P1 molecule.
NK cells play a pivotal role in protective immunity. Especially, NK cells are involved in the earliest stage of immune response because they can exhibit cytotoxicity or cytokine production without prior sensitization (1). Therefore, it is important to elucidate the mechanism of antigen recognition by NK cells to understand innate immunity. Recent analyses have revealed that various types of receptors are involved in the regulation of NK cell activation (2, 3). Among these, the NKR-P1 molecule is thought to be one of NK receptors which mediates NK cell activation because cross-linking of NKR-P1 induces various activation signals such as, Ca2+ flux, PI turnover and lymphokine production (4, 5). The NKR-P1 was originally cloned from rat NK cells (4, 5), and subsequently cloned from mouse and human (6, 7). In mouse NK cells, NK1.1, a specific marker of mouse NK cells, is one of NKR-P1 family molecules, NKR-P1C (8).
NK cells exhibit cytotoxicity and IFN-γ production upon cross-linking of NKR-P1 molecule with specific mAb (4, 5, 9, 10). Indeed, introduction of NKR-P1 into a NKR-P1-deficient NK cell line restored cytotoxicity against certain tumor cell targets (11). Furthermore, anti–NKR-P1 mAb blocked the cytotoxicity by the NKR-P1–transfected NK cells (11). Therefore, NKR-P1 seems to be involved in the antigen recognition by NK cells. Although the exact ligand for NKR-P1 remains to be determined, NKR-P1 belongs to the C-type lectin superfamily and certain carbohydrates expressed on target cells are postulated to be recognized by the NKR-P1 molecule (12).
In respect to the signaling pathway for NKR-P1, it has been shown that NKR-P1 crosslinking stimulates PI turnover and raises intracellular Ca2+ concentration (13). In addition, NKR-P1 possesses a p56lck-binding motif which is represented by YxxL sequence and indeed the association of p56lck to NKR-P1 was recently reported (14). However, essential components for NK cell activation via the NKR-P1 molecule have not been elucidated. In the present study, we analyzed the cell surface complex associated with the NKR-P1 molecule and found that the FcRγ chain is associated with NKR-P1. Furthermore, the analysis of NK cells from FcRγ-deficient mice revealed that the FcRγ chain is essential for mediating activation signals via NKR-P1.
Materials and Methods
Mice.
FcRγ-deficient (γ−/−)1 mice with C57BL/6 background were established by gene targeting using a C57BL/6 origin ES cell line BL6/III (15) in our laboratory (Park, S.Y. et al., manuscript submitted, 16, 17) and maintained in our animal facility in SPF condition.
NK Cell Preparation.
NK cells were purified as previously described (18). In brief, NK cells were purified from spleen of 6–8-wk-old C57BL/6 mice. Splenocytes were mixed with anti-CD4 mAb (GK1.5) and anti-CD8 mAb (53.6.7), and incubated with magnetic beads (Advanced Magnetics, Inc., Cambridge, MA) coupled with goat anti–mouse IgG Ab and goat anti–rat IgG Ab (Cappel, Organon Teknika Co., West Chester, PA) to remove surface Ig (sIg)+ B cells and CD4+ and CD8+ T cells. The residual cells were then stained with PE-anti-NK1.1 (PK136) mAb and FITC-anti-CD3ε (145-2C11) mAb (Pharmingen, San Diego, CA) and NK1.1+ CD3− cells were sorted by FACStarplus® (Becton Dickinson, Mountain View, CA). The purified NK cells were cultured in RPMI-1640 supplemented with 10% FCS, kanamycin (100 μg/ml) and 5 × 10−5 M 2-ME in the presence of 1,000 U/ml human recombinant IL-2 (kindly provided by Dr. Junji Hamuro, Ajinomoto Co. Inc., Kawasaki, Japan) for 5 d.
Flow Cytometric Analysis of NK Cells.
CD4−CD8−sIg− splenocytes prepared as described above were cultured for 4 d in the presence of 1,000 U/ml IL-2. The cultured cells were first stained with FITC–anti-FcγRIII (2.4G2) mAb. Then, the cells were mixed with Biotin–anti-CD3ε and PE–anti-NK1.1 mAbs followed by Quantum-red streptavidin (Sigma Chem. Co., St. Louis, MO).
NK1.1+ T Cell Preparation.
Basically, NK1.1+ T cells were prepared as previously described (10). Thymocytes were treated with anti-HSA mAb (J11d) followed by complement treatment. The resultant cells were incubated with anti-CD8 and anti–MEL-14 mAb. Thereafter, CD8+ MEL-14+ cells were removed by use of magnetic beads coupled with goat anti–rat IgG Ab. The residual cells were cultured for 3 d in the presence of 1,000 U/ml IL-2. Thereafter, cells were stained with PE–anti-NK1.1 mAb and FITC–anti-CD3ε mAb. The stained cells were sorted into NK1.1+CD3+ cells. The sorted cells were further cultured for 2 d in the presence of 1,000 U/ml IL-2 and used for functional analysis.
Surface Biotinylation, Immunoprecipitation and Western Blotting.
Cells were surface biotinylated as previously described (19). Biotinylated cells were lysed with a lysis buffer containing 1% digitonin, 50 mM Tris-HCl (pH 7.6), 150 mM NaCl, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mM PMSF, 10 mM iodoacetamide, at a concentration of 1 × 107 cell/ml. Immunoprecipitation was performed with anti-CD3ε or anti-NK1.1 mAbs. Immunoprecipitates were separated on two-dimensional nonreducing and reducing SDS-PAGE and transferred onto a polyvinylidene difluoride (PVDF) membrane (Immobilon-P; Millipore Corporation, Bedford, MA). The biotinylated proteins were detected using streptavidin-peroxidase (VECSTAIN Elite ABC kit; Vector Laboratories Incorporated, Burlingame, CA), the ECL system (Amersham International, Buckinghamshire, England), and autoradiography. After termination of chemiluminescence, the membrane was blotted with anti-FcRγ Ab followed by peroxidase-labeled anti–rabbit Ab (Amersham) and detected by the ECL system.
NKR-P1 and TCR Stimulation.
(Fab)′2 fragment of anti-NK1.1 mAb was immobilized on a 96-well flat-bottomed culture plate (Coaster, Cambridge, MA) by incubating for 2 h at 37°C at a concentration of 50 μg/ml in PBS. Similarly, anti–TCR-β mAb (H57-597) was immobilized at a concentration of 100 μg/ml. 1 × 105 NK cells cultured for four days in the presence of IL-2 were stimulated with the immobilized anti-NK1.1 mAb, or with recombinant mouse IL-12 (4.9 × 106 U/mg, generously supplied from Genetics Institute, Inc., Cambridge, MA) for 2 d.
Measurement of IFN-γ and IL-4.
IFN-γ and IL-4 in the culture supernatant was measured by ELISA with standard protocol. Anti–IFN-γ (R4-6A2) and anti–IL-4 (BVD4-1D11) mAb was coated to capture IFN-γ and IL-4 and the captured IFN-γ and IL-4 were detected with biotinylated anti–IFN-γ (XMG1.2) and anti–IL-4 (BVD6-24G2) mAbs. These antibodies were purchased from Pharmingen. Concentrations of IFN-γ and IL-4 were determined with recombinant IFN-γ and IL-4 as a standard.
Redirected Cytotoxicity of NK Cells.
Cytotoxic assay was done basically as previously described (20). Briefly, freshly isolated CD4− CD8− HSA− sIg− population was used as NK cell enriched population. About 50% of this population expressed the NK1.1 molecule. These NK cells were incubated at 4°C for 30 min in the presence or absence of anti-NK1.1 mAb. Thereafter, 51Cr-labeled FcR-expressing P815 cells were added and cultured at 37°C for 4 h and analyzed the amount of Cr51 released in the culture supernatant. Specific cytotoxicity was calculated as previously described (20).
Results
To elucidate signaling pathways for NK cell activation through NKR-P1, we analyzed the NKR-P1-associated complex on the cell surface using an NK1.1-expressing T cell line CTLL-2. CTLL-2 cells were surface-biotinylated and the cell lysate was immunoprecipitated with anti-NK1.1 or anti-CD3ε mAbs, followed by analysis on two dimensional nonreducing and reducing SDS-PAGE.
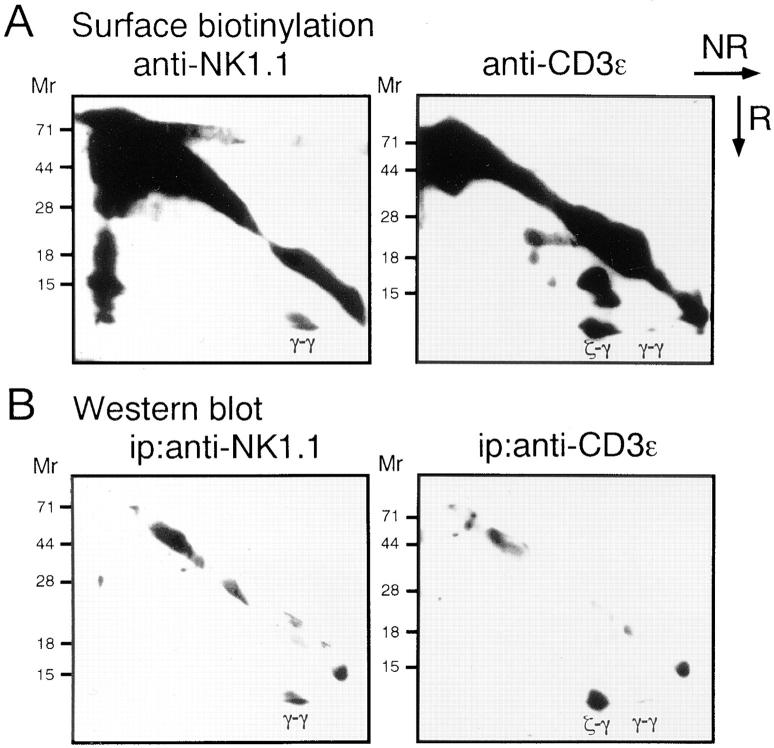
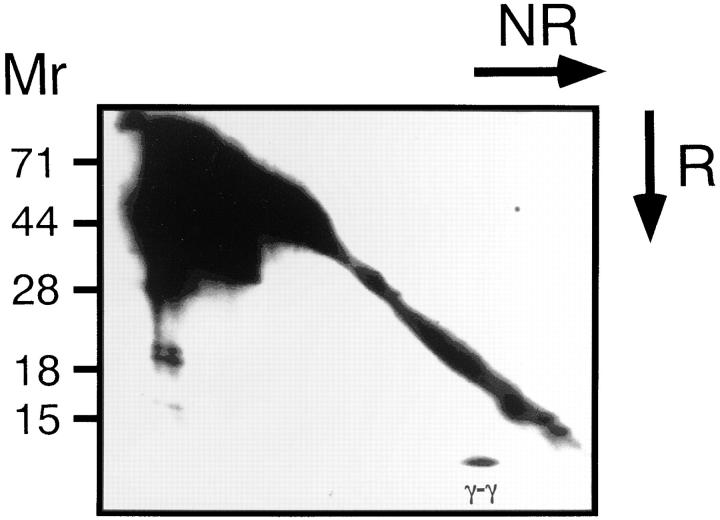
As shown in Fig. 1 A, immunoprecipitation with anti-NK1.1 mAb revealed that a 9-kD homodimer was coprecipitated with NK1.1 molecule. In contrast, when the TCR complex was precipitated with anti-CD3ε mAb, CD3ζ homodimers, CD3ζ-FcRγ heterodimers and a small amount of FcRγ homodimers were observed as previously reported (21). Interestingly, the 9-kD homodimers coprecipitated with NK1.1 appeared to be identical to FcRγ within the TCR complex. Indeed, the homodimers coprecipitated with NK1.1 were blotted with anti-FcRγ Ab similarly to FcRγ observed in the CD3ε-immunoprecipitates (Fig. 1 B). The association of the FcRγ chain with the NKR-P1 molecule was also observed in a rat NK cell line, RNK-16 (data not shown). We next analyzed normal NK cells to generalize the physical association between FcRγ and NKR-P1 in normal NK cell population. Similar to CTLL-2, when NK1.1 was immunoprecipitated from IL-2 activated NK cells, the FcRγ homodimer was coprecipitated with NK1.1 (Fig. 2). Collectively, these data indicate that the association of FcRγ with NKR-P1 is generally observed for NK cells in vivo.
Figure 1.
Association of the FcRγ chain with the NKR-P1 molecule on the cell surface of CTLL-2 cell line. (A) CTLL-2 cells were surface-biotinylated and the cell lysate was immunoprecipitated with anti-NK1.1 mAb or anti-CD3ε mAb and analyzed on two-dimensional nonreducing (NR) and reducing (R) SDS-PAGE. Biotinylated proteins were detected by an ECL. (B) FcRγ chain was detected by blotting the membrane with anti-FcRγ Ab after immunoprecipitation with anti-NK1.1 or anti-CD3ε mAbs. FcRγ homodimer (γ-γ) and CD3ζ-FcRγ heterodimers (ζ-γ) were indicated.
Figure 2.
Association of the FcRγ chain with the NKR-P1 molecule in IL-2–expanded NK cells. NK cells from normal mice expanded for 1 wk in the presence of IL-2 and were surface-biotinylated. The cell lysate of them was immunoprecipitated with anti-NK1.1 mAb ((Fab)′2 fragment) and analyzed on two-dimensional nonreducing (NR) and reducing (R) SDS-PAGE. Biotinylated proteins were detected as Fig. 1 A. FcRγ homodimer (γ-γ) was indicated.
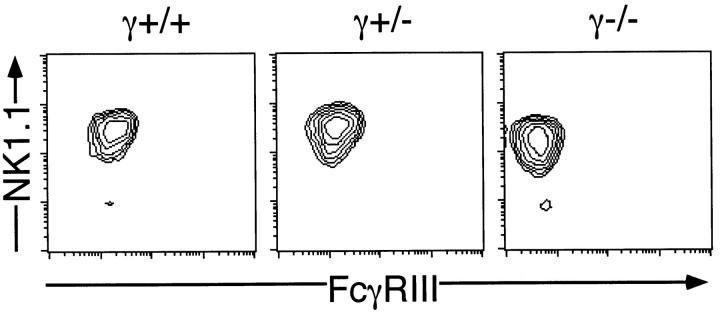
To elucidate the physiological role of the association between FcRγ and NKR-P1 particularly on signal transduction in NK cells, NK cells were prepared from γ−/− mice and analyzed for NKR-P1 expression and NKR-P1–mediated NK cell activation. When CD4−CD8−sIg− splenocytes from γ−/− mice were stained with anti-NK1.1 and anti-CD3ε mAbs, normal development of NK cells (NK1.1+CD3−) was observed (data not shown). When expression of FcγRIII on IL-2–expanded NK cells was analyzed, NK cells from γ−/− mice did not express FcγRIII (Fig. 3). In addition, the expression level of NK1.1 on the cell surface of IL-2–expanded NK cells from γ−/− mice was significantly lower than that of NK cells from γ+/+ and γ+/− mice. However, the difference of NK1.1 expression between γ−/− and γ+/− mice was marginal when freshly isolated NK cells were analyzed (data not shown). This suggested that FcRγ is not absolutely required for but affects the surface expression of the NKR-P1 molecule.
Figure 3.
Loss of the cell surface expression of FcγRIII on NK cells from γ−/− mice. CD4−CD8−HSA−sIg− splenocytes from γ+/+, γ+/− and γ−/− mice were cultured for 4 d in the presence of 1,000 U/ml IL-2. The cultured cells were stained with FITC–anti-FcγRIII, PE–anti-NK1.1 and Quantum red-anti-CD3ε mAbs and the expression of FcγRIII and NK1.1 on the CD3− population was illustrated. Representative data from five independent experiments are shown.
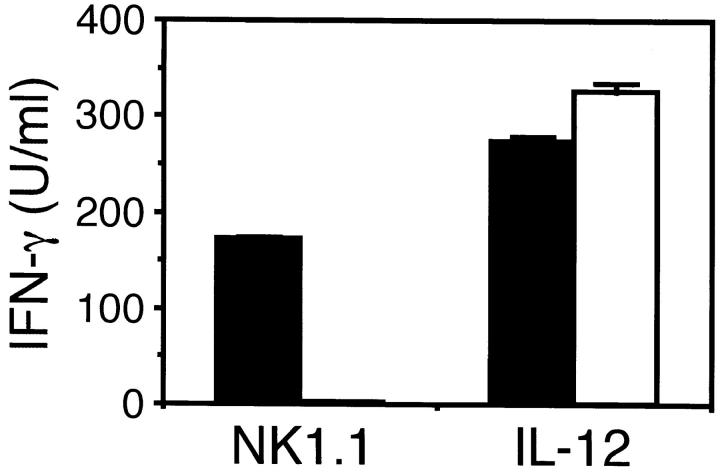
Recently, we have shown that cross-linking of NKR-P1 on NK cells with anti-NK1.1 mAb induced not only cytotoxicity but also IFN-γ production (10). Thus, we stimulated purified NK cells from γ−/− mice with immobilized F(ab)′2 fragment of anti-NK1.1 mAb in order to avoid the binding of the Ab to Fc receptors, and measured the amount of IFN-γ produced in the culture supernatant. Surprisingly, NK cells from γ−/− mice did not produce IFN-γ at all upon NKR-P1 cross-linking, whereas NK cells from γ+/+ mice produced a large amount (Fig. 4). In contrast, NK cells from both γ−/− and γ+/+ mice produced almost equal amount of IFN-γ upon IL-12 stimulation. This suggests that the defect of IFN-γ production upon NKR-P1 cross-linking by NK cells from γ−/− mice is not due to the inability of IFN-γ production but the signaling defect via NKR-P1.
Figure 4.
Defect of IFN-γ production by NK cells from γ−/− mice upon stimulation with NK1.1 cross-linking. NK cells from γ+/+ (closed bar) and γ−/− (open bar) mice were stimulated with immobilized (Fab)′2 fragment of anti-NK1.1 mAb or 1 ng/ml IL-12 and IFN-γ produced in the culture supernatant was measured. Data are presented as mean ± SD of triplicate culture.
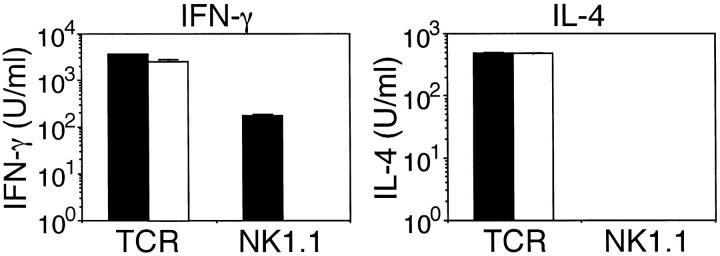
Because NK1.1+ T cells also produce IFN-γ but not IL-4 upon NKR-P1 cross-linking (10), we also analyzed the function of NK1.1+ T cells in γ−/− mice. As we have previously reported (22), NK1.1+ T cells develop normally in γ−/− mice. NK1.1+ T cells were prepared from γ+/+ and γ−/− mice and stimulated with immobilized anti-NK1.1 mAb (Fig. 5). Similar to NK cells, NK1.1+ T cells from γ−/− mice did not produce IFN-γ upon NKR-P1 cross-linking whereas these cells from γ+/+ mice produced a significant amount of IFN-γ. In contrast to the stimulation through NKR-P1, NK1.1+ T cells from both γ+/+ and γ−/− mice produced IFN-γ and IL-4 upon TCR crosslinking. These observations demonstrate that the FcRγ chain is required for IFN-γ production via NKR-P1 cross-linking both in NK cells and NK1.1+ T cells.
Figure 5.
Defect of IFN-γ production by NK1.1+ T cells from γ−/− mice upon stimulation with NK1.1 cross-linking. NK1.1+ T cells from γ+/+ (closed bar) and γ−/− (open bar) mice were stimulated with immobilized anti-NK1.1 mAb or anti-TCRβ mAb and IFN-γ and IL-4 produced in the culture supernatant was measured. Data are presented as mean ± SD of triplicate culture.
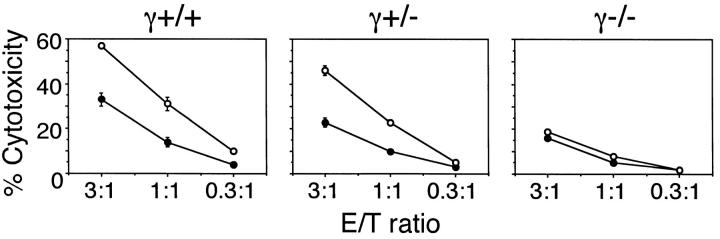
We also investigated the involvement of FcRγ in the NKR-P1–mediated cytotoxicity of NK cells. Since NKR-P1 is known to induce redirected cytotoxicity against FcR-expressing cells, we analyzed cytotoxicity of NK cells against FcR-expressing P815 cells in the presence of anti-NK1.1 mAb (Fig. 6). NK cells from γ+/+ and γ+/− mice showed increased cytotoxicity against P815 cells in the presence of anti-NK1.1 mAb, whereas NK cells from γ−/− mice failed to enhance cytotoxicity. These results demonstrate that FcRγ is involved in both IFN-γ production and cytotoxicity upon stimulation through the NKR-P1 molecule.
Figure 6.
Defect of NK1.1 mediated cytotoxicity by NK cells from γ−/− mice. Cytotoxicity of NK cells from γ+/+, γ+/−, and γ−/− mice against P815 cell line was analyzed in the presence (open circles) or absence (closed circles) of anti-NK1.1 mAb. Data are presented as mean ± SD of triplicate assay.
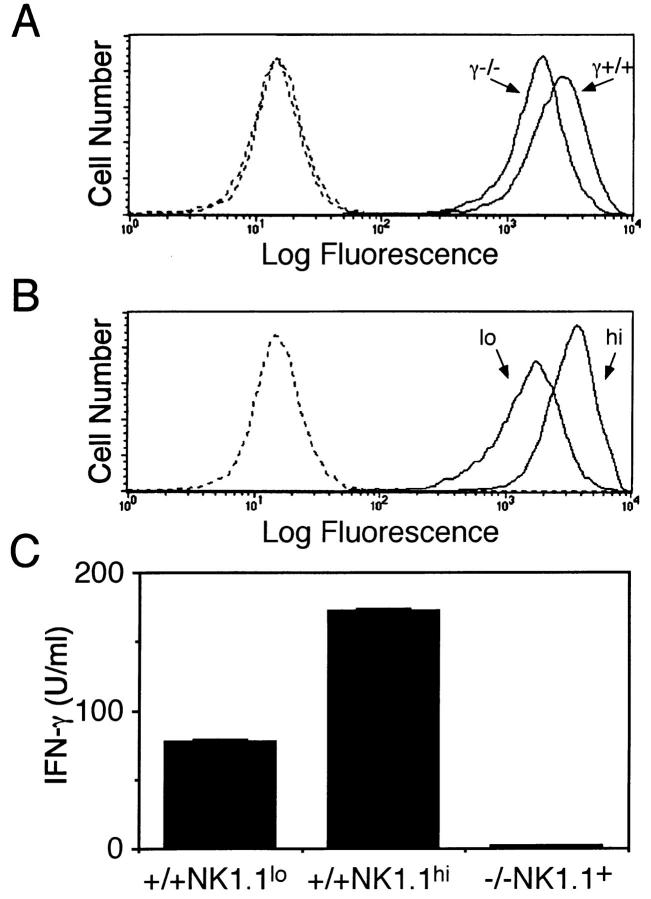
The failure of NK cell activation through NKR-P1 in γ−/− mice is likely due to a defect in signaling pathway of the NKR-P1 molecule. However, since the NK1.1 expression on the cell surface of NK cells from γ−/− mice is slightly lower than that of γ+/− and γ+/+ mice, it might be attributed to the low expression of the cell surface NKR-P1. To clarify these possibilities, we prepared two populations of NK cells from normal mice expressing low or high level of NKR-P1 (NKR-P1lo and NKR-P1hi, respectively) by cell sorting. The expression level of NKR-P1 on the NKR-P1lo population was almost identical to that of NK cells from γ−/− mice. (Fig. 7, A and B). Then, we stimulated these NK cell populations with immobilized anti-NK1.1 mAb. NKR-P1lo NK cells from γ+/+ mice produced significant amount of IFN-γ upon NKR-P1 cross-linking, although the amount of IFN-γ was lower than that by NKR-P1hi NK cells (Fig. 7 C). In contrast, NK cells from γ−/− mice did not produce any detectable level of IFN-γ upon NKR-P1 cross-linking. These observations confirm the crucial role of FcRγ in signaling pathway through NKR-P1 for NK activation and also demonstrate that FcRγ is partly involved in the expression of the cell surface NKR-P1.
Figure 7.
Activation defect of NK cells from FcRγ-deficient mice upon NKR-P1 crosslinking is not due to the low level of NKR-P1 expression. (A) NK1.1 expression on NK cells from γ+/+ and γ−/− mice. NK cells from γ+/+ or γ−/− mice were stained with anti-NK1.1 mAb (solid line) or control Ab (dotted line). (B) Preparation of NK cell population expressing high (hi) and low (lo) level of NKR-P1 from wild-type mice. (C) Production of IFN-γ by NK cell population expressing low NKR-P1 upon stimulation with immobilized anti-NK1.1 mAb. Two NK cell populations expressing high (NK1.1hi) and low (NK1.1lo) level of NK1.1 as shown in B from γ+/+ mice and NK cells from γ−/− mice were stimulated with immobilized (Fab)′2 fragment anti-NK1.1 mAb and IFN-γ production was measured.
Discussion
FcRγ was originally identified in the FcεRI complex and found to play an essential role in the expression and signaling of FcεRI (23). FcRγ was also found to be required for the expression and function of FcγRI, FcγRIII and FcαR (24, 25). On the other hand, we found that FcRγ also associates with the TCR complexes in two types of T cells. One is T cells in epithelia such as CD8αα+ TCR-γ/δ+ intestinal intraepitherial lymphocytes and TCR γ/δ+ dendritic epidermal cells and the association with FcRγ in these cells was shown in vivo from the analysis of CD3ζ-deficient mice (26). The second population is NK1.1+ TCR-α/β+ T cells. (22). However, these T cells obtained from γ−/− mice showed no clear defect in function upon TCR activation, although the expression level of the TCR complex was slightly decreased (27). Accordingly, it is striking that NK cells and NK1.1+ T cells from γ−/− mice show no response upon NKR-P1 cross-linking in spite of normal development. This finding suggests that CD3ζ expressed in NK cells and NK1.1+ T cells can not be replaced for FcRγ in signaling through NKR-P1, because NKR-P1 bind only to FcRγ dimers but not to CD3ζ homodimers or CD3ζ-FcRγ heterodimers. In contrast, T cells show no functional defect in γ−/− mice probably because the TCR complexes can utilize both CD3ζ and FcRγ and CD3ζ can replace for FcRγ in the TCR complexes of γ−/− mice.
FcγRIII which is associated with FcRγ is known to activate NK cells upon cross-linking. Therefore, there is a possibility that NKR-P1 associates with FcγRIII on the cell surface and delivers activating signal through FcRγ which is coupled with FcγRIII. However, this is unlikely because of two reasons. One is that FcγRIII was not expressed on the cell surface of CTLL-2 (data not shown), whereas the association between FcRγ and NKR-P1 was readily detected in the CTLL-2 (Fig. 1). Second, we used the (Fab)′2 fragment of anti-NK1.1 mAb for cross-linking of NKR-P1 and thus it is unlikely that immobilized anti-NK1.1 mAb activated FcγRIII. Taken together, the association of FcRγ with NKR-P1 is essentially independent of FcγRIII.
Recent analysis revealed that activation of NK cells is negatively regulated by several killer inhibitory receptors (KIR) possessing immunoreceptor tyrosine-based inhibition motif (ITIM) (28, 29). These receptors such as p58, NKG2, and Ly49 recognize specific MHC class I molecule and deliver inhibitory signal for the cytotoxicity by NK cells (30–32). Furthermore, association of SHP-1 phosphatase with ITIM is important for the inhibition of NK cell activation (28, 29). Although the coprecipitation of CD3ζ or FcRγ with p58 was suggested, functional significance remained unclear (33). On the other hand, only a few receptors which deliver positive signal for NK cell activation have been identified such as NKR-P1 and CD94/NKG2-C (34) and the molecular mechanism of NK cell activation through these receptors remains unclear. Under these circumstances, it is noteworthy that FcRγ, a signal transducing chain possessing immunoreceptor tyrosine-based activation motif (ITAM), is involved in signal transduction through NKR-P1. Because Syk tyrosine kinase interacts with the phosphorylated ITAM of FcRγ and is involved in NK cell activation through FcR (35, 36), it is likely that Syk also mediates the activating signal through NKR-P1 in NK cells. Furthermore, it has been shown that Lck directly associates with NKR-P1 (14). Taken together, both Syk and Lck seem to play an important role in mediating activation signals through NKR-P1 in NK cells.
Although the NKR-P1 molecule belongs to the C-type lectin superfamily and has been shown to have affinity to specific carbohydrates (12), the exact function of NKR-P1 has remained unclear. This is partly because antibodies against mouse or rat NKR-P1 do not block cytotoxicity of NK cells against YAC-1, a NK-sensitive target cell line. In addition, NK cells from γ−/−− mice showed normal cytotoxicity against representative NK targets (data not shown). However, the failure of blocking with anti–NKR-P1 mAb can be explained by the possible redundancy of NKR-P1 family molecules on NK cells. Indeed, recent observation that introduction of NKR-P1 into a NKR-P1–deficient NK cell line restored cytotoxicity against specific tumor cell targets and the restored cytotoxicity was blocked by anti– NKR-P1 mAb (11) strongly suggested that NKR-P1 is involved in the recognition of specific target molecules on NK cells. A couple of reports demonstrating that anti-NK1.1 mAb blocked cytotoxicity of NK cells against specific targets also support this idea (37, 38). Further analysis of NK cells in γ−/− mice may elucidate novel function of NKR-P1 molecule in the host defence.
Acknowledgments
We would like to thank Dr. J. Hamuro for providing IL-2, Ms. C. Sakuma and Ms. R. Siina for technical assistance, and Ms. H. Yamaguchi for secretarial assistance.
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education to H. Arase and T. Saito.
Footnotes
The first two authors contributed equally to this work.
Abbreviations used in this paper: γ−/−, FcRγ-deficient; ITAM, immunoreceptor tyrosine-based activation motif; ITIM, immunoreceptor tyrosine-based inhibition motif; KIR, killer inhibitory receptors.
References
- 1.Scott P, Trinchieri G. The role of natural killer cells in host-parasite interactions. Curr Opin Immunol. 1995;7:34–40. doi: 10.1016/0952-7915(95)80026-3. [DOI] [PubMed] [Google Scholar]
- 2.Yokoyama WM. Natural killer cell receptors. Curr Opin Immunol. 1995;7:110–120. doi: 10.1016/0952-7915(95)80036-0. [DOI] [PubMed] [Google Scholar]
- 3.Raulet DH, Held W. Natural killer cell receptors: the offs and ons of NK cell recognition. Cell. 1995;82:697–700. doi: 10.1016/0092-8674(95)90466-2. [DOI] [PubMed] [Google Scholar]
- 4.Chambers WH, Vujanovic NL, DeLeo AB, Olszowy MW, Herberman RB, Hiserodt JC. Monoclonal antibody to a triggering structure expressed on rat natural killer cells and adherent lymphokine-activated killer cells. J Exp Med. 1989;169:1373–1389. doi: 10.1084/jem.169.4.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giorda R, Rudert WA, Vavassori C, Chambers WH, Hiserodt JC, Trucco M. NKR-P1, a signal transduction molecule on natural killer cells. Science. 1990;249:1298–1300. doi: 10.1126/science.2399464. [DOI] [PubMed] [Google Scholar]
- 6.Giorda R, Trucco M. Mouse NKR-P1: a family of genes selectively coexpressed in adherent lymphokine-activated killer cells. J Immunol. 1991;147:1701–1708. [PubMed] [Google Scholar]
- 7.Lanier LL, Chang C, Phillips JH. Human NKR-P1A. A disulfide-linked homodimer of the C-type lectin superfamily expressed by a subset of NK and T lymphocytes. J Immunol. 1994;153:2417–2428. [PubMed] [Google Scholar]
- 8.Ryan JC, Turck J, Niemi EC, Yokoyama WM, Seaman WE. Molecular cloning of the NK1.1 antigen, a member of the NKR-P1 family of natural killer cell activation molecules. J Immunol. 1992;149:1631–1635. [PubMed] [Google Scholar]
- 9.Karlhofer FM, Yokoyama WM. Stimulation of murine natural killer (NK) cells by a monoclonal antibody specific for the NK1.1 antigen. IL-2 activated NK cells possess additional specific stimulation pathways. J Immunol. 1991;146:3662–3673. [PubMed] [Google Scholar]
- 10.Arase H, Arase N, Saito T. Interferon-γ production by natural killer cells and NK1.1+T cells upon NKR-P1 cross-linking. J Exp Med. 1996;183:2391–2396. doi: 10.1084/jem.183.5.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryan JC, Niemi EC, Nakamura MC, Seaman WE. NKR-P1A is a target-specific receptor that activates natural killer cell cytotoxicity. J Exp Med. 1995;181:1911–1915. doi: 10.1084/jem.181.5.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bezouska K, Yuen C-T, O'Brien J, Childs RA, Chai W, Lawson AM, Drbal K, Fiserova A, Pospisil M, Feizi T. High affinity oligosaccharide ligands for NKR-P1 protein that elicits natural killer cell activation and cytotoxicity. Nature. 1994;372:150–157. doi: 10.1038/372150a0. [DOI] [PubMed] [Google Scholar]
- 13.Ryan JC, Niemi EC, Goldfien RD, Hiserodt JC, Seaman WE. NKR-P1, an activating molecule on rat natural killer cells, stimulates phosphoinositide turnover and a rise in intracellular calcium. J Immunol. 1991;147:3244–3250. [PubMed] [Google Scholar]
- 14.Campbell KS, Giorda R. The cytoplasmic domain of rat NKR-P1 receptor interacts with the N-terminal domain of p56(lck) via cysteine residues. Eur J Immunol. 1997;27:72–77. doi: 10.1002/eji.1830270111. [DOI] [PubMed] [Google Scholar]
- 15.Ledermann B, Burki K. Establishment of a germ-line competent C57BL/6 embryonic stem cell line. Exp Cell Res. 1991;197:254–258. doi: 10.1016/0014-4827(91)90430-3. [DOI] [PubMed] [Google Scholar]
- 16.van Vugt MJ, Heijnen AF, Capel PJ, Park SY, Ra C, Saito T, Verbeek JS, van de Winkel JG. FcRγ-chain is essential for both surface expression and function of human FcγRI (CD64) in vivo. Blood. 1996;87:3593–3599. [PubMed] [Google Scholar]
- 17.Poole A, Gibbins JM, Turner M, van Vugt MJ, van de Winkel JGJ, Saito T, Tybulewicz VLJ, Watson SP. The Fc receptor γ-chain and the tyrosine kinase syk are essential for activation of mouse platelets by collagen. EMBO (Eur Mol Biol Organ) J. 1997;16:2333–2341. doi: 10.1093/emboj/16.9.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arase H, Arase N, Saito T. Fas-mediated cytotoxicity by freshly isolated natural killer cells. J Exp Med. 1995;181:1235–1238. doi: 10.1084/jem.181.3.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ono S, Ohno H, Saito T. Rapid turnover of the CD3ζ chain independent of the TCR-CD3 complex in normal T cells. Immunity. 1995;2:639–644. doi: 10.1016/1074-7613(95)90008-x. [DOI] [PubMed] [Google Scholar]
- 20.Arase H, Arase-Fukushi N, Good RA, Onoé K. Lymphokine-activated killer cell activity of CD4− CD8− TCRαβ+thymocytes. J Immunol. 1993;151:546–555. [PubMed] [Google Scholar]
- 21.Orloff DG, Ra C, Frank SJ, Klausner RD, Kinet JP. Family of disulfide-linked dimers containing the ζ and η chains of the T-cell receptor and the γ chain of Fc receptors. Nature. 1990;347:189–191. doi: 10.1038/347189a0. [DOI] [PubMed] [Google Scholar]
- 22.Arase H, Ono S, Arase N, Park SY, Wakizaka K, Watanabe H, Ohno H, Saito T. Developmental arrest of NK1.1+ T cell receptor (TCR)-α/β+ T cells and expansion of NK1.1+ TCR-γ/δ+T cell development in CD3ζ-deficient mice. J Exp Med. 1995;182:891–895. doi: 10.1084/jem.182.3.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blank U, Ra C, Miller L, White K, Metzger H, Kinet JP. Complete structure and expression in transfected cells of high affinity IgE receptor. Nature. 1989;337:187–189. doi: 10.1038/337187a0. [DOI] [PubMed] [Google Scholar]
- 24.Ra C, Jouvin M-H, Blank U, Kinet JP. A macrophage Fcγ receptor and the mast cell receptor for IgE share an identical subunit. Nature. 1989;341:752–754. doi: 10.1038/341752a0. [DOI] [PubMed] [Google Scholar]
- 25.Ravetch JV, Kinet JP. Fc receptors. Ann Rev Immunol. 1991;9:457–492. doi: 10.1146/annurev.iy.09.040191.002325. [DOI] [PubMed] [Google Scholar]
- 26.Ohno H, Ono S, Hirayama N, Shimada S, Saito T. Preferential usage of the Fc receptor γ chain in the T cell antigen receptor complex by γ/δ T cells localized in epithelia. J Exp Med. 1994;179:365–369. doi: 10.1084/jem.179.1.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park SY, Arase H, Wakizaka K, Hirayama N, Masaki S, Sato S, Ravetch JV, Saito T. Differential contribution of the FcR-γ chain to the surface expression of the T cell receptor among T cells localized in epithelia: analysis of FcR-γ-deficient mice. Eur J Immunol. 1995;25:2107–2110. doi: 10.1002/eji.1830250746. [DOI] [PubMed] [Google Scholar]
- 28.Olcese L, Lang P, Vely F, Cambiaggi A, Marguet D, Blery M, Hippen KL, Biassoni R, Moretta A, et al. Human and mouse killer-cell inhibitory receptors recruit PTP1C and PTP1D protein tyrosine phosphatases. J Immunol. 1996;156:4531–4534. [PubMed] [Google Scholar]
- 29.Burshtyn DN, Scharenberg AM, Wagtmann N, Rajagopalan S, Berrada K, Yi T, Kinet JP, Long EO. Recruitment of tyrosine phosphatase HCP by the killer cell inhibitory receptor. Immunity. 1996;4:77–85. doi: 10.1016/s1074-7613(00)80300-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colonna M, Samaridis J. Cloning of immunoglobulin-superfamily members associated with HLA-C and HLA-B recognition by human natural killer cells. Science. 1995;268:405–408. doi: 10.1126/science.7716543. [DOI] [PubMed] [Google Scholar]
- 31.Phillips JH, Gumperz JE, Parham P, Lanier LL. Superantigen-dependent, cell-mediated cytotoxicity inhibited by MHC class I receptors on T lymphocytes. Science. 1995;268:403–405. doi: 10.1126/science.7716542. [DOI] [PubMed] [Google Scholar]
- 32.Karlhofer FM, Ribaudo RK, Yokoyama WM. MHC class I alloantigen specificity of Ly-49+IL-2 activated natural killer cells. Nature. 1992;358:66–70. doi: 10.1038/358066a0. [DOI] [PubMed] [Google Scholar]
- 33.Bottino C, Vitale M, Olcese L, Sivori S, Morelli L, Augugliaro A, Ciccone E, Moretta L, Moretta A. The human natural killer cell receptor for major histocompatibility complex class I molecules. Surface modulation of p58 molecules and their linkage to CD3ζ chain, FcεRIγ chain and the p56lckkinase. Eur J Immunol. 1994;24:2527–2534. doi: 10.1002/eji.1830241040. [DOI] [PubMed] [Google Scholar]
- 34.Mason LH, Anderson SK, Yokoyama WM, Smith HR, Winkler-Pickett R, Ortaldo JR. The Ly-49D receptor activates murine natural killer cells. J Exp Med. 1996;184:2119–2128. doi: 10.1084/jem.184.6.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ting AT, Dick CJ, Schoon RA, Karnitz LM, Abraham RT, Leibson PJ. Interaction between lck and syk family tyrosine kinases in Fcγ receptor-initiated activation of natural killer cells. J Biol Chem. 1997;270:16415–16421. doi: 10.1074/jbc.270.27.16415. [DOI] [PubMed] [Google Scholar]
- 36.Shiue L, Zoller MJ, Brugge JS. Syk is activated by phosphotyrosine-containing peptides representing the tyrosine-based activation motifs of the high affinity receptor for IgE. J Biol Chem. 1995;270:10498–10502. doi: 10.1074/jbc.270.18.10498. [DOI] [PubMed] [Google Scholar]
- 37.Kung SKP, Miller RG. The NK1.1 antigen in NK-mediated F1antiparent killing in vitro. J Immunol. 1995;154:1624–1633. [PubMed] [Google Scholar]
- 38.Takeda K, Dennert G. Demonstration of MHC class I-specific cytolytic activity in IL-2-activated NK1+CD3+cells and evidence of usage of T and NK cell receptors. Transplantation. 1994;58:496–504. doi: 10.1097/00007890-199408270-00017. [DOI] [PubMed] [Google Scholar]