Abstract
LMP2, LMP7, and MECL are interferon γ–inducible catalytic subunits of vertebrate 20S proteasomes, which can replace constitutive catalytic subunits (delta, X, and Z, respectively) during proteasome biogenesis. We demonstrate that MECL requires LMP2 for efficient incorporation into preproteasomes, and preproteasomes containing LMP2 and MECL require LMP7 for efficient maturation. The latter effect depends on the presequence of LMP7, but not on LMP7 catalytic activity. This cooperative mechanism favors the assembly of homogeneous “immunoproteasomes” containing all three inducible subunits, suggesting that these subunits act in concert to enhance proteasomal generation of major histocompatibility complex class I–binding peptides.
Proteasomes are multisubunit multicatalytic proteases that are responsible for the majority of nonlysosomal protein degradation within eukaryotic cells (1), and have a central role in the generation of peptides presented by MHC class I molecules (2). The 20S catalytic core (20S proteasome) is composed of 28 subunits assembled in four stacked seven-membered rings (3). The outer rings contain seven different noncatalytic α-type subunits, and the inner rings contain seven different β-type subunits, three of which are catalytic (delta, X, and Z; reference 4) (alternative nomenclature for vertebrate proteasome subunits [3]: iota, α1; C3, α2; C9, α3; C6, α4; zeta, α5; C2, α6; C8, α7; delta, Y or β1; LMP2, β1i; Z, β2; MECL, β2i; C10, β3; C7, β4; X, MB1 or β5; LMP7, β5i; C5, β6; N3, beta or β7). In addition to seven constitutively synthesized β subunits, vertebrates have three IFN-γ–inducible β subunits (LMP2, LMP7, and MECL), the former two being encoded in the MHC (5–9). All three inducible subunits have removable presequences and are catalytically active (7–11). Each inducible subunit is homologous with a constitutive catalytic subunit (LMP2/delta, LMP7/X, and MECL/Z), and can replace its homologue during proteasome assembly (7–9, 12). The inducible subunits appear to be responsible for altered peptidase specificities in IFN-γ–treated cells (13–15), transfected cells (16–18), and cells from LMP7−/− and LMP2−/− mice (19, 20). Presentation of certain antigens is diminished in LMP2−/− and LMP7−/− mice (20, 21), and in the case of LMP7−/− mice, MHC class I expression is reduced (21). These results support a role for inducible subunits in enhancing proteasomal generation of MHC class I–binding peptides.
The assembly of 20S proteasomes and the mechanism by which inducible subunits replace constitutive homologues are poorly understood. We have recently characterized proteasome assembly in mouse cells expressing both inducible and constitutive catalytic subunits using an antibody to an α subunit, anti-C8, that immunoprecipitates only 12-16S preproteasomes (22). These catalytically inactive precursor complexes (∼300 kD) contain all seven α subunits and some unprocessed β subunits. They appear to assemble in two stages, with certain unprocessed β subunits (pre-Z, pre-LMP2, pre-MECL, C10, and C7) being incorporated before others (pre-C5, pre-delta, and pre-LMP7). Maturation of preproteasomes to 20S proteasomes (∼700 kD) involves the juxtaposition of two preproteasomes at the β ring interface (3), with β subunit presequences being removed coincident with completion of assembly (23, 24). It is unknown whether the incorporation of inducible subunits and their homologues into proteasomes depends only on relative expression levels, or whether certain proteasome forms are assembled preferentially.
Materials and Methods
Episomal Expression Vectors.
pCEP4 (ampicillinr, hygromycinr) and pREP9 (ampicillinr, neomycinr) were purchased from Invitrogen (Carlsbad, CA). pCEP9 (ampicillinr, neomycinr) was constructed from three DNA fragments: SalI–XbaI (1,377 to 2) from pREP9, XbaI–BamHI (1 to 405) from pCEP4, and BamHI–SalI (405 to 1,315) from pCEP4. pCEP9 is similar to pCEP4 except the hygromycin resistance gene replaces the neomycin resistance gene. pCEP9.LMP2 was constructed by inserting at HindIII– BamHI a full-length human LMP2 cDNA, obtained from H.O. McDevitt (Stanford University School of Medicine, Stanford, CA) (25). pCEP4.LMP7 was constructed by inserting at KpnI– BamHI a full-length human LMP7 cDNA, obtained from T. Spies (Fred Hutchinson Cancer Research Center, Seattle, WA) (10). pCEP4.LMP7E1 was constructed using synthetic oligonucleotides to change only the presequence of LMP7E2. The promoter and translation control sequences upstream of the start codon were unchanged; thus, transcription and translation efficiencies were expected to be similar to LMP7E2. pCEP4.LMP7(T1A), pCEP4.LMP7(K33A), pCEP9.LMP2(T1A), and pCEP9.LMP2 (K33A) were constructed by site-directed mutagenesis using the Altered sites® II in vitro mutagenesis system (Promega, Madison, WI) or the Quickchange™ site-directed mutagenesis kit (Stratagene Corp., La Jolla, CA).
Antibodies.
MCP21 is a mouse monoclonal antibody that recognizes human C3 and immunoprecipitates C3-containing 20S proteasomes and 12-16S preproteasomes (26). MCP21 ascites fluid was obtained from K.B. Hendil and the hybridoma expressing MCP21 was obtained from the European Collection of Animal Cell Cultures (Salisbury, Wiltshire, UK). Polyclonal antisera recognizing LMP2, LMP7, MECL, delta, Z, C8, C9, or TAP2 were from rabbits immunized with recombinant mouse subunits (or rat for anti-C9). Rabbit polyclonal antisera raised against human LMP2 and LMP7 were obtained from H.O. McDevitt (Stanford University School of Medicine, Stanford, CA) (25). Anti-X (P93250), obtained from K.B. Hendil (August Krogh Institute, University of Copenhagen, Copenhagen, Denmark), is a rabbit polyclonal antiserum raised against human X (18).
Cell Culture.
T2 cells, obtained from P. Cresswell (Howard Hughes Medical Institute, Yale University School of Medicine, New Haven, CT) (27), were transfected with episomal expression vectors by electroporation (250 V and 500 μFd) in serum-free RPMI at 2.5 μg DNA/5 × 106 cells. Transfected cells were grown in RPMI with 10% FCS (R10) for 48 h before the addition of selection antibiotic(s). Selected cells were maintained in RPMI with 5% calf serum with either G418 at 0.8 mg activity/ ml (GIBCO BRL, Gaithersburg, MD) for pCEP9 and/or hygromycin at 360 U/ml (Calbiochem Corp., La Jolla, CA) for pCEP4. Con A–stimulated T cell blasts were prepared by growing mouse spleen lymphocytes, isolated using lympholyte M, in R10 with Con A (5 μg/ml) for 72 h.
Immunoprecipitation and Immunoblotting.
T2 cells were lysed with 1% NP-40 in 20 mM Tris, pH 7.6, 10 mM EDTA, and 100 mM NaCl. Proteasomes were immunoprecipitated from postnuclear supernatants with MCP21 (1 μg/106 cells) and protein G–Sepharose (250 μg/106 cells). Immunoprecipitates were boiled in 2 × SDS sample buffer, subjected to 12.5% SDS-PAGE, and electroblotted onto polyvinylidene difluoride paper at 500 mA (milliamps) for 90 min. Specific proteins were detected using antisera or MCP21 as primary antibodies, alkaline phosphatase conjugated goat anti–rabbit or goat anti–mouse as secondary antibodies, and 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium for color development. Mouse spleen Con A–stimulated T cell blasts were lysed with 1% digitonin and postnuclear supernatants were boiled in 6 × SDS sample buffer and immunoblotted as above.
Sucrose Gradient Centrifugation.
NP-40 cell lysates (1 ml) were layered onto 10-ml sucrose gradients (15–35% in NP-40 lysis buffer) and ultracentrifuged in a rotor (SW40 Ti; Beckman, Fullerton, CA) at 40,000 rpm for 20 h at 4°C. Fractions (1 ml × 11) were collected from the bottom of the tubes.
Metabolic Labeling, Immunoprecipitation, and Two-dimensional Gel Electrophoresis.
Con A–stimulated T cell blasts derived from mouse spleens (∼20 × 106 cells) were labeled with 0.5 mCi [35S]methionine/cysteine for 45 min. Half the cells were lysed with 0.5% NP-40 and the rest were chased for 8 h in R10 containing 4 mM cold methionine and then lysed. 12-16S preproteasomes were immunoprecipitated with anti-C8, whereas 20S proteasomes and a small subset of 12-16S preproteasomes were immunoprecipitated with anti-C9 (22). Immunoprecipitates were resuspended in nonequilibrium pH gradient electrophoresis (NEPHGE)1 sample buffer and subjected to two-dimensional NEPHGE-PAGE and autoradiography (22).
Results
LMP7 Mediates Efficient LMP2 Processing in Transfected T2 Cells.
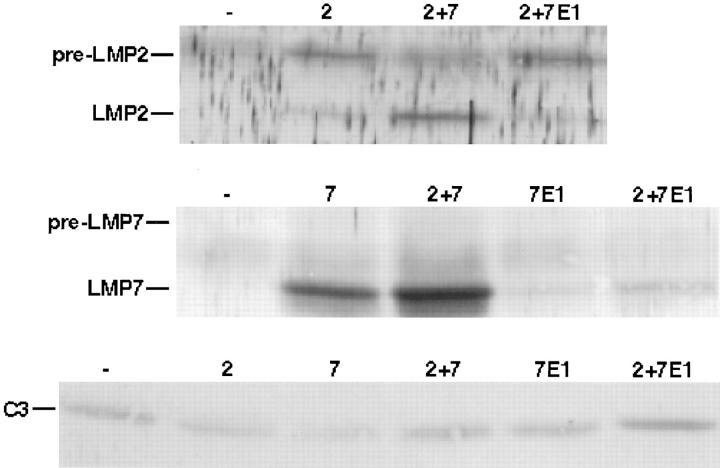
T2 is a human lymphoblastoid cell line with a single copy of chromosome 6, which has a deletion in the MHC class II region that includes the genes for LMP2 and LMP7 (27, 28). We observed that proteasomes from T2 cells transfected with LMP2 (T2.LMP2) contained primarily unprocessed LMP2 (pre-LMP2) (Fig. 1). The relative amount of pre-LMP2 decreased when cells were overgrown; however, we always detected more of the precursor than the mature form (data not shown), suggesting that processing of LMP2 was inefficient in these cells. A similar observation has been made using .174 lymphoblasts (which also lack LMP2 and LMP7) transfected with LMP2 (16). To determine whether LMP7 could mediate efficient processing of LMP2, T2 cells were transfected with both LMP2 and LMP7 (T2.LMP2/LMP7). Proteasomes from these double transfectants contained predominantly processed LMP2 (Fig. 1), indicating that LMP7 can indeed enhance the efficiency of LMP2 processing. In contrast, there was no effect of LMP2 on the efficiency of LMP7 processing, as proteasomes from both double transfectants as well as T2 cells transfected with LMP7 alone (T2.LMP7) contained primarily processed LMP7. Immunoblotting with polyclonal antisera raised against human LMP2 and LMP7 produced identical results but greater background staining (data not shown). Consequently, all of the anti-LMP2 and anti-LMP7 immunoblots presented in this report use antisera raised against mouse subunits. Recently, Groettrup et al. did not observe that efficient LMP2 processing required LMP7 in transfected T2 cells (29). Their results may differ from ours because they used an expression vector (pSG5) that leads to significant overexpression of LMP2, and they used mouse LMP2 and LMP7 in this human cell line.
Figure 1.
Effect of LMP7 on processing of LMP2 in transfected T2 cells. Proteasomes were immunoprecipitated with MCP21 from lysates (postnuclear supernatants) of T2 cells transfected with LMP2 (2), LMP2 and LMP7 (2 + 7 ), LMP2 and LMP7E1 (2 + 7E1), LMP7 (7), and LMP7E1 (7E1). Specific subunits were visualized by immunoblotting after SDS-PAGE. Anti-LMP2 and anti-LMP7 antisera were raised against mouse subunits and cross-react with human subunits. Immunoprecipitates from 5 × 106 cells were loaded per lane. The C3 immunoblot demonstrates the relative amounts of proteasomes in each sample.
LMP7E1 Is Inefficiently Incorporated into Proteasomes and Fails to Mediate Efficient LMP2 Processing in Transfected T2 Cells.
There are two forms of human LMP7 (E1 and E2) which result from alternative first exon usage (10). These two forms have different amino acid sequences only in their presequences (NH2 terminus to residue −24), with the mature proteins being identical. LMP7E2 is the predominant form expressed in tissues and cell lines (10), and this form of LMP7 is used throughout our work except where noted. It has been shown that LMP7E1 is inefficiently incorporated into proteasomes in transfected HeLa cells (24). We obtained a similar result using transfected T2 cells (T2.LMP7E1; Fig. 1). Interestingly, LMP2 was inefficiently processed in T2.LMP2/LMP7E1 cells where LMP7E1 was incorporated inefficiently. These results indicate that the nature of the LMP7 presequence is important for LMP7 incorporation into proteasomes, and LMP7 incorporation is necessary for efficient LMP2 processing. We also found low steady state levels of free LMP7E1 in transfected T2 cells (data not shown), suggesting that LMP7E1 is relatively unstable when not incorporated into proteasomes.
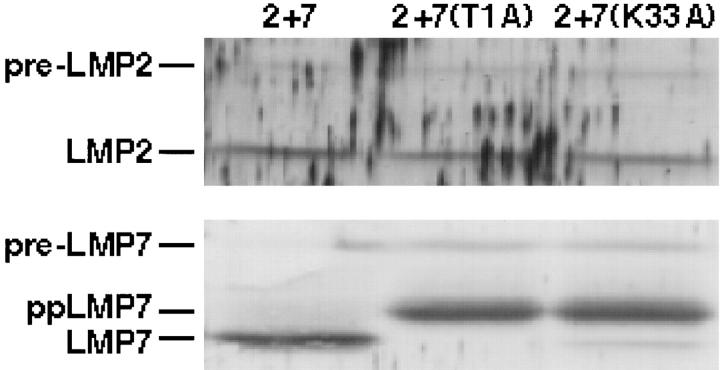
LMP7 Catalytic Activity Is Not Required for Efficient LMP2 Processing in Transfected T2 Cells.
To determine whether catalytic activity of LMP7 is required for its effect on LMP2 processing, catalytically inactive forms of LMP7 were produced by mutating active site residues threonine 1 or lysine 33 to alanine (T1A or K33A). Neither of these point mutations diminished incorporation of LMP7 into proteasomes, nor did they reduce the ability of LMP7 to enhance LMP2 processing (Fig. 2). Both mutants failed to undergo complete autocatalytic processing (30–32), resulting in higher molecular weight partially processed forms, as seen with active site mutations of other catalytic β subunits (31, 32). We also produced catalytically inactive forms of LMP2 (T1A or K33A), which were unprocessed in the absence of LMP7 (like wild-type LMP2) and partially processed in the presence of either LMP7 or its catalytically inactive forms. LMP2 inactivity did not affect LMP7 processing (data not shown). Taken together, these results demonstrate that catalytic activity of LMP7 is not required for its effect on LMP2 processing, nor is LMP2 catalytic activity required for this effect.
Figure 2.
Effect of LMP7 active site mutations on LMP2 processing. Proteasomes were immunoprecipitated with MCP21 from lysates of T2 cells transfected with LMP2 and LMP7 (2 + 7), LMP2 and LMP7(T1A) [2 + 7(T1A)], and LMP2 and LMP7(K33A) [2 + 7(K33A)]. Specific subunits were visualized by immunoblotting after SDS-PAGE. Immunoprecipitates from 107 cells were loaded per lane. ppLMP7, partially processed LMP7. Partial loss of the pre-LMP7 band in lane 2 + 7 is due to poor transfer as a result of an air bubble.
Pre-LMP2 Accumulates in Preproteasomes in the Absence of LMP7 in Transfected T2 Cells.
Removal of β subunit presequences is associated with maturation of 20S proteasomes (23, 24). Therefore, we hypothesized that pre-LMP2, which accumulates in T2.LMP2 cells, remains in preproteasomes. To test this, cell extracts were fractionated on sucrose gradients and C3-containing proteasomes (both 20S proteasomes and 12-16S preproteasomes) were immunoprecipitated from each fraction and analyzed by immunoblotting for specific subunits (Fig. 3). Indeed, pre-LMP2 was found exclusively in lower mol wt fractions 4 and 5, comigrating with a portion of C3, consistent with α subunit–containing preproteasomes. The small amount of processed LMP2 in these cells (fraction 2) comigrated with other processed β subunits (delta and X), as well as the majority of C3, consistent with 20S proteasomes. These results agree with previous demonstrations of pre-LMP2 in preproteasomes from transfected T2 cells overexpressing mouse LMP2 (23, 32). Of note, there was no detectable pre-delta or pre-X in the pre-LMP2–containing fractions. In contrast, when LMP7 was coexpressed with LMP2 (T2.LMP2/ LMP7), processed LMP2 predominated and comigrated with processed LMP7 and the bulk of C3, consistent with 20S proteasomes (Fig. 3). These results demonstrate that preproteasomes containing pre-LMP2 accumulate in the absence of LMP7, despite the presence of its homologue, X. In contrast, pre-LMP2–containing preproteasomes do not accumulate in the presence of LMP7, suggesting a strong preference for the subsequent incorporation of LMP7 rather than X into these assembling proteasomes. It is important to note that LMP7 can be incorporated into 20S proteasomes in the absence of LMP2 (Fig. 1 and data not shown), suggesting that preproteasomes lacking pre-LMP2 do not have a strong preference for the incorporation of X or LMP7.
Figure 3.
Sucrose gradient fractionation of proteasomes from transfected T2 cells. Lysates of 4 × 107 cells were separated on sucrose gradients, and then mature and precursor proteasomes were immunoprecipitated from individual fractions with MCP21. Specific subunits were visualized by immunoblotting after SDS-PAGE, with one-quarter of each immunoprecipitation loaded per lane. Fractions 1–6 out of 11 are shown, with fraction 1 representing the bottom of the gradient.
Pre-LMP2 and Pre-MECL Accumulate in Spleen Cells from LMP7−/− Mice; The Level of MECL Is Reduced in Spleen Cells from LMP2−/− Mice.
Since T2 cells lack a large portion of the MHC class II region (27), they are deficient in not only LMP2 and LMP7, but also other genes involved in antigen processing (TAP1 and TAP2) and potentially other as yet undiscovered genes. Thus, we were interested in determining whether LMP7 is required for efficient LMP2 processing in other model systems, such as mice with targeted deletions of LMP2 or LMP7. Indeed, consistent results were obtained using Con A–stimulated T cell blasts from spleens of B10 (wild type), LMP7−/−, and LMP2−/− mice (Fig. 4). Immunoblots reflecting steady-state subunit levels demonstrated that all of the detectable LMP2 in wild-type cells was processed, whereas the majority of LMP2 in cells from LMP7−/− mice was unprocessed. Comparable to T2 transfectants, LMP2 deficiency had no effect on LMP7 processing. Interestingly, using an antibody against mouse MECL, we found that the level of processed MECL was dramatically reduced in the absence of either LMP7 or LMP2. Furthermore, unprocessed MECL accumulated in the absence of LMP7, similar to the accumulation of unprocessed LMP2 in these cells, indicating that absence of LMP7 results in inefficient processing of not only LMP2 but also MECL. Groettrup et al. have also noted that MECL incorporation into proteasomes is enhanced by overexpression of mouse LMP2 in transfected T2 cells, and LMP2 incorporation is enhanced by overexpression of MECL in transfected mouse B8 fibroblasts (29). The reduced level of MECL in cells from LMP2−/− mice is possibly a consequence of increased degradation of free MECL secondary to its inefficient incorporation into proteasomes in the absence of LMP2. Notably, reduced levels of each processed inducible subunit correlated with increased levels of each respective constitutive homologue. For example, the amount of delta was increased not only in the absence of LMP2 but also when LMP2 was processed inefficiently in cells from LMP7−/− mice. Similarly, the amount of Z was increased when processed MECL was reduced due to lack of either LMP7 or LMP2. The amount of X was increased only when LMP7 was absent.
Figure 4.
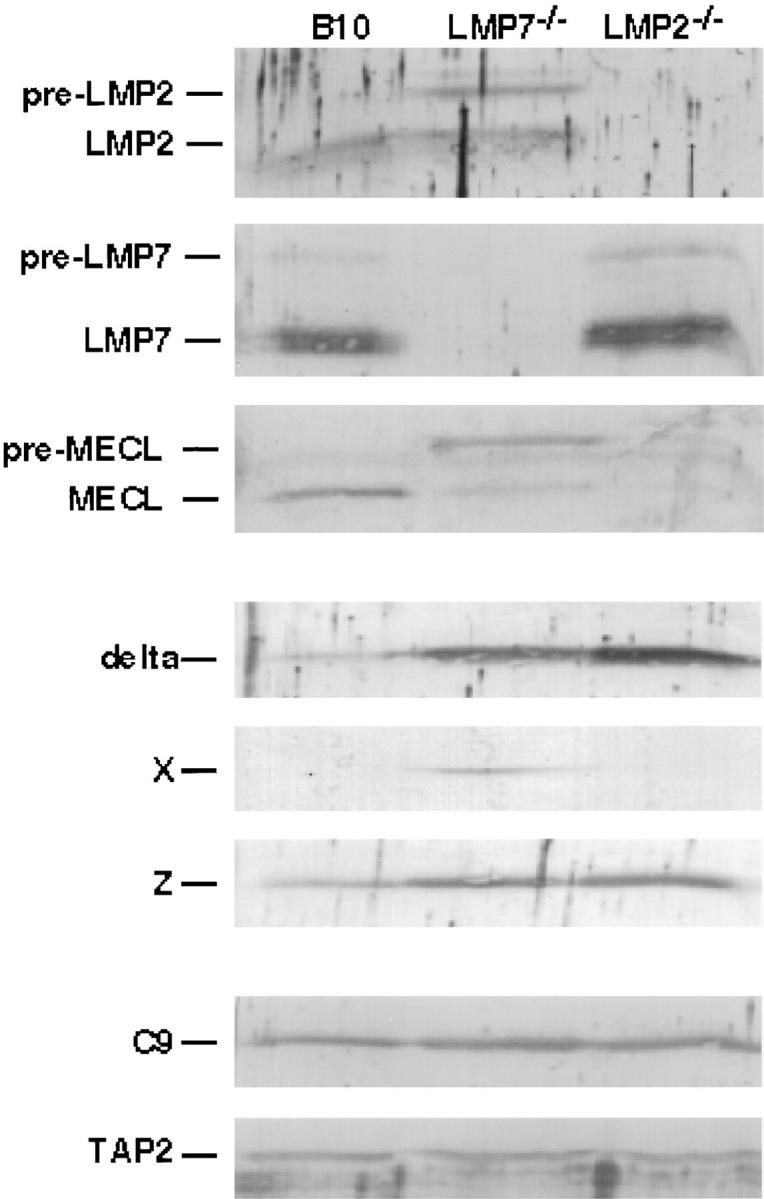
Relative levels of proteasome subunits in spleen cells from B10 (wild type), LMP7−/−, and LMP2−/− mice. Lysates of mouse spleen Con A–stimulated T cell blasts (1.5 × 106/lane) were subjected to SDS-PAGE and specific proteasome subunits were visualized by immunoblotting. The C9 and TAP2 immunoblots demonstrate the relative amounts of proteasomes and total protein present in each sample.
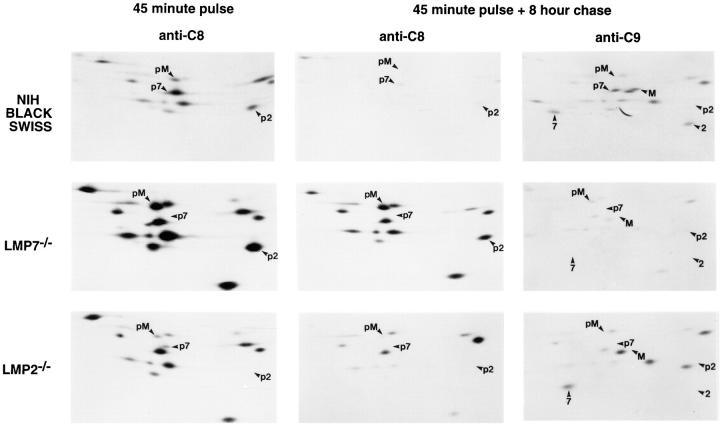
Proteasomes Are Assembled Inefficiently in Spleen Cells from LMP7−/− and LMP2−/− Mice.
To confirm the presence of unprocessed LMP2 and unprocessed MECL in preproteasomes from LMP7−/− mice, and to assess kinetics of proteasome assembly in both LMP7−/− and LMP2−/− mice, Con A–stimulated T cell blasts derived from spleens of NIH Black Swiss (wild type), LMP7−/−, and LMP2−/− mice were subjected to metabolic pulse-chase labeling (Fig. 5). 12-16S preproteasomes and 20S proteasomes were analyzed on two-dimensional gels after immunoprecipitation with anti-C8 or anti-C9, respectively (22). Several salient observations can be made from this experiment. First, overall proteasome assembly in cells from LMP7−/− mice was inefficient. Despite an apparent increase in the synthesis of proteasome subunits in these cells (45 min pulse, anti-C8), the percentage of radioactive material chasing into 20S proteasomes (45 min pulse + 8 h chase, anti-C9) was small as compared to wild type. Second, preproteasomes synthesized in cells from LMP7−/− mice appeared to be longer lived than wild type (45 min pulse + 8 h chase, anti-C8). Third, preproteasomes that accumulated in these cells contained significant amounts of pre-LMP2 (spot p2) and pre-MECL (spot pM) (45 min pulse + 8 h chase, anti-C8), yet very little of these particular subunits were processed and chased into 20S proteasomes (spots 2 and M) (45 min pulse + 8 h chase, anti-C9), indicating inefficient completion of assembly of preproteosomes containing pre-LMP2 and pre-MECL in the absence of LMP7, consistent with immunoblotting results (Fig. 4). The increased synthesis of proteasome subunits and relatively inefficient assembly of 20S proteasomes in cells from LMP7−/− mice has been observed in three separate experiments (data not shown). Fourth, preproteasomes in cells from LMP2−/− mice contained a small amount of pre-MECL (spot pM) (45 min pulse), and were longer lived than wild type (45 min pulse + 8 h chase, anti-C8). However, MECL did not chase into 20S proteasomes in these cells (spot M) (45 min pulse + 8 h chase, anti-C9), whereas processed Z, which is the dark spot just to the left of the MECL position, is incorporated into 20S proteasomes. This indicates that MECL, unlike its homologue Z, is inefficiently incorporated into proteasomes in the absence of LMP2, consistent with immunoblotting results (Fig. 4) and recent studies of Groettrup et al. (29).
Figure 5.
Pulse-chase labeling of proteasomes synthesized in spleen cells from NIH Black Swiss (wild type), LMP7−/−, and LMP2−/− mice. Equal numbers of mouse spleen Con A–stimulated T cell blasts were labeled with [35S]methionine/cysteine for 45 min and then harvested (45 minute pulse) or chased for 8 h (45 minute pulse + 8 hour chase). At the end of each incubation, cells were lysed and proteasomes were immunoprecipitated from half of each lysate with anti-C8, which recognizes only 12-16S preproteasomes (anti-C8), and from the other half of each lysate with anti-C9, which recognizes 20S proteasomes and a small subset of 12-16S preproteasomes (anti-C9) (22). (Anti-C9 immunoprecipitates after a 45-min pulse demonstrated little radioactivity in 20S proteasomes and are not shown.) Proteasome subunits were separated by two dimensional NEPHGE-PAGE and visualized by autoradiography. Spots corresponding to inducible subunits and their precursors, as identified previously (22), are labeled (p2, pre-LMP2; 2, LMP2; p7, pre-LMP7; 7, LMP7; pM, pre-MECL; M, MECL). Note that pre-LMP7 is partially obscured by a larger spot (iota) in NIH Black Swiss preproteasomes. LMP7, LMP2, and their precursors have slightly different isoelectric points in NIH Black Swiss mice due to known polymorphisms (34). Immunoprecipitates from 5 × 106 cells were loaded on each gel.
Discussion
Based on our results, we propose a cooperative model for immunoproteasome assembly (Fig. 6). When both IFN-γ–inducible and constitutive catalytic β subunits are synthesized, either pre-LMP2 and pre-MECL are cooperatively incorporated early into preproteasomes or, alternatively, pre-Z is incorporated early. The relative levels of pre-LMP2 and pre-MECL compared to pre-Z may determine which major pathway is favored (33), although differences in affinity of each of these precursor subunits for preproteasomes could also be important. Nevertheless, preproteasomes containing pre-LMP2 and pre-MECL favor the incorporation of LMP7, since these preproteasomes do not efficiently mature when LMP7 is absent and its homologue X is present. This cooperative incorporation of LMP7 into preproteasomes containing pre-LMP2 and pre-MECL leads to 20S immunoproteasomes containing all three IFN-γ–inducible subunits. Cooperative incorporation of constitutive catalytic subunits may also occur, with preproteasomes containing pre-Z favoring the subsequent incorporation of delta and X. Exclusion of X from the immunoproteasome pathway may increase its availability for incorporation into constitutive proteasomes. Evidence supporting this comes from Gaczynska et al. who have shown that overexpression of X increases the content of delta and decreases the content of LMP2 in proteasomes (18). Despite cooperativity, some mixed proteasomes may form, since LMP7 can be incorporated into proteasomes in the absence of LMP2.
Figure 6.
Cooperative model for proteasome assembly. One major pathway leads to immunoproteasomes containing all three IFN-γ–inducible catalytic β subunits (LMP2, LMP7, and MECL). Another major pathway leads to constitutive proteasomes containing all three constitutive catalytic β subunits (delta [δ], X, and Z). Minor pathways lead to mixed proteasomes containing assortments of both inducible and constitutive catalytic subunits. Appendages represent removable presequences. Subunit positions are based on the structure of the yeast 20S proteasome and assume that inducible subunits occupy the same sites as their constitutive homologues (3).
Cooperative incorporation of inducible subunits may serve several important functions. First, it favors the assembly of homogeneous “immunoproteasomes”, which would be important if certain antigen-processing functions of inducible subunits occur in concert. This is supported by the observation that LMP7−/− mice, which have reduced levels of all three processed inducible subunits, have a more profound phenotype (reduced MHC class I expression) than LMP2−/− mice, which have reduced levels of only LMP2 and MECL. Second, it discourages assembly of heterogeneous “mixed” proteasomes containing mass-action random assortments of inducible and constitutive catalytic subunits. This may be important in the antiviral response where IFN-γ upregulation of inducible subunits occurs without downregulation of constitutive subunits (7–9, 12), which could result in the assembly of mixed proteasomes that generate peptides not normally produced during T cell development and lead to autoimmune reactivity. Third, it increases the likelihood that at least some homogeneous “constitutive” proteasomes are assembled in lymphoid tissues where inducible subunits predominate. This would be important if certain vital housekeeping functions of constitutive proteasomes are not served by immunoproteasomes.
In summary, we demonstrate a mechanism at the protein level which favors the assembly of immunoproteasomes containing all three IFN-γ–inducible catalytic β subunits in the same 20S complex. This mechanism depends on the nature of the inducible subunits, including presequences, but not on their catalytic activity. Incorporation of these subunits into proteasomes does not appear to be simply a function of relative levels of expression. Our cooperative model for proteasome assembly suggests an important role for ensuring the assembly of either homogeneous immunoproteasomes and/or homogeneous constitutive proteasomes in cells and tissues expressing both inducible and constitutive catalytic subunits to varying degrees.
Acknowledgments
We thank Klavs B. Hendil for generous gifts of MCP21 ascites fluid and anti–human X antiserum, and David B. Ginsburg for technical assistance.
This work was supported in part by a Trustee Grant from the Children's Hospital Research Foundation of Cincinnati, the Schmidlapp Foundation, and the National Institutes of Health. T.A. Griffin was supported by an Arthritis Foundation Postdoctoral Fellowship. D. Nandi was supported by a fellowship from the Irvington Institute for Medical Research. M. Cruz received support from the Hospital de Especialidades, Centro Medico Nacional Siglo XXI, Instituto Mexicano del Seguro Social (Mexico City, Mexico). R.A. Colbert received support as a Pfizer Scholar. The Basel Institute for Immunology was founded and is supported by F. Hoffmann-La Roche (Basel, Switzerland).
Footnotes
Abbreviation used in this paper: NEPHGE, nonequilibrium pH gradient electrophoresis.
References
- 1.Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 2.Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 3.Groll M, Ditzel L, Löwe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 4.Seemüller E, Lupas A, Stock D, Löwe J, Huber R, Baumeister W. Proteasome from Thermoplasma acidophilum: a threonine protease. Science. 1995;268:579–582. doi: 10.1126/science.7725107. [DOI] [PubMed] [Google Scholar]
- 5.Martinez CK, Monaco JJ. Homology of proteasome subunits to a major histocompatibility complex–linked LMP gene. Nature. 1991;353:664–667. doi: 10.1038/353664a0. [DOI] [PubMed] [Google Scholar]
- 6.Glynne R, Powis SH, Beck S, Kelly A, Kerr L-A, Trowsdale J. A proteasome-related gene between the two ABC transporter loci in the class II region of the human MHC. Nature. 1991;353:357–360. doi: 10.1038/353357a0. [DOI] [PubMed] [Google Scholar]
- 7.Hisamatsu H, Shimbara N, Saito Y, Kristensen P, Hendil KB, Fujiwara T, Takahashi E, Tanahashi N, Tamura T, Ichihara A, Tanaka K. Newly identified pair of proteasomal subunits regulated reciprocally by interferon-γ. J Exp Med. 1996;183:1807–1816. doi: 10.1084/jem.183.4.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nandi D, Jiang H, Monaco JJ. Identification of MECL-1 (LMP-10) as the third IFN-γ–inducible proteasome subunit. J Immunol. 1996;156:2361–2364. [PubMed] [Google Scholar]
- 9.Groettrup M, Kraft R, Kostka S, Standera S, Stohwasser R, Kloetzel P-M. A third interferon-γ–induced subunit exchange in the 20S proteasome. Eur J Immunol. 1996;26:863–869. doi: 10.1002/eji.1830260421. [DOI] [PubMed] [Google Scholar]
- 10.Fruh K, Yang Y, Arnold D, Chambers J, Wu L, Waters JB, Spies T, Peterson PA. Alternative exon usage and processing of the major histocompatibility complex–encoded proteasome subunits. J Biol Chem. 1992;267:22131–22140. [PubMed] [Google Scholar]
- 11.Martinez CK, Monaco JJ. Post-translational processing of a major histocompatibility complex–encoded proteasome subunit, LMP-2. Mol Immunol. 1993;30:1177–1183. doi: 10.1016/0161-5890(93)90136-y. [DOI] [PubMed] [Google Scholar]
- 12.Akiyama K, Yokota K, Kagawa S, Shimbara N, Tamura T, Akioka H, Nothwang HG, Noda C, Tanaka K, Ichihara A. cDNA cloning and interferon γ down-regulation of proteasomal subunits X and Y. Science. 1994;265:1231–1234. doi: 10.1126/science.8066462. [DOI] [PubMed] [Google Scholar]
- 13.Gaczynska M, Rock KL, Goldberg AL. γ-interferon and expression of MHC genes regulate peptide hydrolysis by proteasomes. Nature. 1993;365:264–267. doi: 10.1038/365264a0. [DOI] [PubMed] [Google Scholar]
- 14.Boes B, Hengel H, Ruppert T, Multhaup G, Koszinowski UH, Kloetzel P-M. Interferon γ stimulation modulates the proteolytic activity and cleavage site preference of 20S mouse proteasomes. J Exp Med. 1994;179:901–909. doi: 10.1084/jem.179.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehring B, Meyer TH, Eckerskorn C, Lottspeich F, Tampé R. Effects of major-histocompatibility-complex–encoded subunits on the peptidase and proteolytic activities of human 20S proteasomes. Eur J Biochem. 1996;235:404–415. doi: 10.1111/j.1432-1033.1996.00404.x. [DOI] [PubMed] [Google Scholar]
- 16.Gaczynska M, Rock KL, Spies T, Goldberg AL. Peptidase activities of proteasomes are differentially regulated by the major histocompatibility complex–encoded genes for LMP2 and LMP7. Proc Natl Acad Sci USA. 1994;91:9213–9217. doi: 10.1073/pnas.91.20.9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuckelkorn U, Frentzel S, Kraft R, Kostka S, Groettrup M, Kloetzel P-M. Incorporation of major histocompatibility complex–encoded subunits LMP2 and LMP7 changes the quality of the 20S proteasome polypeptide processing products independent of interferon-γ. Eur J Immunol. 1995;25:2605–2611. doi: 10.1002/eji.1830250930. [DOI] [PubMed] [Google Scholar]
- 18.Gaczynska M, Goldberg AL, Tanaka K, Hendil KB, Rock KL. Proteasome subunits X and Y alter peptidase activities in opposite ways to the interferon-γ–induced subunits LMP2 and LMP7. J Biol Chem. 1996;271:17275–17280. doi: 10.1074/jbc.271.29.17275. [DOI] [PubMed] [Google Scholar]
- 19.Stohwasser R, Kuckelkorn U, Kraft R, Kostka S, Kloetzel P-M. 20S proteasome from LMP7 knock out mice reveals altered proteolytic activities and cleavage site preferences. FEBS Lett. 1996;383:109–113. doi: 10.1016/0014-5793(96)00110-x. [DOI] [PubMed] [Google Scholar]
- 20.Van Kaer L, Ashton-Rickardt PG, Eichelberger M, Gaczynska M, Nagashima N, Rock KL, Goldberg AL, Doherty PC, Tonegawa S. Altered peptidase and viral-specific T cell response in Lmp2mutant mice. Immunity. 1994;1:533–541. doi: 10.1016/1074-7613(94)90043-4. [DOI] [PubMed] [Google Scholar]
- 21.Fehling HJ, Swat W, Laplace C, Kühn R, Rajewsky K, Müller U, von Boehmer H. MHC class I expression in mice lacking the proteasome subunit LMP-7. Science. 1994;265:1234–1237. doi: 10.1126/science.8066463. [DOI] [PubMed] [Google Scholar]
- 22.Nandi D, Woodward E, Ginsburg DB, Monaco JJ. Intermediates in the formation of mouse 20S proteasomes: implications for the assembly of precursor β subunits. EMBO (Eur Mol Biol Organ) J. 1997;16:5363–5375. doi: 10.1093/emboj/16.17.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frentzel S, Pesold-Hurt B, Seelig A, Kloetzel P-M. 20 S proteasomes are assembled viadistinct precursor complexes. J Mol Biol. 1994;236:975–981. doi: 10.1016/0022-2836(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 24.Yang Y, Früh K, Ahn K, Peterson PA. In vivoassembly of the proteasomal complexes, implications for antigen processing. J Biol Chem. 1995;270:27687–27694. doi: 10.1074/jbc.270.46.27687. [DOI] [PubMed] [Google Scholar]
- 25.Patel SD, Monaco JJ, McDevitt HO. Delineation of the subunit composition of human proteasomes using antisera against the major histocompatibility complex–encoded LMP2 and LMP7 subunits. Proc Natl Acad Sci USA. 1994;91:296–300. doi: 10.1073/pnas.91.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hendil KB, Kristensen P, Uerkvitz W. Human proteasomes analysed with monoclonal antibodies. Biochem J. 1995;305:245–252. doi: 10.1042/bj3050245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salter RD, Howell DN, Cresswell P. Genes regulating HLA class I antigen expression in T–B lymphoblastoid hybrids. Immunogenetics. 1985;21:235–246. doi: 10.1007/BF00375376. [DOI] [PubMed] [Google Scholar]
- 28.Ortiz-Navarette V, Seelig A, Gernold M, Frentzel S, Kloetzel P-M, Hämmerling GJ. Subunit of the ‘20S' proteasome (multicatalytic proteinase) encoded by the major histocompatibility complex. Nature. 1991;353:662–664. doi: 10.1038/353662a0. [DOI] [PubMed] [Google Scholar]
- 29.Groettrup M, Standera S, Stohwasser R, Kloetzel P-M. The subunits MECL-1 and LMP2 are mutually required for incorporation into the 20S proteasome. Proc Natl Acad Sci USA. 1997;94:8970–8975. doi: 10.1073/pnas.94.17.8970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seemüller E, Lupas A, Baumeister W. Autocatalytic processing of the 20S proteasome. Nature. 1996;382:468–470. doi: 10.1038/382468a0. [DOI] [PubMed] [Google Scholar]
- 31.Chen P, Hochstrasser M. Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell. 1996;86:961–972. doi: 10.1016/s0092-8674(00)80171-3. [DOI] [PubMed] [Google Scholar]
- 32.Schmidtke G, Kraft R, Kostka S, Henklein P, Frömmel C, Löwe J, Huber R, Kloetzel P-M, Schmidt M. Analysis of mammalian 20S proteasome biogenesis: the maturation of β-subunits is an ordered two-step mechanism involving autocatalysis. EMBO (Eur Mol Biol Organ) J. 1996;15:6887–6898. [PMC free article] [PubMed] [Google Scholar]
- 33.Stohwasser R, Standera S, Peters I, Kloetzel P-M, Groettrup M. Molecular cloning of the mouse proteasome subunits MC14 and MECL-1: reciprocally regulated tissue expression of interferon-γ–modulated proteasome subunits. Eur J Immunol. 1997;27:1182–1187. doi: 10.1002/eji.1830270520. [DOI] [PubMed] [Google Scholar]
- 34.Nandi D, Iyer MN, Monaco JJ. Molecular and serological analysis of polymorphisms in the murine major histocompatibility complex–encoded proteasome subunits, LMP-2 and LMP-7. Exp Clin Immunogenet. 1996;13:20–29. [PubMed] [Google Scholar]