Abstract
Formation of major histocompatibility complex class I–associated peptides from membrane proteins has not been thoroughly investigated. We examined the processing of an HLA-A*0201–associated epitope, YMDGTMSQV, that is derived from the membrane protein tyrosinase by posttranslational conversion of the sequence YMNGTMSQV. Only YMDGTMSQV and not YMNGTMSQV was presented by HLA-A*0201 on cells expressing full-length tyrosinase, although both peptides have similar affinities for HLA-A*0201 and are transported by TAP. In contrast, translation of YMNGTMSQV in the cytosol, as a minigene or a larger fragment of tyrosinase, led to the presentation of the unconverted YMNGTMSQV. This was not due to overexpression leading to saturation of the processing/conversion machinery, since presentation of the converted peptide, YMDGTMSQV, was low or undetectable. Thus, presentation of unconverted peptide was associated with translation in the cytosol, suggesting that processing of the full-length tyrosinase occurs after translation in the endoplasmic reticulum. Nevertheless, presentation of YMDGTMSQV in cells expressing full-length tyrosinase was TAP (transporter associated with antigen processing) and proteasome dependent. After inhibition of proteasome activity, tyrosinase species could be detected in the cytosol. We propose that processing of tyrosinase involves translation in the endoplasmic reticulum, export of full-length tyrosinase to the cytosol, and retransport of converted peptides by TAP for association with HLA-A*0201.
CD8+ T cells recognize peptides in association with class I MHC proteins on the surface of cells. In general, these MHC class I–associated peptides are derived from intracellular proteins (1). In the classical pathway for processing of class I–associated peptides, cytosolic proteases such as the proteasome degrade proteins to generate peptides that are transported into the endoplasmic reticulum (ER)1 by the transporter associated with antigen processing (TAP; 2–6). Upon entry into the ER, the peptides are bound to “empty” or peptide-free MHC class I molecules that are associated with TAP (7, 8) via an intermediary protein, tapasin (9). After binding peptide, the MHC class I heterotrimer dissociates from TAP and proceeds through the ER, Golgi, and exocytic pathway to the cell surface (10). The peptides in association with the MHC class I molecule are then available for recognition on the cell surface by CTLs.
Because membrane and secreted proteins are normally cotranslationally translocated into the ER, they would appear to bypass the cytosolic proteases of the classical pathway. Nevertheless, a number of MHC class I–associated peptides that originate from membrane proteins have been identified, and the pathways by which they are produced have been the object of several recent studies. Peptides from the signal sequences of IP-30, HLA-E, Signal Sequence Receptor Protein–α (SSR-α), and calreticulin (11, 12), as well as peptides from more internal sequences of the HIV env (13) and Epstein-Barr virus Latent Membrane Protein 2 (LMP2) proteins (14), and a peptide epitope of uncertain location (15) are presented by HLA-A*0201 in cells that lack expression of TAP. Independence of TAP indicates that the source proteins for these peptides are produced in the ER, and that complete proteolytic processing occurs in the ER or distal vesicular compartments, and not the cytosol. Although the signal peptidase is likely to be involved in the generation of signal sequence–derived peptides, it is unlikely to account for the production of peptides from more internal sequences. In addition, none of the peptides derived from signal sequences is full-length, raising the possibility that additional proteases are involved in secondary proteolytic events. In support of this possibility, it has been shown that the production of some, but not all, of these peptides is sensitive to high concentrations of the protease inhibitor LLnL (16). In addition, several vaccinia virus constructs containing peptide epitopes embedded in a larger sequence that is in turn linked to a signal sequence can be processed for presentation in a TAP-independent manner, presumably via ER resident proteases (17–20).
An alternative pathway for the processing of membrane protein–derived epitopes has been suggested by the observation that the presentation of peptides from the measles virus transmembrane (21) and the HIV env (22) proteins as well as peptides from the signal sequence of some MHC class I molecules (23, 24) and the LCMV gp33 protein (25) are dependent on TAP function. Roelse et al. (26) demonstrated that in vitro, peptides transported into the ER that are too long to bind to class I MHC molecules could be exported to the cytosol for further processing, and the products then retransported to the ER by TAP. Although a similar mechanism has not been demonstrated in vivo, partial proteolysis in the ER followed by final proteolysis in the cytosol could account for the TAP-dependent presentation of these epitopes. Alternatively, their presentation may occur after mistranslation of the source protein in the cytosol (27). This could occur either as the result of incomplete translational blockade by signal sequences on cytosolic ribosomes, or by the use of an alternate start codon internal to the signal sequence, as has been shown to occur for several class I–associated epitopes (28–34). Support for such a cytosolic mistranslation mechanism has been provided by observations with an HLA-B*3501–restricted HIV env epitope. This epitope contains a site for N-linked glycosylation that is modified during cotranslational translocation of the full-length protein into the ER (35). However, the epitope presented by HLA-B*3501 has not undergone either glycosylation or deglycosylation (22, 36). Thus, it appears that HIV env protein that gives rise to this epitope has been mistranslated in the cytosol and processed there.
A final possible explanation for the TAP-dependent presentation of peptides derived from membrane proteins is that the source protein itself is exported from the ER for proteolysis in the cytosol. Recently, the reverse translocation of several apparently full-length membrane proteins from the ER to the cytosol has been reported (37–48). Visualization of these proteins has generally been possible only after inhibition by protease inhibitors, in most cases those which are effective against the proteasome (38–48). However, no evidence supporting the involvement of this pathway in the production of MHC class I–associated peptide antigens has been presented. Thus, at this point in time, the only in vivo data available for the processing of membrane proteins for class I presentation support a cytosolic mistranslation mechanism.
Recently, we described an epitope from the membrane-associated protein tyrosinase that is presented by HLA-A*0201 to CTL reactive with human melanomas (49). The sequence of this peptide, YMDGTMSQV, differed from the corresponding primary sequence of the tyrosinase protein, YMNGTMSQV, by the substitution of aspartic acid for asparagine at position 3. This was shown to be due to a posttranslational conversion, and neither to spontaneous deamidation nor RNA editing. The asparagine in YMNGTMSQV is part of an N-linked glycosylation site and a mammalian enzyme, peptide-N4-(N acetyl-β-glucosaminyl) asparagine amidase (PNGase), has been isolated, which removes the N-linked oligosaccharide side chains from glycopeptides. This process converts the modified asparagine residues to aspartic acid (50). Our working hypothesis is that glycosylation in the ER and subsequent deglycosylation at an unknown site are responsible for the conversion of YMNGTMSQV to YMDGTMSQV. Regardless of whether this hypothesis is correct, it focused our attention on how tyrosinase is processed. Since tyrosinase is a melanosomal membrane protein, mistranslation of tyrosinase in the cytosol could lead to proteolysis of the unconverted protein, generating peptides that would be transported by TAP into the ER and converted before or after binding to HLA-A*0201. Alternatively, conversion could occur during normal cotranslational translocation into the ER and subsequent degradation either in the ER, after export of partially proteolysed fragments, or in the cytosol after reverse translocation of the source protein. In this paper, we have examined the processing and conversion of this epitope in detail.
Materials and Methods
Cell Lines.
NA8 Mel + Tyr was derived by transfection of the tyrosinase negative melanoma NA8 Mel with the full-length tyrosinase gene and was a gift from Vincent Brichard and Thierry Boon (Ludwig Institute for Cancer, Université Catholique de Louvain, Brussels, Belgium). NA8 Mel + T3.1 was derived from transfection of NA8 Mel with the truncated tyrosinase cDNA T3.1 (51). JY is a human HLA-A*0201+ cell line. T2 is an HLA-A*0201+ human B lymphoblastoid cell line with a deletion in the MHC including the genes for TAP1, TAP2, LMP2, and LMP7 (52, 53). All cell lines were maintained in RPMI 1640 supplemented with 5% FCS/SerXtend (Irvine Scientific, Santa Ana, CA) in a humidified 5% CO2 atmosphere at 37°C.
Peptides.
Synthetic peptides were made by standard Fmoc chemistry using a model AMS422 peptide synthesizer (Gilson Co. Inc., Middleton, WI). All peptides were purified to >98% purity by reverse-phase HPLC on a C-8 column (VYDAC, Hesperia, CA). Purity and identity were established using a triple quadropole mass spectrometer (model TSQ-7000; Finnigan, San Jose, CA).
Recombinant Vaccinia Viruses.
The minigene recombinant vaccinia were constructed using synthetic oligonucleotides (5′ GTACCACCATGTATATGAATGGAACAATGTCCCAGGTATA 3′ and 5′ AGCTTATACCTGGGACATTGTTCCATTCATATACATGGTG 3′ for peptide (M)YMNGTMSQV and 5′ GTACCACCATGTATATGGATGGAACAATGTCCCAGGTATA 3′ and 5′ AGCTTATACCTGGGACATTGTTCCATCCATATACATGGTG 3′ for peptide (M)YMDGTMSQV) ligated directionally into the plasmid pSC11.3 (54) at the Acc65I and HindIII sites. In addition to appropriate overhang sequences for ligation, the synthetic nucleotide sequences contained a favorable Kozac sequence for translation initiation, a methionine start codon, the nucleotides encoding the T cell epitope, and a translational stop signal. The full-length tyrosinase was excised from the pcDNA1/amp-123.b2 vector using HindIII and XbaI and subcloned into pSC11.3 at the HindIII and SpeI sites. Recombinant vaccinia viruses were produced from these vectors using standard methods (55). Purified vaccinia stocks were titered and tested for proper expression of tyrosinase or minigene using specific murine HLA-A2–restricted CTL. Vaccinia viruses encoding the TAP1 and TAP2 genes were a gift from Drs. Jon Yewdell and Jack Bennink (Laboratory of Viral Diseases, National Institutes of Allergy and Infectious Diseases, Bethesda, MD).
CTL Lines and Cytotoxicity.
CTL lines were generated by intraperitoneal injection of 5 × 108 PFUs of recombinant vaccinia encoding either YMDGTMSQV or YMNGTMSQV into C57Bl/6 mice expressing a chimeric MHC class I with the α1 and α2 domains from HLA-A2.1 and the α3 domain from Kd (56). 3 wk after priming, splenocytes were removed and stimulated with autologous, irradiated splenocytes that had been pulsed with YMDGTMSQV or YMNGTMSQV peptide. After the first week in culture, autologous, irradiated peptide-pulsed splenocytes and 10 U/ml IL-2 were added. IL-2 was also added on day 4 of every weekly stimulation. Standard 51Cr-release assays were performed to determine CTL recognition of tyrosinase peptides. For peptide dose response assays, T2 cells were 51Cr-labeled in the presence of 10 μg/ml MA2.1. and then incubated with the indicated concentrations of synthetic tyrosinase peptides for 3 h at 37°C. For vaccinia-infected target cells, 2 × 107 PFUs vaccinia were added to 106 targets in HBSS for 1 h. RPMI with 10% FCS, 15 mM Hepes, 50 μM β-mercaptoethanol, 2 mM glutamine, and essential and nonessential amino acids was then added to the infected cells for 9 h to allow for expression. Targets were labeled in 100 μCi Na51CrO4 for the final 2 h. CTLs were added at an effector: target ratio of 10:1 unless otherwise noted.
HLA-A*0201-associated Peptide Isolation.
Peptides were acid eluted from affinity-purified HLA-A*0201 molecules as previously described (49, 57). The peptide extracts from 2 × 109 NA8 Mel + Tyr or 3 × 109 NA8 Mel + T3.1 cells were separated on a narrow bore reverse phase column (RP-18 Spheri-5, 2.1 × 3 mm). Buffer A was 0.1% TFA in water and buffer B was 0.085% TFA in 80% acetonitrile. The gradient consisted of 100% buffer A (0–20 min), 0–15% buffer B (20–25 min) and 15–67% buffer B (25–80 min) at a flow rate of 200 μl/min. The synthetic peptides YMDGTMSQV and YMNGTMSQV were separated on the same column under identical conditions immediately after the extracts, and their elution positions were used to identify fractions from the extracts that contained the naturally processed forms of these peptides. These fractions were loaded onto a C18 microcapillary column (75 μm intradermally × 12 cm) and eluted using a 2%/min increasing gradient of acetonitrile in 0.1 M acetic acid into a triple quadropole mass spectrometer (model TSQ-7000; Finnigan) equipped with an electrospray ion source. Scans were acquired every 1.5 s over a mass range m/z (mass/charge ratio) 300:1400 and then plotted with intensities for m/z 1,031: 1,032. For the NA8 Mel + Tyr peptide extract, 10% of the sample was analyzed by mass spectrometry. For the NA8 Mel + T3.1 peptide extract, 20% of the sample was analyzed by mass spectrometry. The loaded sample amounts were standardized using an unrelated peptide of m/z 541 found in fractions 28, 29, and 30. We determined the cell surface copy numbers of YMDGTMSQV peptides using synthetic YMDGTMSQV peptides as standards.
Inhibitors.
Lactacystin (gift of Dr. S. Omura of the Kitasato Institute, Tokyo, Japan) is a Streptomyces metabolite which irreversibly inhibits proteasomes via covalent binding to the active sites of the catalytically active β subunits (58). LLnL, also known as Calpain Inhibitor I, was purchased from Calbiochem (La Jolla, CA). LLnL reversibly inhibits proteasomes as well as several other classes of proteases, including cysteine proteases, calpain, and cathepsin B (59).
Epitope Reexpression Assay.
Tumor targets (1–2 × 107) were grown in RPMI 1640 supplemented with 5% FCS/SerXtend and then centrifuged. The pellet was gently resuspended in 500 μl of 300 mM glycine, pH 2.5, 1% (wt/vol) BSA and incubated for 3 min at 37°C. The suspension was diluted with 40 ml of RPMI 1640 medium supplemented with 5% FCS/SerXtend and then centrifuged. Cells (2 × 106) were aliquoted into 1 ml of RPMI 1640 medium supplemented with 5% FCS/SerXtend and 100 μCi Na 51CrO4 in the presence or absence of 10 μg/ml of Brefeldin A (BFA), 10 μM Lactacystin, or 250 μM LLnL, and incubated at 37°C to allow epitope reexpression for 5 h. After washing four times, all targets were resuspended in RPMI 1640 medium supplemented with 5% FCS/SerXtend and 10 μg/ml BFA to inhibit further epitope expression, and used in a standard 4-h 51Cr-release assay.
Cell Fractionation.
107 NA8 Mel and DM93 melanoma cells were frozen twice in liquid N2 and thawed in 1 ml of hypoosmotic lysis buffer consisting of 20 mM Hepes, 2 mM EDTA, and a protease inhibitor cocktail containing 4 mM PMSF, 10 μg/ml aprotinin, 10 μM pepstatin A, 10 μg/ml leupeptin, and 100 μM iodoacetamide. The lysate was centrifuged at 16,000 g at 4°C for 15 min. The supernatant was collected as the cytosolic fraction. The pellet was washed twice with the hypoosmotic lysis buffer before being solubilized in 1 ml of 0.5% deoxycholate, 1% NP-40, 5 mM EDTA, 10 mM Tris, pH 7.5, and the protease inhibitor cocktail described earlier. This lysate was spun at 16,000 g for 15 min at 4°C. The supernatant was collected as the membrane fraction. Whole cell lysate was obtained using 1 ml of the 0.5% deoxycholate, and 1% NP-40 lysis buffer for 107 cells. The suspension was incubated for 1 h at 4°C and spun at 30,000 g for 30 min at 4°C, and then the supernatant was collected. As a control to determine the separation efficiency of the membrane and cytosolic fractions, each fraction was immunoblotted for TAP, which was only present in the membrane fraction (data not shown).
Immunoblotting.
106 cell equivalents per lane were separated by SDS-PAGE (60) on 10% gels. Gels were transferred to Immobilon P and blocked with 5% nonfat dried milk in PBS with 0.05% Tween 20. Blots were probed with either the antityrosinase mAb T311 (61; gift from Drs. E. Stockert and L. Old, Ludwig Institute for Cancer Research, Memorial Sloan Kettering, New York), at a 1:10 dilution of culture supernatant, or the anti-TAP mAb (gift from Robert Tampe, Max-Planck-Institute, Martinsried, Germany), at a 1:30 dilution. After 10 h, blots were washed and probed with horseradish peroxidase–conjugated sheep anti–mouse IgG, and developed according to the Amersham ECL protocol (Amersham Corp., Arlington Heights, IL).
Deglycosylation of Tyrosinase.
Membrane or cytosolic fractions from 5 × 105 cells generated as described above were precipitated via the addition of 8 volumes of acetone and incubating at −20°C for 3 h. The precipitate was resuspended in 25 μl 0.5% SDS/0.1 M β-mercaptoethanol and boiled for 5 min. After cooling to room temperature, 25 μl sodium phosphate, pH 7.0, 10 μl 1,10 phenanthroline, 10 μl 10% NP-40, and 5 μl of PNGase F 300 mU/ml (Sigma Chemical Co., St. Louis, MO) were added, and the reactions were incubated for the indicated times at 37°C.
Results
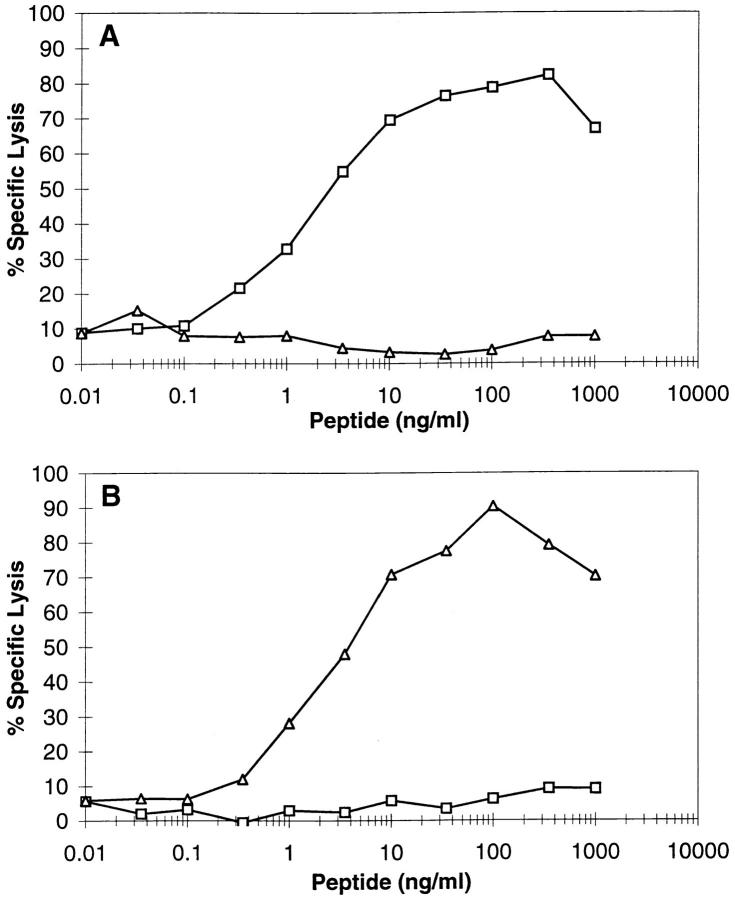
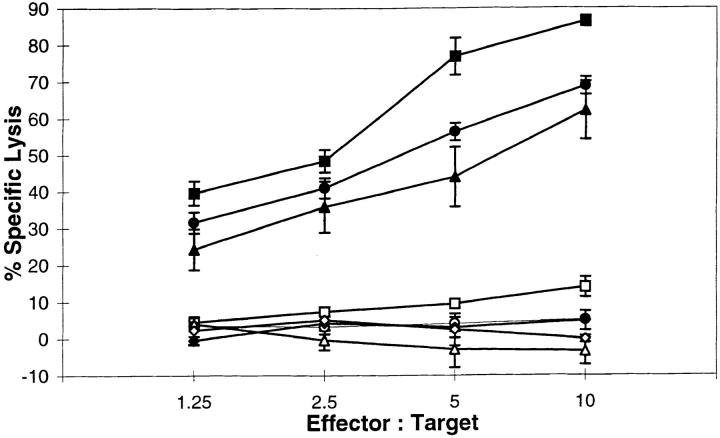
To detect either the YMDGTMSQV or the YMNGTMSQV epitopes, we developed CTLs specific for each peptide from transgenic mice expressing a chimeric HLA-A*0201/H2Dd molecule. CTLs generated against the YMDGTMSQV peptide (D-specific CTLs) required 5–6 logs less of the YMDGTMSQV peptide than the YMNGTMSQV peptide to give equivalent lysis of peptide-pulsed target cells (Fig. 1 A). Conversely, CTLs generated against the YMNGTMSQV peptide (N-specific CTLs) required 5–6 logs less YMNGTMSQV than YMDGTMSQV for recognition (Fig. 1 B). Since YMNGTMSQV and YMDGTMSQV bind to HLA-A2.1 with similar affinities (49), these differences reflect the ability of the CTLs to discriminate between these two peptides. Nevertheless, HLA-A*0201+ melanomas expressing tyrosinase were lysed by D-specific CTLs but not by N-specific CTLs at similar effector:target ratios (Fig. 2). The lack of reactivity of N-specific CTLs with these tumors confirms our earlier conclusion based on mass spectrometry data (49) that there is no YMNGTMSQV peptide on the cell surface, although the tyrosinase gene encodes this sequence.
Figure 1.
Recognition of tyrosinase-derived peptides by D-specific and N-specific CTLs. Indicated concentrations of YMDGTMSQV (squares) or YMNGTMSQV (triangles) were incubated with 51Cr-labeled T2 cells in the presence of 10 μg/ml MA2.1 Ab for 3 h at 37°C. D-specific (A) or N specific (B) CTL were added at an E:T ratio of 10:1 for a 4-h 51Cr-release assay.
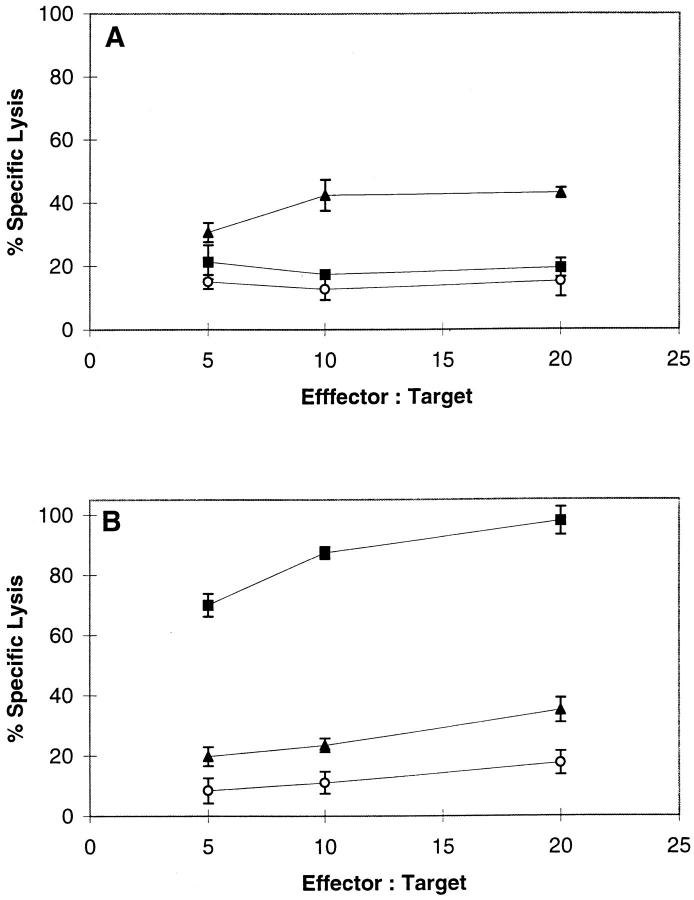
Figure 2.
Recognition of melanoma tumor lines by D-specific CTLs (solid symbols) and N-specific CTLs (open symbols) in a 4-h 51Cr-release assay. DM6 (circles), DM93 (squares), and NA8 Mel + Tyr (triangles) express full-length tyrosinase while NA8 Mel (crosses) does not.
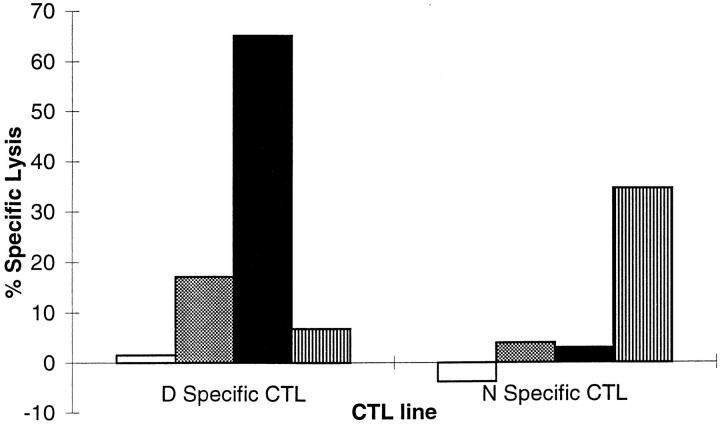
One possible explanation for the lack of expression of the unconverted YMNGTMSQV peptide on the surface of melanoma cells was that it was not transported via TAP after production in the cytosol. Androlewicz (62) recently demonstrated that this peptide was transported in an in vitro system. We used a recombinant vaccinia virus expressing a minigene encoding YMNGTMSQV to address this question in vivo. HLA-A*0201+ lymphoblastoid JY cells infected with this vaccinia construct were lysed by N-specific CTL but not D-specific CTL (Fig. 3). These results demonstrate that the YMNGTMSQV peptide produced in the cytosol can be expressed in association with HLA-A*0201 molecules at the cell surface. In addition, this expression is dependent upon the expression of TAP by the target cell (data not shown). Interestingly, however, expression of this minigene-encoded form of the YMNGTMSQV sequence did not result in conversion and presentation of the YMDGTMSQV peptide at the surface. This contrasts to the presentation of only the YMDGTMSQV peptide in cells expressing full-length tyrosinase.
Figure 3.
Recognition of JY cells infected with various vaccinia-encoded minigene constructs. JY cells were incubated with no vaccinia (white bars), an irrelevant vaccinia control encoding YLEPGPVTA (gray bars), D minigene vaccinia encoding YMDGTMSQV (black bars), or N minigene vaccinia encoding YMNGTMSQV (striped bars).
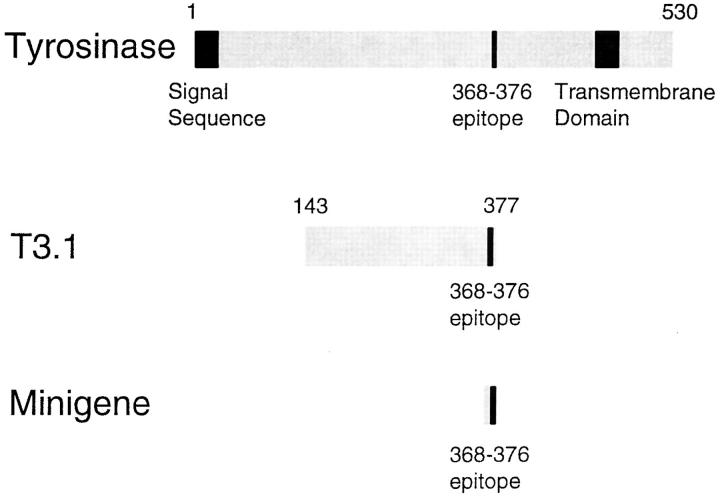
One explanation for the exclusive production of YMDGTMSQV from full-length tyrosinase and of YMN from the minigene is that the former is translated in the ER, where it undergoes conversion, whereas the latter is translated in the cytosol, where it does not. However, the minigene product was preprocessed to the ideal length to bind to HLA-A*0201, while the full-length tyrosinase requires proteolysis before binding. Therefore, it was also possible that immediate binding of the minigene product to HLA-A*0201 after entry into the ER prevented conversion, whereas longer peptides derived from full-length tyrosinase were converted before being trimmed to the optimal length for binding. To distinguish between these two possibilities, we transfected NA8 Mel, a tyrosinase-negative melanoma cell line, with a truncated tyrosinase gene fragment called T3.1 (51). The translation product of T3.1 spans residues 143–377 of the full-length tyrosinase sequence (Fig. 4). Because it lacks the NH2-terminal end of full-length tyrosinase, including the signal sequence, it should be translated in the cytosol, as is the minigene, rather than the ER. However, the YMNGTMSQV epitope sequence in T3.1 (residues 368–376) is bounded by 225 residues at its NH2 terminus and 1 residue at its COOH terminus. To be presented by HLA-A*0201, it requires additional processing as compared to the minigene product. If the failure of the minigene product to undergo conversion were due to its expression in the cytosol, regardless of length, then we would expect that cells expressing T3.1 would also express the unconverted peptide epitope. This was found to be the case (Fig. 5 A). We conclude from this that the production of the unconverted epitope is associated with translation in the cytosol, and that its absence from cells expressing full-length tyrosinase indicates that the converted epitope is produced from this protein after it is translated into the ER, rather than mistranslated in the cytosol. Although the presence of the unconverted epitope was clearly associated with the translation of protein in the cytosol, the absence of the converted epitope was not. Surprisingly, we found that cells expressing T3.1 presented the converted epitope as well as the unconverted form (Fig. 5 B). This low but reproducible level of lysis by the D-specific CTL indicates that cytosolic proteins can give rise to epitopes that have undergone conversion.
Figure 4.
Schematic of tyrosinase peptide encoding constructs.
Figure 5.
Recognition of NA8 Mel and transfectants by N-specific (A) and D-specific (B) CTLs in a 4-h 51Cr-release assay. Targets used were the tyrosinase negative melanoma NA8 Mel (circles), its full-length tyrosinase transfectant NA8 Mel + Tyr (squares), or its T3.1 transfectant NA8 Mel + T3.1 (triangles). The results are representative of five independent experiments.
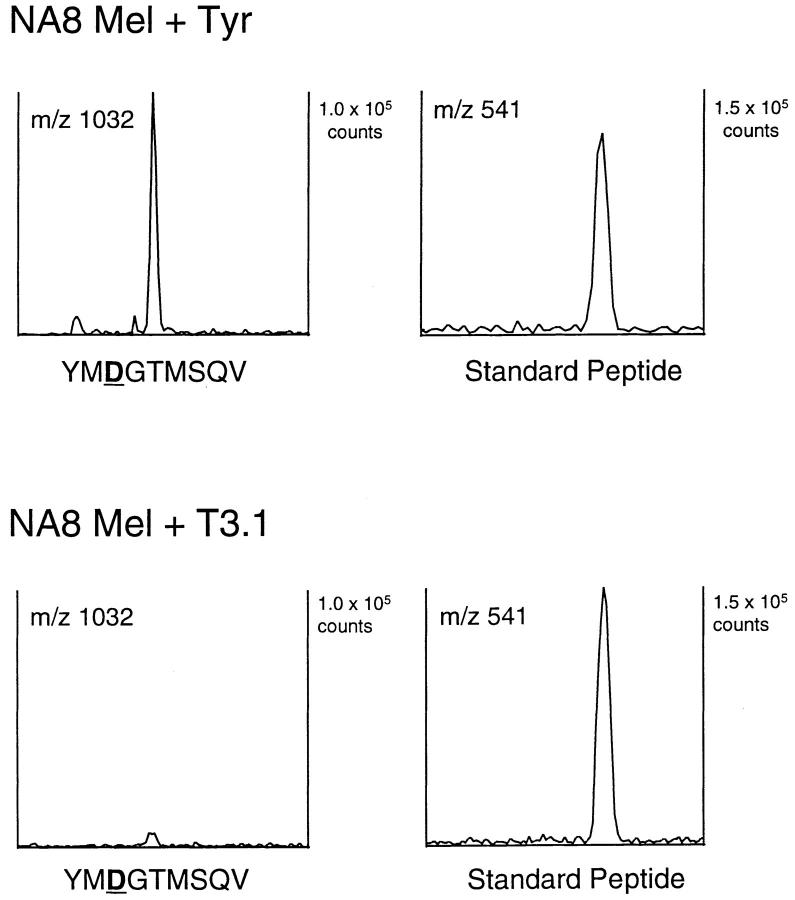
One possible caveat to the conclusion that the presence of the unconverted epitope was associated with cytosolic translation was the possibility that the amount of T3.1 protein in the cells, or its inability to fold appropriately, led to the accumulation of a much higher level of the YMNGTMSQV peptide or a precursor than would be produced in cells expressing full-length tyrosinase. This could lead to a saturation of the processing and conversion machinery, resulting in a failure to convert all of the peptides produced in the T3.1 transfectant to YMDGTMSQV. However, if this were so, then the T3.1 transfectant should express an amount of the converted YMDGTMSQV peptide equal to or higher than that found on the full-length transfectant. Consequently, we immunoaffinity purified the HLA-A2.1 molecules from comparable amounts of NA8 Mel + T3.1 and NA8 Mel + Tyr, and extracted the associated peptides. After HPLC separation, we used tandem mass spectrometry to determine the quantity of the YMDGTMSQV peptide in these extracts (Fig. 6). A coeluting species at m/z 541 was found in both extracts and was used to normalize the amount of cell extract injected into the mass spectrometer. By using known amounts of synthetic peptide as a standard, we determined the quantity of this peptide in each extract, and normalized to the number of cell equivalents of material injected. The density of YMDGTMSQV on the full-length tyrosinase transfectant was determined to be 1,400 peptides per cell, in good agreement with a previously reported value of 1,200 copies per cell (49). However, the density of this peptide on the T3.1 transfectant was only 50 copies per cell, or 4% of that seen on the NA8 + Tyr cells. Although detected by N-specific CTL, the density of YMNGTMSQV on the NA8 Mel + T3.1 transfectant was too low to be detected by mass spectrometry with the amount of cell extract analyzed (<25 copies per cell). These results demonstrate that the T3.1 transfectant generated less YMDGTMSQV than did the full-length tyrosinase transfectant, and indicate that the presence of the unconverted peptide in the T3.1 transfectant was not due to saturation of the processing and conversion machinery. We conclude that the processing of the T3.1 translation product and full-length tyrosinase must differ in some respect. Since T3.1 has no signal sequence or transmembrane domain, it should be produced in the cytosol and processed by the classical antigen-processing pathway. Since full-length tyrosinase does not give rise to the unconverted peptide that is characteristic of cytosolic translation of both T3.1 and the minigene, it must therefore be translated in the ER, and undergo processing in a distinct manner.
Figure 6.
Ion chromatograms recorded on peptides from fractions 28– 30 of the first dimension separation of NA8 Mel + Tyr (A) and NA8 Mel + T3.1 (B) extracts. Sample preparation and mass spectrometric analysis are described in Materials and Methods. After mass spectra were acquired, a computer search was conducted to determine the abundance of ions in a 2-mass unit window centered on either m/z 1032, which corresponds to the (M+H)+ ion species of YMDGTMSQV or m/z 541, an unknown peptide which was found in these pooled fractions in both extracts. The 541 peptide was used to normalize the number of cell equivalents loaded onto the mass spectrometer for each sample.
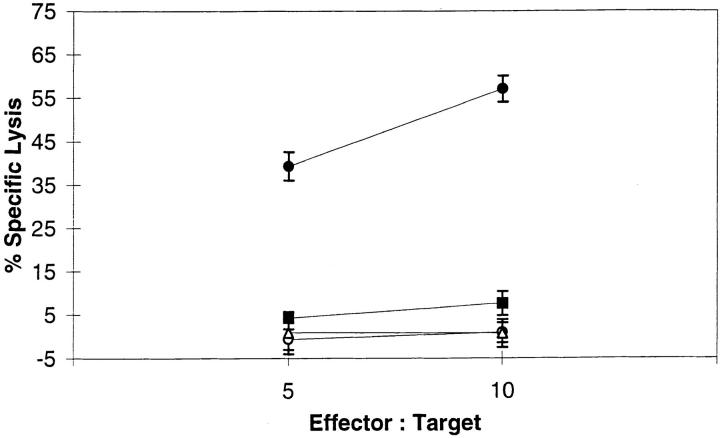
Several epitopes from membrane proteins are generated by a pathway that is independent of the TAP protein, suggesting that after translation in the ER, their processing can be completed in this compartment (37, 63–65). We were interested in whether the formation of YMDGTMSQV from full-length tyrosinase, which involves both translation in the ER and conversion at an unknown site, was also TAP-independent. T2 cells, which are HLA-A*0201+ but have deleted the genes encoding TAP1 and TAP2, were infected with a vaccinia vector encoding full-length tyrosinase and screened with the D-specific CTLs. There was no lysis of the vaccinia-tyrosinase–infected T2 cells. However, when T2 cells were infected with a mixture of recombinant vaccinia viruses expressing tyrosinase and the genes encoding TAP1 and TAP2, the YMDGTMSQV epitope was restored on the cell surface (Fig. 7). These results demonstrate that the production of the YMDGTMSQV epitope is TAP-dependent, and indicates that it, or a precursor that contains the epitope, is in the cytosol before binding HLA-A2.1 in the ER.
Figure 7.
TAP dependence of YMDGTMSQV epitope formation. T2 cells were infected with recombinant vaccinia viruses expressing either tyrosinase (squares), TAP1+2 (triangles), or both tyrosinase and TAP1+2 (closed circles), or else left uninfected (open circles), and used in a 51Cr-release assay with D-specific CTLs. The results are representative of three independent experiments.
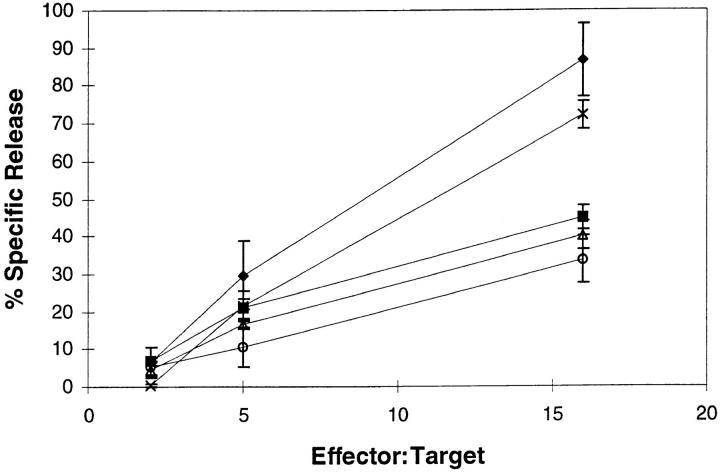
Because of the TAP dependence of the YMDGTMSQV epitope, we were interested in determining whether the proteasome is involved in epitope formation. Proteasome-dependent processing was evaluated by brief exposure of target cells to pH 2.5 medium to denature all cell surface MHC class I molecules, followed by incubation to allow reexpression of the epitope in the presence or absence of lactacystin or LLnL as described elsewhere (Luckey, C.J., G.M. King, J.A. Marto, B.F. Maier, V.L. Crotzer, T.A. Colella, J. Shabanowitz, D.F. Hunt, and V.H. Engelhard, manuscript submitted for publication). CTL recognition of acid-treated target cells was abolished, but recovered to control levels if the targets were incubated for 5 h at 37°C (Fig. 8). This recovery of epitope expression was inhibited by >50% in the presence of BFA, which blocks the egress of newly synthesized peptide MHC class I complexes from the ER (66–68). Although the failure of BFA to completely inhibit reexpression of the epitope is not understood, the difference in recognition between the acid-washed, untreated cells and the acid-washed, BFA-treated cells allowed us to determine whether protease inhibitors also blocked the generation of the YMDGTMSQV epitope. Incubation of acid-washed targets with 20 μM LLnL, a concentration known to selectively inhibit the proteasome (59), inhibited reexpression of the YMDGTMSQV epitope on the cell surface to the same extent as BFA (Fig. 8). In addition, treatment of the acid-washed cells with lactacystin, a highly specific inhibitor of the proteasome (58), also blocked reexpression of the YMDGTMSQV epitope. It has been shown elsewhere that the effects of these proteasome inhibitors are not due to nonspecific effects on class I biosynthesis or susceptibility of target cells to CTL-mediated lysis (Luckey, C.M.J., G.M. King, J.A. Marto, B.F. Maier, V.L. Crotzer, T.A. Colella, J. Shabanowitz, D.F. Hunt, and V.H. Engelhard, manuscript submitted for publication). Together these results indicate that the processing of intact tyrosinase to generate the YMDGTMSQV epitope is dependent upon the proteasome.
Figure 8.
Recognition of the melanoma line DM93 by D Tyr CTL. DM93 was left untreated (crosses) or acid-treated as described in Materials and Methods and then incubated at 37°C for 5 h either in the absence of any inhibitor (diamonds) or in the presence of one of the follwing: 10 μg/ml BFA (squares), 20 μM lactacystin (triangles) or 20 μM LLnL (circles). All targets were then washed and used in a 4-h 51Cr-release assay in the presence of 10 μg/ml BFA to block any further expression of newly synthesized peptide–HLA-A*0201 complexes. These results are representative of four independent experiments.
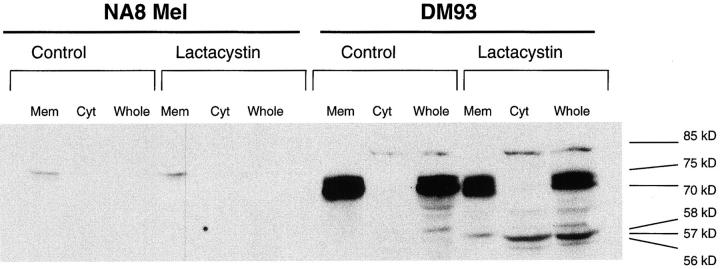
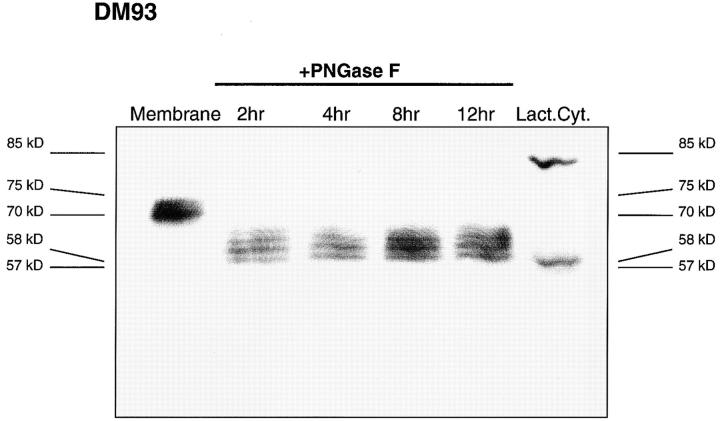
The results obtained thus far suggested that initial translation of tyrosinase in the ER was followed by the export of the full-length protein or a fragment to the cytosol for processing by the proteasome. To investigate the nature of the tyrosinase species exported from the ER to the cytosol, we used lactacystin to inhibit proteasome activity in DM93, a tyrosinase-expressing melanoma, and NA8 Mel, a tyrosinase-negative melanoma. After 5 h the cells were divided into cytosolic and membrane fractions, separated by SDS-PAGE, and immunoblotted for tyrosinase (Fig. 9). In untreated DM93 cells, we observed a strong doublet of 70–75 kD that was located exclusively in the membrane fraction. This doublet represents two major glycosylated species of tyrosinase, and correlate well with those previously identified by others (61, 69). In DM93 cells that had been treated with lactacystin, three additional bands were observed at 56–58 kD, similar in size to the unglycosylated full-length tyrosinase. The band of lowest mobility among this group was located exclusively in the membrane fraction, whereas the other two were found only in the cytosol. To further identify the potential nature of these lower molecular weight bands, the membrane fraction from DM93 cells was treated with PNGase F to deglycosylate asparagine-linked glycosylations (Fig. 10). Human tyrosinase contains seven potential glycosylation sites (70, 71) yet it appears that only four or five sites are used in DM93 cells. Although deglycosylation was not complete after 12 h, the fastest migrating species comigrated with the cytosolic 57-kD band, which appears after lactacystin treatment of DM93 cells. One additional species with an apparent molecular weight of 85-kD was found in the cytosol of untreated cells, but increased in intensity upon lactacystin treatment. The size of this band is too high to be attributed to glycosylation, but is consistent with the attachment of four ubiquitin chains to the unglycosylated full-length tyrosinase polypeptide chain. After long exposure times, two additional low intensity bands of 61 and 65 kD were also found in the cytosol. None of these bands were observed in extracts of NA8 Mel (Fig. 9), whereas a similar pattern was observed in NA8 Mel + Tyr cells (data not shown), indicating that all of the species are derived from tyrosinase. These results are consistent with the hypothesis that the export of a full-length or almost full-length tyrosinase from the ER for degradation by the proteasome provides source material for the production of the YMDGTMSQV peptide epitope.
Figure 9.
Tyrosinase can be detected in the cytosol of lactacystin treated melanoma. NA8 Mel (lanes 1–6), a tyrosinase-negative melanoma, and DM93 (lanes 7–12), a tyrosinase-expressing melanoma, were left untreated (lanes 1–3, 7–9) or treated (lanes 4–6, 10–12) with 10 μg/ml lactacystin for 5 h, after which time they were separated into membrane (lanes 1, 4, 7, 10) or cytosolic (lanes 2, 5, 8, 11) fractions or used as whole cell lysates (lanes 3, 6, 9, 12) as described in Materials and Methods. Lysate equivalent to 106 cells was run in each lane of a 10% SDS-PAGE denaturing gel. The blot from this gel was probed with T311, a tyrosinase specific mAb. These results are representative of four independent experiments.
Figure 10.
Deglycosylation of membrane tyrosinase produces a band which comigrates or nearly comigrates with the tyrosinase species found in the cytosol. Lane 1 contains the membrane fraction from 5 × 105 cells mock treated with PNGase F. Lanes 2–5 contain equivalent DM93 membrane fractions treated with 1.5 mU PNGase for 2, 4, 8 and 12 h, respectively. Lane 6 contains the cytosolic fraction of 5 × 105 DM93 cells treated with lactacystin for 5 h.
Discussion
In a previous paper, we demonstrated that the peptide YMDGTMSQV is formed by the processing and post-translational modification of the sequence YMNGTMSQV located in the full-length tyrosinase protein sequence (49). Since the proposed posttranslational modification responsible for this conversion involved glycosylation in the endoplasmic reticulum, and since tyrosinase is a membrane protein, the objective of this study was to determine whether processing to produce this epitope occurred after normal translation of tyrosinase into the endoplasmic reticulum, or after aberrant translation in the cytosol. Using either a vaccinia-encoded minigene or a transfected fragment of tyrosinase that is missing the signal sequence, we have demonstrated that translation of the sequence YMNGTMSQV in the cytosol results in detectable quantities of this unconverted peptide on the cell surface in association with HLA-A*0201. In contrast, this peptide is not observed in cells that express the full-length form of tyrosinase. This suggests that the routes by which the full-length and truncated protein forms are processed are different, and that the full-length form is not processed after mistranslation in the cytosol.
It is not immediately clear why expression of the unconverted YMNGTMSQV peptide should be a marker of protein translation in the cytosol. It is also unclear why this is the only form detectable on the surface of cells expressing the peptide epitope as a vaccinia minigene, whereas in cells expressing the T3.1 fragment of tyrosinase, both this and the posttranslationally converted epitopes are detected. We believe that the most likely explanation lies in the length and heterogeneity of the epitope-containing fragments produced from these two translation products. The vaccinia minigene product contains the unconverted YMNGTMSQV peptide in “preprocessed” form with the exception of an NH2-terminal methionine residue that is likely to be rapidly removed by cytosolic aminopeptidases (72). As a result, the epitope-containing peptides that are located in the cytosol and transported into the ER by TAP are of the optimal size to bind to newly synthesized HLA-A*0201 molecules. Androlewicz has demonstrated TAP-dependent transport of the YMNGTMSQV peptide from the cytosol to the ER, and has further shown that a fraction of these HLA-A*0201–associated peptides become glycosylated (62). However, we hypothesize that rapid peptide binding prevents further posttranslational conversion of asparagine to aspartic acid by deglycosylation or another mechanism, either because of steric hindrance or because trafficking of class I–peptide complexes does not bring them into contact with all of the enzymes necessary to mediate this process. As a result, the converted form of the peptide is not expressed at the cell surface.
In contrast, the epitope-containing fragments produced after translation and the initial processing of the T3.1 protein could contain an additional COOH-terminal residue and up to 225 additional residues at the NH2 terminus. Based on the size selectivity of TAP (73), it is likely that only those epitope-containing peptides of 9–16 amino acids will be efficiently transported into the ER. As with the minigene-encoded peptides, those 9mers with the sequence YMNGTMSQV would immediately bind to HLA-A*0201 and be protected from posttranslational conversion. However, those peptides of 10–16 residues would be unable to bind to HLA-A*0201 because the dominant motif residues are no longer positioned appropriately with respect to the peptide termini. These peptides would be substrates for enzymes that mediate the conversion of asparagine to aspartic acid. After additional proteolytic processing, they would give rise to the YMDGTMSQV peptides associated with HLA-A*0201.
Regardless of whether the above hypothesis is correct in all aspects, the absence of the unconverted peptide and the exclusive presence of the posttranslationally converted form in cells expressing full-length tyrosinase indicates that the unconverted 9mer peptide is not produced at all, or at least is not produced in a compartment that allows it to bind to HLA-A*0201 before conversion. One possibility is that the normal translation of full-length tyrosinase in the ER results in quantitative glycosylation of the asparagine residue in the context of the intact protein, thus precluding the possibility of producing the unconverted peptide. Deglycosylation before, after, or during proteolysis would lead to conversion of asparagine to aspartic acid, and result in the production of the converted YMDGTMSQV peptide. Alternatively, if glycosylation is incomplete, the unconverted N residue may be a site for proteolysis in the ER, resulting in epitope destruction. In this scenario, the production of the unconverted peptide from T3.1 would be a result of incomplete glycosylation or incomplete proteolysis in the ER, presumably as a result of association with HLA-A*0201. Thus, the differential expression of unconverted and converted forms of this tyrosinase-derived peptide is most likely to be due to the site of translation, the extent of glycosylation, the length of the fragments produced during initial proteolytic processing, and protection from further processing by association with HLA-A*0201.
Given the initial translation of full-length tyrosinase in the ER, a second important result of this study is that the expression of the converted peptide epitope in association with HLA-A*0201 is dependent on TAP and the proteasome. Although it was formally possible that this peptide is fully processed in the ER, and that the role of TAP was to enable efficient peptide loading by interaction with an HLA-A*0201–tapasin complex (64), the inhibition of epitope formation by the specific proteasomal inhibitor lactacystin provides strong evidence that processing occurs at least in part in the cytosol. In fact, in the presence of this inhibitor, tyrosinase species of molecular weights that were consistent with a full-length deglycosylated polypeptide chain were detected in the cytosol. These results are consistent with observations in several other laboratories of a reverse translocation mechanism for the export of misfolded membrane proteins from the ER for degradation by the proteasome (37–48). In addition, tyrosinase expressed in amelanotic melanomas was recently shown to be trapped in the ER and then degraded by a lactacystin-dependent pathway (74). Our results do not allow us to completely exclude a mechanism in which tyrosinase is partially degraded by proteases in the ER, followed by export of the fragments to the cytosol for processing by the proteasome. Nonetheless, we believe that the most likely mechanism involves the reverse translocation of the full-length protein into the cytosol, accompanied by deglycosylation, and followed by proteasomal processing. Previous studies have provided suggestive evidence for two mechanisms of membrane protein processing that involve partial or complete proteolysis in the ER (11–20), or mistranslation of the protein in the cytosol (21–25, 27– 35). The results of this study provide strong evidence for a third pathway that involves reverse translocation of the intact protein from the ER to the cytosol for proteolysis. If, as seems likely, this is a normal pathway for the degradation of misfolded membrane and secreted proteins, then we think it likely that it will also be the major pathway for processing and presentation of the epitopes from these proteins as well.
Acknowledgments
This work was supported by United States Public Health Grants AI-20963 and AI-21393 (V.H. Engelhard); CA-57653 (C.L. Slinghuff, Jr.); GM-37537 and AI-33993 (D.F. Hunt); and by American Cancer Society Grant IM768 (C.L. Slinghuff, Jr.). C.A. Mosse, C.J. Luckey, and E.L. Huczko were supported by Medical Scientist Training Program Grant GM07267. D.J. Kittlesen was supported by a fellowship from the Cancer Research Foundation of America. C.L. Slinghuff, Jr. is a recipient of the Cancer Research Institute's Elaine R. Shepard Clinical Investigator Award in Cancer Immunology.
Footnotes
Abbreviations used in this paper: BFA, brefeldin A; ER, endoplasmic reticulum; m/z, mass/charge ratio; TAP, the transporter associated with antigen.
References
- 1.Engelhard VH. Structure of peptides associated with MHC class I molecules. Curr Opin Immunol. 1994;6:13–23. doi: 10.1016/0952-7915(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 2.Goldberg AL, Rock KL. Proteolysis, proteasomes and antigen presentation. Nature. 1992;357:375–379. doi: 10.1038/357375a0. [DOI] [PubMed] [Google Scholar]
- 3.Bevan MJ. Antigen presentation to cytotoxic T lymphocytes in vivo. J Exp Med. 1995;182:639–641. doi: 10.1084/jem.182.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hill A, Ploegh H. Getting the inside out: the transporter associated with antigen processing (TAP) and the presentation of viral antigen. Proc Natl Acad Sci USA. 1995;92:341–343. doi: 10.1073/pnas.92.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Germain RN. MHC-dependent antigen processing and peptide presentation: providing ligands for T lymphocyte activation. Cell. 1994;76:287–299. doi: 10.1016/0092-8674(94)90336-0. [DOI] [PubMed] [Google Scholar]
- 6.Monaco JJ. Pathways for the processing and presentation of antigens to T cells. J Leukocyte Biol. 1995;57:543–547. doi: 10.1002/jlb.57.4.543. [DOI] [PubMed] [Google Scholar]
- 7.Ortmann B, Androlewicz MJ, Cresswell P. MHC class I/β2-microglobulin complexes associate with TAP transporters before peptide binding. Nature. 1994;368:864–867. doi: 10.1038/368864a0. [DOI] [PubMed] [Google Scholar]
- 8.Suh WK, Cohen-Doyle MF, Fruh K, Wang K, Peterson PA, Williams DB. Interaction of MHC class I molecules with the transporter associated with antigen processing. Science. 1994;264:1322–1326. doi: 10.1126/science.8191286. [DOI] [PubMed] [Google Scholar]
- 9.Sadasivan B, Lehner PJ, Ortmann B, Spies T, Cresswell P. Roles for calreticulin and a novel glycoprotein, tapasin, in the interaction of MHC class I molecules with TAP. Immunity. 1996;5:103–114. doi: 10.1016/s1074-7613(00)80487-2. [DOI] [PubMed] [Google Scholar]
- 10.Degen E, Williams DB. Participation of a novel 88-kD protein in the biogenesis of murine class I histocompatibility molecules. J Cell Biol. 1991;112:1099–1115. doi: 10.1083/jcb.112.6.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henderson RA, Michel H, Sakaguchi K, Shabanowitz J, Appella E, Hunt DF, Engelhard VH. HLA-A2.1 associated peptides from a mutant cell line: a second pathway of antigen presentation. Science. 1992;255:1264–1266. doi: 10.1126/science.1546329. [DOI] [PubMed] [Google Scholar]
- 12.Wei ML, Cresswell P. HLA-A2 molecules in an antigen-processing mutant cell contain signal sequence–derived peptides. Nature. 1992;356:443–446. doi: 10.1038/356443a0. [DOI] [PubMed] [Google Scholar]
- 13.Hammond SA, Bollinger RC, Toberty TW, Siliciano RF. Transport-independent processing of HIV-1 envelope protein for recognition by CD8+T cells. Nature. 1993;364:158–161. doi: 10.1038/364158a0. [DOI] [PubMed] [Google Scholar]
- 14.Lee SP, Thomas WA, Blake NW, Rickinson AB. Transporter (TAP)-independent processing of a multiple membrane-spanning protein, the Epstein-Barr virus latent membrane protein 2. Eur J Immunol. 1996;26:1875–1883. doi: 10.1002/eji.1830260831. [DOI] [PubMed] [Google Scholar]
- 15.Henderson RA, Cox AL, Sakaguchi K, Appella E, Shabanowitz J, Hunt DF, Engelhard VH. Direct identification of an endogenous peptide recognized by multiple HLA-A2.1 specific cytotoxic T cells. Proc Natl Acad Sci USA. 1993;90:10275–10279. doi: 10.1073/pnas.90.21.10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hughes EA, Ortmann B, Surman M, Cresswell P. The protease inhibitor, N-acetyl-l-leucyl-l-leucyl-leucyl-l-norleucinal, decreases the pool of major histocompatibility complex class I–binding peptides and inhibits peptide trimming in the endoplasmic reticulum. J Exp Med. 1996;183:1569–1578. doi: 10.1084/jem.183.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elliott T, Willis A, Cerundolo V, Townsend A. Processing of major histocompatibility class I–restricted antigens in the endoplasmic reticulum. J Exp Med. 1995;181:1481–1491. doi: 10.1084/jem.181.4.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bacik I, Cox JH, Anderson R, Yewdell JW, Bennink JR. TAP (transporter associated with antigen processing)–independent presentation of endogenously synthesized peptides is enhanced by endoplasmic reticulum insertion sequences located at the amino- but not carboxyl-terminus of the peptide. J Immunol. 1994;152:381–387. [PubMed] [Google Scholar]
- 19.Anderson K, Cresswell P, Gammon M, Hermes J, Williamson A, Zweerink H. Endogenously synthesized peptide with an endoplasmic reticulum signal sequence sensitizes antigen processing mutant cells to class I–restricted cell-mediated lysis. J Exp Med. 1991;174:489–492. doi: 10.1084/jem.174.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snyder HL, Yewdell JW, Bennink JR. Trimming of antigenic peptides in an early secretory compartment. J Exp Med. 1994;180:2389–2394. doi: 10.1084/jem.180.6.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Binnendijk RS, van Baalen CA, Poelen MC, de Vries P, Boes J, Cerundolo V, Osterhaus AD, Uyt de Haag FG. Measles virus transmembrane fusion protein synthesized de novo or presented in immunostimulating complexes is endogenously processed for HLA class I– and class II–restricted cytotoxic T cell recognition. J Exp Med. 1992;176:119–128. doi: 10.1084/jem.176.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hammond SA, Johnson RP, Kalams SA, Walker BD, Takiguchi M, Safrit JT, Koup RA, Siliciano RF. An epitope-selective, transporter associated with antigen presentation (TAP)-1/2–independent pathway and a more general TAP-1/2–dependent antigen-processing pathway allow recognition of the HIV-1 envelope glycoprotein by CD8+CTL. J Immunol. 1995;154:6140–6156. [PubMed] [Google Scholar]
- 23.Aldrich CJ, Waltrip R, Hermel E, Attaya M, Fischer K, Lindahl, Monaco JJ, Forman J. T cell recognition of Qa-1bantigens on cells lacking a functional TAP-2 transporter. J Immunol. 1992;149:3773–3777. [PubMed] [Google Scholar]
- 24.Aldrich CJ, DeCloux A, Woods AS, Cotter RJ, Soloski MJ, Forman J. Identification of a TAP-dependent leader peptide recognized by alloreactive T cells specific for a class Ib antigen. Cell. 1994;79:649–658. doi: 10.1016/0092-8674(94)90550-9. [DOI] [PubMed] [Google Scholar]
- 25.Hombach J, Pircher H, Tonegawa S, Zinkernagel RM. Strictly transporter of antigen presentation (TAP)- dependent presentation of an immunodominant cytotoxic T lymphocyte epitope in the signal sequence of a virus protein. J Exp Med. 1995;182:1615–1619. doi: 10.1084/jem.182.5.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roelse J, Gromme M, Momburg F, Hammerling G, Neefjes J. Trimming of TAP-translocated peptides in the endoplasmic reticulum and in the cytosol during recycling. J Exp Med. 1994;180:1591–1597. doi: 10.1084/jem.180.5.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yewdell JW, Bennink JR. Cell biology of antigen processing and presentation to major histocompatibility complex class I molecule–restricted T lymphocytes. Adv Immunol. 1992;52:1–123. doi: 10.1016/s0065-2776(08)60875-5. [DOI] [PubMed] [Google Scholar]
- 28.Lurquin C, Van Pel A, Mariame B, De Plaen E, Szikora JP, Janssens C, Reddehase MJ, Lejeune J, Boon T. Structure of the gene of tum- transplantation antigen P91A: the mutated exon encodes a peptide recognized with Ld by cytolytic T cells. Cell. 1989;58:293–303. doi: 10.1016/0092-8674(89)90844-1. [DOI] [PubMed] [Google Scholar]
- 29.Boon T, Van Pel A. T cell–recognized antigenic peptides derived from the cellular genome are not protein degradation products but can be generated directly by transcription and translation of short subgenic regions. A hypothesis [see comments] Immunogenetics. 1989;29:75–79. doi: 10.1007/BF00395854. [DOI] [PubMed] [Google Scholar]
- 30.Sibille C, Chomez P, Wildmann C, Van Pel A, De Plaen E, Maryanski JL, de Bergeyck V, Boon T. Structure of the gene of tum- transplantation antigen P198: a point mutation generates a new antigenic peptide. J Exp Med. 1990;172:35–45. doi: 10.1084/jem.172.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chomez P, De Plaen E, Van Pel A, de Smet C, Szikora JP, Lurquin C, Lebacq-Verheyden AM, Boon T. Efficient expression of tum- antigen P91A by transfected subgenic fragments. Immunogenetics. 1992;35:241–252. doi: 10.1007/BF00166829. [DOI] [PubMed] [Google Scholar]
- 32.Scott DM, Ehrmann IE, Ellis PS, Simpson E, Agulnik AI, Bishop CE, Mitchell MJ. Identification of a mouse male-specific transplantation antigen, H-Y. Nature. 1995;376:695–698. doi: 10.1038/376695a0. [DOI] [PubMed] [Google Scholar]
- 33.Wang RF, Parkhurst MR, Kawakami Y, Robbins PF, Rosenberg SA. Utilization of an alternative open reading frame of a normal gene in generating a novel human cancer antigen. J Exp Med. 1996;183:1131–1140. doi: 10.1084/jem.183.3.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bullock TNJ, Eisenlohr LC. Ribosomal scanning past the primary initiation codon as a mechanism for expression of ctl epitopes encoded in alternative reading frames. J Exp Med. 1996;184:1319–1329. doi: 10.1084/jem.184.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leonard CK, Spellman MW, Riddle L, Harris RJ, Thomas JN, Gregory TJ. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J Biol Chem. 1990;265:10373–10382. [PubMed] [Google Scholar]
- 36.Ferris RL, Buck C, Hammond SA, Woods AS, Cotter RJ, Takiguchi M, Igarashi Y, Ichikawa Y, Siliciano RF. Class I–restricted presentation of an HIV-1 gp41 epitope containing an N-linked glycosylation site. Implications for the mechanism of processing of viral envelope proteins. J Immunol. 1996;156:834–840. [PubMed] [Google Scholar]
- 37.McCracken AA, Brodsky JL. Assembly of ER-associated protein degradation in vitro: dependence on cytosol, calnexin, and ATP. J Cell Biol. 1996;132:291–298. doi: 10.1083/jcb.132.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Werner ED, Brodsky JL, McCracken AA. Proteasome-dependent endoplasmic reticulum–associated protein degradation: an unconventional route to a familiar fate. Proc Natl Acad Sci USA. 1996;93:13797–13801. doi: 10.1073/pnas.93.24.13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hiller MM, Finger A, Schweiger M, Wolf DH. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- 40.Jensen TJ, Loo MA, Pind S, Williams DB, Goldberg AL, Riordan JR. Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell. 1995;83:129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- 41.Ward CL, Omura S, Kopito RR. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 42.Hughes EA, Hammond C, Cresswell P. Misfolded major histocompatibility complex class I heavy chains are translocated into the cytoplasm and degraded by the proteasome. Proc Natl Acad Sci USA. 1997;94:1896–1901. doi: 10.1073/pnas.94.5.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huppa JB, Ploegh HL. The alpha chain of the T cell antigen receptor is degraded in the cytosol. Immunity. 1997;7:113–122. doi: 10.1016/s1074-7613(00)80514-2. [DOI] [PubMed] [Google Scholar]
- 44.Kopito RR. ER quality control: the cytoplasmic connection. Cell. 1997;88:427–430. doi: 10.1016/s0092-8674(00)81881-4. [DOI] [PubMed] [Google Scholar]
- 45.Yu H, Kaung G, Kobayashi S, Kopito RR. Cytosolic degradation of T-cell receptor alpha chains by the proteasome. J Biol Chem. 1997;272:20800–20804. doi: 10.1074/jbc.272.33.20800. [DOI] [PubMed] [Google Scholar]
- 46.Brodsky JL, McCracken AA. ER-associated and proteasome-mediated protein degradation: how two topologically restricted events came together. Trends Cell Biol. 1997;7:151–156. doi: 10.1016/S0962-8924(97)01020-9. [DOI] [PubMed] [Google Scholar]
- 47.Wiertz EJ, Tortorella D, Bogyo M, Yu J, Mothes W, Jones TR, Rapoport TA, Ploegh HL. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature. 1996;384:432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]
- 48.Wiertz EJ, Jones TR, Sun L, Bogyo M, Geuze HJ, Ploegh HL. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell. 1996;84:769–779. doi: 10.1016/s0092-8674(00)81054-5. [DOI] [PubMed] [Google Scholar]
- 49.Skipper JCA, Hendrickson RC, Gulden PH, Brichard V, Van Pel A, Chen Y, Shabanowitz J, Wolfel T, Slingluff CL, Boon T, Hunt DF, Engelhard VH. An HLA-A2–restricted tyrosinase antigen on melanoma cells results from posttranslational modification and suggests a novel processing pathway for membrane proteins. J Exp Med. 1996;183:527–534. doi: 10.1084/jem.183.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suzuki T, Seko A, Kitajima K, Inoue Y, Inoue S. Identification of peptide: N-glycanase activity in mammalian-derived cultured cells. Biochem Biophys Res Commun. 1993;194:1124–1130. doi: 10.1006/bbrc.1993.1938. [DOI] [PubMed] [Google Scholar]
- 51.Wolfel T, Van Pel A, Brichard V, Schneider J, Seliger B, Meyer zum Buschenfelde KH, Boon T. Two tyrosinase nonapeptides recognized on HLA-A2 melanomas by autologous cytolytic T lymphocytes. Eur J Immunol. 1994;24:759–764. doi: 10.1002/eji.1830240340. [DOI] [PubMed] [Google Scholar]
- 52.Salter RD, Howell DN, Cresswell P. Genes regulating HLA class I antigen expression in T–B lymphoblast hybrids. Immunogenetics. 1985;21:235–246. doi: 10.1007/BF00375376. [DOI] [PubMed] [Google Scholar]
- 53.Henderson RA, Michel H, Sakaguchi K, Shabanowitz J, Appella E, Hunt DF, Engelhard VH. HLA-A2.1–associated peptides from a mutant cell line: a second pathway of antigen presentation. Science. 1992;255:1264–1266. doi: 10.1126/science.1546329. [DOI] [PubMed] [Google Scholar]
- 54.Hahn YS, Braciale VL, Braciale TJ. Presentation of viral antigen to class I major histocompatibility complex–restricted cytotoxic T lymphocyte. Recognition of an immunodominant influenza hemagglutinin site by cytotoxic T lymphocyte is independent of the position of the site in the hemagglutinin translation product. J Exp Med. 1991;174:733–736. doi: 10.1084/jem.174.3.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mackett M, Smith GL, Moss B. General method for product on and selection of infectious vaccinia virus recombinants expressing foreign genes. J Virol. 1984;49:857–864. doi: 10.1128/jvi.49.3.857-864.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Newberg MH, Ridge JP, Vining DR, Salter RD, Engelhard VH. Species specificity in the interaction of CD8 with the α3 domain of MHC class I molecules. J Immunol. 1992;149:136–142. [PubMed] [Google Scholar]
- 57.Hunt DF, Henderson RA, Shabanowitz J, Sakaguchi K, Michel H, Sevilir N, Cox A, Appella E, Engelhard VH. Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science. 1992;255:1261–1263. doi: 10.1126/science.1546328. [DOI] [PubMed] [Google Scholar]
- 58.Fenteany G, Standaert RF, Lane WS, Choi S, Corey EJ, Schreiber SL. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 59.Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 60.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 61.Chen YT, Stockert E, Tsang S, Coplan KA, Old LJ. Immunophenotyping of melanomas for tyrosinase: implications for vaccine development. Proc Natl Acad Sci USA. 1995;92:8125–8129. doi: 10.1073/pnas.92.18.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Androlewicz MJ. An N-glycosylated tyrosinase epitope associates with newly synthesized MHC class I molecules in melanoma cells. Hum Immunol. 1996;51:81–88. doi: 10.1016/s0198-8859(96)00237-6. [DOI] [PubMed] [Google Scholar]
- 63.Deverson EV, Gow IR, Coadwell WJ, Monaco JJ, Butcher GW, Howard JC. MHC class II region encoding proteins related to the multidrug resistance family of transmembrane transportors. Nature. 1990;348:738–741. doi: 10.1038/348738a0. [DOI] [PubMed] [Google Scholar]
- 64.Sadasivan B, Lehner PJ, Ortmann B, Spies T, Cresswell P. Roles for calreticulin and a novel glycoprotein, tapasin, in the interaction of MHC class I molecules with TAP. Immunity. 1996;5:103–114. doi: 10.1016/s1074-7613(00)80487-2. [DOI] [PubMed] [Google Scholar]
- 65.Lee SP, Thomas WA, Blake NW, Rickinson AB. Transporter (TAP)-independent processing of a multiple membrane-spanning protein, the Epstein-Barr virus latent membrane protein 2. Eur J Immunol. 1996;26:1875–1883. doi: 10.1002/eji.1830260831. [DOI] [PubMed] [Google Scholar]
- 66.Yewdell JW, Bennink JR. Brefeldin A specifically inhibits presentation of protein antigens to cytotoxic T lymphocytes. Science. 1989;244:1072–1075. doi: 10.1126/science.2471266. [DOI] [PubMed] [Google Scholar]
- 67.Nuchtern JG, Bonifacino JS, Biddison WE, Klausner RD. Brefeldin A implicates egress from endoplasmic reticulum in class I–restricted antigen presentation. Nature. 1989;339:223–226. doi: 10.1038/339223a0. [DOI] [PubMed] [Google Scholar]
- 68.Lippincott-Schwartz J, Yuan LC, Bonifacino JS, Klausner RD. Rapid redistribution of Golgi proteins in the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell. 1989;56:801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jimenez M, Maloy WL, Hearing VJ. Specific identification of an authentic clone for mammalian tyrosinase. J Biol Chem. 1989;264:3397–3403. [PubMed] [Google Scholar]
- 70.Kwon BS, Haq AK, Pomerantz SH, Halaban R. Isolation and sequence of a cDNA for human tyrosinase that maps at the mouse c-albino locus. Proc Natl Acad Sci USA. 1987;84:7473–7477. doi: 10.1073/pnas.84.21.7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bouchard B, Fuller BB, Vijayasaradhi S, Houghton AN. Induction of pigmentation in mouse fibroblasts by expression of human tyrosinase cDNA. J Exp Med. 1989;169:2029–2042. doi: 10.1084/jem.169.6.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arfin SM, Kendall RL, Hall L, Weaver LH, Stewart AE, Matthews BW, Bradshaw RA. Eukaryotic methionyl aminopeptidases: two classes of cobalt-dependent enzymes. Proc Natl Acad Sci USA. 1995;92:7714–7718. doi: 10.1073/pnas.92.17.7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schumacher TN, Kantesaria DV, Heemels MT, Ashton-Rickardt PG, Shepherd JC, Fruh K, Yang Y, Peterson PA, Tonegawa S, Ploegh HL. Peptide length and sequence specificity of the mouse TAP1/TAP2 translocator. J Exp Med. 1994;179:533–540. doi: 10.1084/jem.179.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Halaban R, Cheng E, Zhang Y, Moellmann G, Hanlon D, Michalak M, Setaluri V, Hebert DN. Aberrant retention of tyrosinase in the endoplasmic reticulum mediates accelerated degradation of the enzyme and contributes to the dedifferentiated phenotype of amelanotic melanoma cells. Proc Natl Acad Sci USA. 1997;94:6210–6215. doi: 10.1073/pnas.94.12.6210. [DOI] [PMC free article] [PubMed] [Google Scholar]