Abstract
The addition of new telomeres to the ends of broken chromosomes, termed chromosome healing, has been extensively studied in unicellular organisms; however, its role in the mammalian cell response to double-strand breaks is unknown. A system for analysis of chromosome healing, which involves the integration of plasmid sequences immediately adjacent to a telomere, has been established in mouse embryonic stem cells. This “marked” telomere contains a neo gene for positive selection in G418, an I-SceI endonuclease recognition sequence for introducing double-strand breaks, and a herpes simplex virus thymidine kinase gene for negative selection with ganciclovir for cells that have lost the telomere. Transient expression of the I-SceI endonuclease results in terminal deletions involving telomeric repeat sequences added directly onto the end of the broken chromosome. The sites of addition of the new telomeres contain short regions of complementarity to telomeric repeat sequences. The most common site of addition is the last A of the ATAA 3′ overhang generated by the I-SceI endonuclease, without the loss of a single nucleotide from the end of the chromosome. The next most frequent site involved 5 bp of complementarity, which occurred after the loss of four nucleotides from the end of the chromosome. The new telomeres are generally much shorter than in the parental cell line, and most increase in size with time in culture. These results demonstrate that chromosome healing is a mechanism for repair of chromosome breaks in mammalian cells.
Telomeres are DNA–protein complexes that contain short repeat sequences that are added on to the ends of chromosomes by the enzyme telomerase (1). Telomerase is a reverse transcriptase that carries an RNA template that is aligned with the end of the chromosome for addition of the repeat sequences. Telomeres serve multiple functions, including protecting the ends of chromosomes (1) and preventing chromosome fusion (2). Telomeres are maintained in the germ line in mammals but shorten with age in most somatic cells (2), which has been proposed to be the signal for senescence (3). Consistent with this hypothesis, the introduction of telomerase into normal human fibroblasts can result in an increase in their capacity to divide in culture (4), although telomerase activity alone is not sufficient to prevent senescence in human keratinocytes (5).
Telomeres lost from the ends of chromosomes can be restored by a mechanism termed chromosome healing, which has been observed in a number of different organisms (6–11). The addition of new telomeres onto broken chromosomes requires little or no complementarity between the telomeric repeat sequences (TRS) and the end of the chromosome in most organisms (6–8). In yeast, chromosome healing has been studied by using the HO endonuclease to generate a specific double-strand break (dsb) within integrated sequences in cells that are deficient in dsb repair (9–11). Unlike many other organisms, chromosome healing in yeast occurs predominantly at locations where TRS are located within the chromosome (9). However, in cells with mutations in PIF1, a DNA helicase, the frequency of chromosome healing increases approximately 1,000-fold, and the location of telomere addition is much more random (11). Thus, like most organisms, yeast telomerase itself requires little sequence complementarity to prime the addition of TRS onto the ends of broken chromosomes.
The most conclusive evidence for chromosome healing in humans comes from studies of terminal deletions associated with genetic disease. Terminal deletions account for approximately 10% of genetic disease associated with chromosome aberrations (12). In six different cases of thalassemia, terminal deletions resulting in the loss of function of the α-globin gene involved TRS being added directly onto the end of the broken chromosome (13). Similarly, a terminal deletion of the end of chromosome 22q that is associated with mental retardation was also shown to involve the addition of a telomere directly to the end of the broken chromosome (14). Little or no complementarity to TRS was evident in the cellular DNA near the sites of chromosome healing on chromosomes 16p or 22q (13, 14). Consistent with this observation, in vitro studies with human cell extracts demonstrated that the addition of TRS onto the ends of oligonuceotides with ends similar to a site of chromosome healing on chromosome 16p required only a few bp of complementarity to TRS (15).
Although chromosome healing has been demonstrated to involve telomerase in Tetrahymena (16), the mechanism(s) of chromosome healing in mammalian cells has not been established. The de novo addition of telomeres is one possible mechanism in cells expressing telomerase; however, chromosome healing could also occur by nonhomologous end joining of preexisting TRS, termed telomere capture. In fact, in one study terminal deletions in cancer cells were shown to be cryptic translocations that were too small to be detected by cytogenetic analysis (17). Another study by using fluorescence in situ hybridization suggested that some chromosome breaks generated with ionizing radiation were associated with telomere capture (18). However, the conclusions of this study were based on the unproven assumption that telomeres added by de novo synthesis would be too short to be detectable by this method. Evidence that capture of preexisting telomeres can occur is also suggested by the observation that telomeres were found on both ends of plasmid DNA (19) after transfection into a cell line that was later found to have no detectable telomerase activity (20). Clearly, a more in-depth study of chromosome healing is required in mammalian cells to determine the mechanism(s) involved, the rate at which it occurs, and its role in the cellular response to dsbs. To address these questions, we have developed mouse embryonic stem (ES) cell lines that contain selectable marker genes and an 18-bp I-SceI endonuclease recognition site immediately adjacent to a telomere. The selection of cells with terminal deletions on the end of this chromosome after the introduction of specific dsbs by transient expression of the I-SceI endonuclease provides a means of studying the sequences and mechanisms involved in chromosome healing in mammalian cells.
MATERIALS AND METHODS
Cell Culture.
The mouse ES cell line, JM-1, used in these studies was obtained from Roger Pedersen (University of California, San Francisco). JM-1 cells were grown on feeder layers consisting of STO cells that had been treated with 50 Gy of ionizing radiation as previously described (21). Leukemia inhibition factor (GIBCO) was added to the medium at 1,000 units/ml. The pNPT-tel plasmid was introduced into the cells by electroporation (Bio-Rad). Clones containing the integrated plasmid were selected with 300 μg/ml G418.
Plasmids.
The plasmid used for analysis of terminal deletions in our studies, pNPT-tel (Fig. 1a), was constructed from the pSXneo-1.6T2AG3 plasmid previously shown to form new telomeres on integration (22). The pSXneo-1.6T2AG3 plasmid contains an ampicillin-resistance gene for selection in bacteria, a neo gene for selection in mammalian cells with G418, as well as TRS to “seed” new telomeres on integration on the end of broken chromosomes. pNPT-tel was created from pSXneo-1.6T2AG3 by insertion of an 18-bp I-SceI endonuclease recognition sequence and a herpes simplex virus thymidine kinase (HSV-tk). After linearization with the NotI restriction enzyme and integration, the HSV-tk gene is immediately adjacent to the telomere, with the I-SceI site between the HSV-tk gene and the neo gene (Fig. 1a). The HSV-tk gene is used for negative selection with ganciclovir (23, 24) and has a phosphoglycerate kinase (pgk) promoter that transcribes efficiently in ES cells (25).
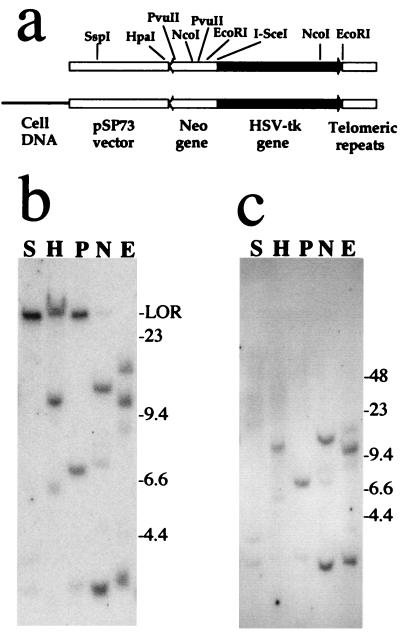
Figure 1.
The characterization of the integrated pNPT-tel plasmid sequences in clone A211. (a) Comparison of the structure of NotI-linearized pNPT-tel plasmid and the integrated plasmid sequences in clone A211 indicate that a single copy of the plasmid is integrated at a telomere. Southern blot analysis of genomic DNA digested with SspI (S), HpaI (H), PvuII (P), NcoI (N), or EcoRI (E), and electrophoresed on (b) conventional agarose or (c) pulse-field gels. Hybridization was performed with the pNTP-Δ plasmid. The location of λ bacteriophage HindIII fragments used as size standards (kb) are shown. The large terminal restriction fragments are at the limit of resolution (LOR) on conventional gels, whereas they are polymorphic on pulse-field gels because of the variation in the length of the TRS.
Treatment of Cells with I-SceI.
Electroporation of plasmids expressing the I-SceI endonuclease gene has been shown to promote recombination in mammalian cells at sites containing the 18-bp I-SceI recognition sequence (26–30). The plasmid used in our study is pCBASce, which contains the I-SceI with a chicken β-actin promoter that was found to provide a high efficiency of cutting at I-SceI sites in mammalian cells (28). pCBASce was electroporated at 30 μg/5 × 105 cells. After waiting 6 days for turnover of existing HSV-tk, previously shown to be the required time for the generation of resistance to ganciclovir (23, 24), the cells were plated in medium containing 2 μM ganciclovir and 300 μg/ml G418. After approximately 2 weeks, individual colonies were selected for analysis.
Fluctuation Analysis.
The analysis of the rate of formation of Ganr cells was determined by first plating 50 cells/well in 24-well tissue culture dishes. After 7 days, the cells were replated in six-well tissue culture dishes. On reaching near confluence, the cells were replated at 200,000 cells/100-mm tissue culture dish in quadruplicate. The cells were plated into 2 μM gancilovir and incubated, with changes every 5 days, for a period of 2 to 3 weeks. The cells were then stained and the colonies counted. The calculations for the rate of appearance of Ganr cells was determined as described by Luria and Delbruck (31) as modified by Capizzi and Jameson (32).
Southern Blot Analysis.
Genomic DNA was purified as previously described (20). Digestion of DNA with restriction enzymes and Bal31 exonuclease, and conventional agarose gel electrophoresis were performed as previously described (19). Pulse-field gel electrophoresis was performed on 1% agarose gels in 0.5× TBE (0.045 M Tris-borate, 0.001M EDTA), at 200 V, 10°C, and pulsed at 2.0- to 3.6- or 1.0- to 3.0-sec intervals. Southern blots and hybridization were performed as previously described (20) by using the pNTP-Δ probe that contains the same sequences as the integrated pNPT-tel plasmid, except that the TRS have been removed.
DNA Analysis.
PCR was performed by using one primer complementary to the HSV-tk promoter on the neo gene (ACCTGCGTGCAATCCATC) and one primer complementary to the TRS (AACCCTAACCCTAACCCT). PCR was performed as previously described (33) by using an initial incubation for 2 min at 95°C, followed by 40 cycles of 95°C for 30 sec, 62°C for 30 sec, and 72°C for 30 sec, followed by one cycle of 95°C for 30 sec, 62°C for 30 sec, and 72°C for 2 min. The PCR products were cloned into the PCRII cloning vector (Invitrogen) by using the protocols provided by the manufacturer. Nucleotide sequence analysis was performed by the Biomolecular Resources Center, University of California, San Francisco, by using universal primers contained within the cloning vector.
RESULTS
Selection of Cell Clones Containing a Plasmid Integrated at a Telomere.
Linearized plasmids containing TRS on one end have been previously demonstrated to form new telomeres when integrated on the ends of truncated chromosomes in mammalian cells (22, 34, 35). After introduction into the cell, these marked telomeres are elongated and maintained similarly to other telomeres within the cell (36). In the case of mouse ES cells, the marked telomeres become very long (34), similar to the endogenous telomeres (37). The mouse ES cell clones used in the present study were established after electroporation of the linearized pNPT-tel plasmid (Fig. 1a). Southern blot analysis was used as an initial screen for clones in which the plasmid was integrated at a telomere, as demonstrated by the presence of large polymorphic fragments containing the TRS.
Four clones have now been identified that have the pNPT-tel plasmid integrated at a telomere. Southern blot analysis of one of these, A211, is consistent with a single copy of the unrearranged plasmid at the integration site (Fig. 1a). SspI that cuts once at the end of the plasmid furthest from the telomere gives a large band on conventional agarose gels that is at the limit of resolution (Fig. 1b), while in pulse-field gels this band is polymorphic in length (Fig. 1c), typical of terminal restriction fragments. A discrete band representing the internal portion of the plasmid is also present. In addition, two lighter bands are also commonly present that represent cross-hybridizing mouse sequences, as shown by hybridization to untransfected mouse ES cell DNA (data not shown). Digestion with HpaI or PvuII, which cut in the middle of the plasmid, also give large bands that are polymorphic on pulse-field gels, as well as additional discrete bands that represent the internal portion of the plasmid. In contrast, restriction enzymes that cut in two locations in the plasmid (NcoI and EcoRI), one of which is near the TRS, both produce two discrete bands. The lengths of one of the bands with NcoI (3.4 kb) and EcoRI (3.3 kb) are consistent with the lengths of the internal fragments in the unrearranged plasmid (Fig. 1a). A single telomeric integration site in clone A211 was also indicated by fluorescence in situ hybridization. By using the pNTP-Δ plasmid as a probe, a single pair of spots was observed at the end of the long arm of an acrocentric chromosome (data not shown).
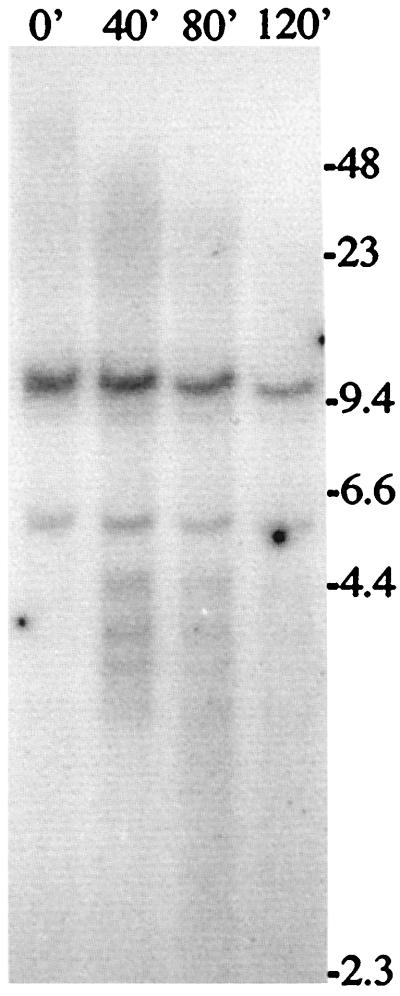
To provide further evidence that the plasmid in clone A211 is located at a telomere, the genomic DNA from clone A211 was treated with Bal31 exonuclease before digestion with the HpaI restriction enzyme (Fig. 2). Bal31 is commonly used to confirm that sequences are located near telomeres, based on the increased susceptibility of DNA located at the ends of chromosomes to exonuclease digestion (19, 34). As shown in Fig. 1, the untreated DNA digestion with HpaI generates two discrete bands in addition to the polymorphic band containing the TRS. The upper dark band is the internal portion of the plasmid, while the lower lighter band is a cross-hybridizing mouse sequence. These internal bands serve as controls for the specificity of digestion with the Bal31 exonuclease. Within 40 min of Bal31 digestion, the polymorphic band is significantly reduced in size, with many of the digestion products smaller than the internal control fragments (Fig. 2). By 120 min, nearly the entire polymorphic band has been reduced to fragments smaller than the internal controls. The larger discrete band, which represents the internal portion of the plasmid, also shows some degree of digestion at 120 min, consistent with its location adjacent to the large terminal restriction fragment. The degradation of the terminal fragment results in bands of discrete sizes, suggesting that some regions within the fragment are resistant to exonuclease digestion. The size of these fragments (4 kb and less) shows that these regions are found within the subtelomeric plasmid sequences (which are 4 kb with HpaI) and not in the TRS.
Figure 2.
Southern blot analysis of genomic DNA from clone A211 digested with Bal31 exonuclease. Genomic DNA was digested for 0, 40, 80, or 120 min with Bal31, followed by complete digestion with HpaI, which cuts once in the middle of the plasmid. The DNA was separated by pulse-field gel electrophoresis, and hybridization was performed with the pNTP-Δ plasmid as a probe.
Selection and Characterization of Spontaneous Ganr Subclones of A211.
Growth in medium containing 2 μM ganciclovir without G418 demonstrated that clone A211 had ganciclovir-resistant (Ganr) cells present at a frequency of approximately 10−3 in the population. Fluctuation analysis of the rate of appearance of Ganr cells in the population demonstrated that the loss of function of the HSV-tk gene occurred at a rate of approximately 10−4 events/cell/generation. However, Southern blot analysis of genomic DNA isolated from the Ganr subclones failed to demonstrate any change in the structure of the integrated plasmid sequences (data not shown). Thus, the high rate of generation of Ganr cells in clone A211 appears to be caused by either silencing or a high rate of point mutations within the HSV-tk gene. Silencing appears to be the most likely explanation, because silencing has previously been found to occur in DNA transfected into mouse ES cells (38). Consistent with this conclusion, clone A211 gradually loses resistance to G418 when grown in medium without G418. As a result, clone A211 is routinely grown in G418 (200 μg/ml) to prevent silencing of the region near the telomere. Because the promoters for both the neo gene and HSV-tk gene are immediately adjacent to one another, preventing silencing of the neo gene should also inhibit silencing of the HSV-tk gene. In fact, coselection with 200 μg/ml G418 in addition to ganciclovir caused a 100-fold decrease in the number of colonies, as compared with ganciclovir alone. Fluctuation analysis of A211 cells in the presence of both ganciclovir and G418 demonstrated that cells resistant to both ganciclovir and G418 (Ganr/G418r) are generated at a rate of less than 10−6 events/cell/generation. Thus, most cells that had lost the function of the HSV-tk gene had also lost the function of the neo gene. However, Southern blot analysis of genomic DNA from spontaneous Ganr/G418r subclones showed no detectable changes in the plasmid (data not shown). In these subclones, the HSV-tk gene either has point mutations or is silenced without affecting transcription of the adjacent neo gene. Regardless of the mechanism, spontaneous rearrangements of the HSV-tk gene have not been observed, demonstrating that the plasmid integration site is highly stable.
Selection and Characterization of dsb-Induced Ganr/G418r Subclones of A211.
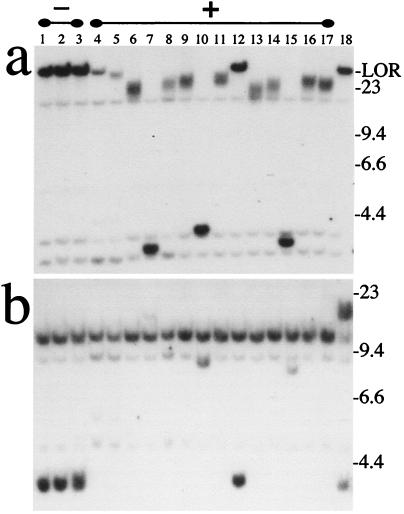
The influence of dsbs on telomere loss and chromosome healing was determined by transiently expressing a plasmid containing the I-SceI endonuclease gene into clone A211 to introduce a dsb at the 18-bp recognition sequence between the neo and HSV-tk genes. I-SceI endonuclease has previously been demonstrated to promote recombination in ES cells in integrated plasmid sequences containing the I-SceI recognition sequence (26, 27, 30). The frequency of Ganr/G418r colonies increased significantly (by as much as 10-fold) when clone A211 was treated with the I-SceI endonuclease. Genomic DNA isolated from mock-treated Ganr/G418r subclones that was digested with SspI (Fig. 3a) showed bands identical to those of the parental clone A211, i.e., a band at the limit of resolution, a smaller discrete band representing the internal portion of the plasmid, and two other light bands caused by cross hybridization with mouse sequences. Thus, the inactivation of the HSV-tk gene in the mock-treated Ganr/G418r subclones resulted from either point mutations or silencing. In contrast, in most Ganr/G418r subclones treated with I-SceI, the SspI fragments containing the TRS are much smaller than in the parental clone (Fig. 3a) and are polymorphic in length typical of terminal restriction fragments. The lighter intensity of these bands in most I-SceI-treated subclones suggested that terminal deletions had occurred. In other Ganr/G418r subclones, the large band containing the TRS is absent and smaller discrete bands appear (lanes 7, 10, and 15), suggesting that the telomere has been lost and that new nontelomeric DNA has been ligated onto the end of the chromosome. Finally, some Ganr/G418r subclones show no detectable changes in the plasmid (lane 12). Approximately one in 10 subclones would be expected to show no change in the structure of the plasmid, because there was up to a 10-fold increase in Ganr/G418r colonies after treatment with I-SceI.
Figure 3.
Southern blot analysis of Ganr/G418r subclones of A211. Genomic DNAs from three mock-treated Ganr/G418r subclones (lanes 1–3), 14 Ganr/G418r subclones isolated after electroporation with the pCBASce plasmid containing the I-SceI gene (lanes 4–17), and a clone of A211 (lane 18) were digested with (a) SspI that cuts once in the plasmid, or (b) EcoRI that cuts the plasmid once in the neo promoter and once between the HSV-tk gene and the TRS. Hybridization was performed with the pNTP-Δ plasmid as a probe.
Further analysis of the structure of the integrated plasmid sequences in the Ganr/G418r subclones was performed by Southern blot analysis after digestion with the EcoRI restriction enzyme. EcoRI cuts the integrated plasmid sequences twice, once in the neo promoter near one end of the HSV-tk gene and once at the other end of the HSV-tk gene near the TRS (see Fig. 1). The digestion of genomic DNA from the mock-treated clones with EcoRI (Fig. 3b) gave bands expected from the intact plasmid. Two plasmid-specific bands are seen, a 3-kb restriction fragment containing the HSV-tk gene and an 11-kb band containing the internal portion of the plasmid. No polymorphic bands are present because the TRS are not contained in the hybridizing fragments. In contrast, most of the I-SceI-treated Ganr/G418r subclones show the loss of the 3-kb EcoRI fragment containing the HSV-tk gene. Combined with the SspI data, the absence of the 3-kb EcoRI fragment demonstrates that these subclones have sustained a terminal deletion on the end of this chromosome, and that a telomere has been added on at a new location. Thus, the terminal fragment containing the HSV-tk gene has been either degraded or lost in these subclones. One I-SceI-treated clone (lane 12) and the mock-treated subclones that showed no changes with SspI retained the 3-kb fragment containing the HSV-tk gene. The three subclones that showed smaller nonpolymorphic bands after SspI digestion (lanes 7, 10, and 15) also showed new light bands after EcoRI digestion, consistent with the interpretation that nontelomeric DNA had been ligated onto the end of the chromosome after the loss of the HSV-tk gene. The genomic DNA from a clone of A211 that was grown for a prolonged period in the absence of G418 (Fig. 3b, lane 18) demonstrated an additional larger band resulting from the partial digestion of the EcoRI site in the promoter for the neo gene. The resistance to digestion at this restriction site corresponds to the loss of resistance to G418 and is not seen when clone A211 is grown continuously in G418 (data not shown). The sequence at this EcoRI site is identical (CGAATTCG) to the sequence at an EcoRI site previously shown to be inhibited by methylation (39), suggesting that the loss of expression of the neo and HSV-tk genes in the plasmid is because of methylation.
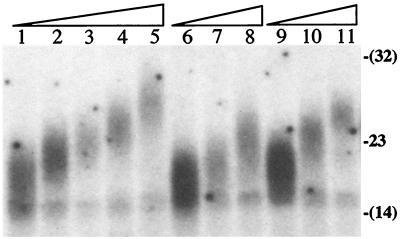
The analysis of telomere length in the I-SceI-induced Ganr/G418r subclones demonstrated that the newly added telomeres increased in length with increasing time in culture (Fig. 4). Based on a cell-cycle time of 17 hr (unpublished observation), the rate of increase in telomere length in subclone +4 (lane 14, Fig. 3) was 38 bp/cell division. A similar rate of increase in telomere length was seen in two other subclones, +11 (lane 13, Fig. 3) and +21 (lane 6, Fig. 3).
Figure 4.
The increase in the length of the marked telomere in I-SceI-induced Ganr/G418r subclones with increasing time in culture. The genomic DNAs from the I-SceI-induced Ganr/G418r subclone +4 initially (1) or after an additional 13 (2), 58 (3), 102 (4), or 153 (5) cell divisions, the I-SceI-induced Ganr/G418r subclone +11 initially (6) or after an additional 83 (7) or 154 (8) cell divisions, and the I-SceI-induced Ganr/G418r subclone +21 initially (9) or after an additional 13 (10) or 58 (11) cell divisions were digested with SspI and separated by pulsed-field gel electrophoresis. Southern blot analysis was performed with the pNTP-Δ plasmid as a probe. The locations are shown for the λ bacteriophage 23-kb HindIII size standard, as well as for 14- and 32-kb DNA fragments (parentheses) estimated from the location of λ bacteriophage HindIII fragments and λ bacteriophage ladder.
Characterization of Sites of Chromosome Healing.
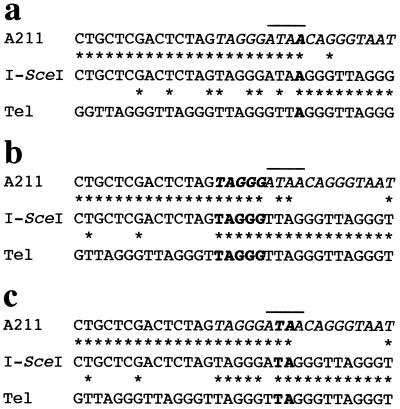
The location of the site of addition of the TRS in the Ganr/G418r subclones was determined by PCR. Because of the selection with G418, the location of the site of addition of the newly added telomere is limited to the region between the neo gene promoter and the I-SceI recognition sequence. By using an 18-bp primer complementary to the neo promoter and another 18-bp primer complementary to TRS, the terminus of the chromosome was amplified from the I-SceI-treated Ganr/G418r subclones. As expected, bands were observed in the I-SceI-treated subclones that were not present in the mock-treated subclones (data not shown). Nucleotide sequence analysis of these PCR products from four separate experiments demonstrated that the new telomeres were added directly onto the end of the chromosome at or near the site of cleavage by the I-SceI endonuclease (Fig. 5). In 19 of the 27 I-SceI-treated Ganr/G418r subclones analyzed, the site of addition contained 1 bp of complementarity, involving the last nucleotide of the 4-bp 3′ overhang of the I-SceI cleavage site (Fig. 5a). Thus, in most subclones chromosome healing occurred without the loss of any nucleotides from the break site. The next most common site of addition of the new telomeres (6 of 27) was a region with 5 bp of complementarity within the I-SceI recognition sequence (Fig. 5b). In these subclones, four nucleotides were lost from the end of the chromosome before healing. Finally, in two subclones, chromosome healing was found to involve 2 bp of complementarity, with two nucleotides lost from the end of the chromosome (Fig. 5c).
Figure 5.
Nucleotide sequence analysis of the site of addition of telomeres to the end of the broken chromosome. The nucleotide sequence of DNA amplified from the Ganr/G418r subclones treated with I-SceI (I-SceI) is compared with the sequence in the parental clone (A211) and with the TRS (Tel). Identical nucleotides (∗), the I-SceI recognition sequence (italics), and the 4-bp ATAA overhang generated by the I-SceI endonuclease (horizontal line) are indicated. The region of homology between the cellular DNA and TRS at the site of addition of the new telomere is also indicated (bold).
DISCUSSION
The results presented in this study clearly show that mammalian cells in culture can efficiently perform chromosome healing after the loss of a telomere. How frequently chromosome healing occurs compared with other forms of dsb repair is difficult to determine, because the efficiency of cutting at the I-SceI site after electroporation of the expression vector containing the I-SceI gene is not known. A previous study concluded that electroporation of an expression vector containing the I-SceI gene could result in the loss of the I-SceI site in approximately 1% of the cells because of recombination or improper end joining (28). Presumably, many of the other I-SceI sites would have also been cut but precisely rejoined. The frequency of chromosome healing after transient treatment with I-SceI (10−4) was 100-fold lower than the maximum frequency of intrachromosomal recombination induced by I-SceI (ref. 26; unpublished observations). However, many of the I-SceI-induced dsbs in our system would be rejoined without the loss of the terminal fragment. Presumably, chromosome healing would occur only in cells that are unable to rejoin the ends of the dsb and thereby lose the end of the chromosome. This possibility would be consistent with our observation that the fragment containing the HSV-tk gene was never observed in any of the subclones in which chromosome healing occurred. Estimating the efficiency of chromosome healing is further complicated by the fact that the frequency of chromosome healing detected in our system is likely to be underestimated. Because of the position of the I-SceI site next to the neo gene promoter, and the requirement for selection with G418 to prevent silencing, selection for the neo gene would eliminate cells in which exonuclease digestion at the break site occurred. In fact, previous studies in yeast have observed that extensive degradation of chromosome ends commonly occurs before chromosome healing (9, 11).
How the rate of chromosome healing in human cells compares to that occurring in yeast cells is also difficult to determine. Without the presence of internal TRS, the rate of chromosome healing in yeast was found to be less than 10−3 (9, 11), while the rate observed in our studies with mouse ES cells was 10−4. However, these rates cannot be compared directly because there was no restriction on the extent of degradation of the chromosome in the yeast studies. In addition, the studies in yeast were performed by using dsb repair-deficient cells, because chromosome healing cannot be detected in repair-proficient cells. Thus, mammalian cells may be more proficient in chromosome healing than yeast, as might be expected from the minimal sequence complementarity required for chromosome healing in humans (13–15) and mice (this study). In this respect, mammalian cells would appear to be more like many other organisms (6–8) and yeast PIF1 mutants (11), which also add new telomeres to sites with little complementarity to TRS.
Although it is clear that chromosome healing occurs in mouse ES cells, the exact mechanism(s) involved has yet to be determined. Based on the analysis of the Ganr/G418r subclones, it would appear that the 3′ overhang generated by the I-SceI endonuclease serves as an excellent substrate for chromosome healing. 3′ overhangs are likely to be found after ionizing radiation-induced or spontaneous dsb, because dsbs have been shown to be resected 5′-to-3′ (40, 41). Our results also demonstrate that short regions of complementarity facilitate chromosome healing in mammalian cells. Studies with extracts of human cells (15) suggest that human telomerase can initiate de novo synthesis of telomeres by using short regions of complementarity similar to those in our study. However, it cannot be concluded that telomerase is involved in our studies, because short regions of complementarity are also seen with both nonhomologous end joining (42). Therefore the new telomeres could have originated by nonhomologous end joining of preexisting short TRS present in the cell. However, the rejoining of the original telomere seems unlikely in view of the extensive unidirectional degradation that would be required to generate the shortened telomeres observed.
The observation that the new telomeres are gradually elongated with time in culture (Fig. 5) suggests that they were much shorter initially, because the Ganr/G418r subclones had been through approximately 20 cell divisions after treatment with I-SceI. However, the large size of the new telomeres (16–30 kb) at the time the subclones are first analyzed (after 20–25 cell divisions) suggests that the rate of elongation must have been much greater initially or that preexisting TRS were added on by nonhomologous end joining. Evidence that an initial period of rapid elongation occurs after the addition of new telomeres in ES cells is provided by the fact that newly seeded telomeres established from transfected plasmid sequences are also quite long at the time the transfected clones are first analyzed (ref. 34; this study) and then continue to gradually increase in length (34).
Chromosome healing was previously proposed by McClintock to be a mechanism for preventing chromosome instability resulting from telomere loss and the initiation of breakage/fusion/bridge cycles (43). By using a system similar to our present study, we have found that spontaneous telomere loss in a human tumor cell line usually results in chromosome fusion and the initiation of breakage/fusion/bridge cycles (unpublished observation). Chromosome healing is also seen, but only about one-tenth as frequently as chromosome fusion. The studies presented here show that the opposite is true in mouse ES cells, where DNA fused to the end of the chromosome after treatment with I-SceI occurs much less frequently than chromosome healing (Fig. 3). These results may mean that chromosome healing is deficient in some human tumor cells, which would promote chromosome instability after telomere loss. Alternatively, chromosome fusion could be selected against in mouse ES cells, which have been shown to have a p53-independent pathway for apoptosis in response to DNA damage (44). The use of this assay system will provide a means of addressing the role that chromosome healing plays in preventing chromosome instability resulting from telomere loss.
Acknowledgments
This work was supported by grant number RO1 ES008427 from the National Institute of Environmental Health Sciences, National Institutes of Health.
ABBREVIATIONS
- HSV-tk
herpes simplex virus thymidine kinase
- TRS
telomeric repeat sequences
- ES
embryonic stem
- dsb
double-strand break
References
- 1.Blackburn E H, Greider C W. Cold Spring Harbor Monograph. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. [Google Scholar]
- 2.de Lange T. In: Telomeres. Blackburn E H, Greider C W, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. pp. 265–293. [Google Scholar]
- 3.Harley C B, Kim N W, Prowse K R, Weinrich S L, Hirsch K S, West M D, Becchetti S, Hirte H W, Counter C M, Greider C W, et al. Cold Spring Harbor Symp Quant Biol. 1994;91:2900–2904. doi: 10.1101/sqb.1994.059.01.035. [DOI] [PubMed] [Google Scholar]
- 4.Bodnar A G, Ouellette M, Frolkis M, Holt S E, Chiu C-P, Morin G B, Harley C B, Shay J W, Lichtsteiner S, Wright W E. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 5.Klyono T, Foster S A, Koop J I, McDougall J K, Galloway D A, Klingelhutz A J. Nature (London) 1998;396:84–88. doi: 10.1038/23962. [DOI] [PubMed] [Google Scholar]
- 6.Melek M, Shippen D E. BioEssays. 1996;18:301–308. doi: 10.1002/bies.950180408. [DOI] [PubMed] [Google Scholar]
- 7.Muller F, Wicky C, Spicher A, Tobler H. Cell. 1991;67:815–822. doi: 10.1016/0092-8674(91)90076-b. [DOI] [PubMed] [Google Scholar]
- 8.Bottius E, Bakhsis N, Scherf A. Mol Cell Biol. 1998;18:919–925. doi: 10.1128/mcb.18.2.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kramer K M, Haber J E. Genes Dev. 1993;7:2345–2356. doi: 10.1101/gad.7.12a.2345. [DOI] [PubMed] [Google Scholar]
- 10.Sandell L L, Zakian V A. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- 11.Schulz V P, Zakian V A. Cell. 1994;76:145–155. doi: 10.1016/0092-8674(94)90179-1. [DOI] [PubMed] [Google Scholar]
- 12.Borgaonkar D S. Chromosomal Variation in Man. New York: Liss; 1989. [Google Scholar]
- 13.Flint J, Craddock C F, Villegas A, Bentley D P, Williams H J, Galanello R, Cao A, Wood W G, Ayyub H, Higgs D R. Am J Hum Genet. 1994;55:505–512. [PMC free article] [PubMed] [Google Scholar]
- 14.Wong A C, Ning Y, Flint J, Clark K, Dumanski J P, Ledbetter D H, McDermid H E. Am J Hum Genet. 1997;60:113–120. [PMC free article] [PubMed] [Google Scholar]
- 15.Morin G B. Nature (London) 1991;353:454–456. doi: 10.1038/353454a0. [DOI] [PubMed] [Google Scholar]
- 16.Yu G, Blackburn E H. Cell. 1991;67:823–832. doi: 10.1016/0092-8674(91)90077-c. [DOI] [PubMed] [Google Scholar]
- 17.Meltzer P S, Guan X-Y, Trent J M. Nat Genet. 1993;4:252–255. doi: 10.1038/ng0793-252. [DOI] [PubMed] [Google Scholar]
- 18.Slijepcevic P, Natarajan A T, Bryant P E. Mutagenesis. 1998;13:45–49. doi: 10.1093/mutage/13.1.45. [DOI] [PubMed] [Google Scholar]
- 19.Murnane J P, Yu L-C. Mol Cell Biol. 1993;13:977–983. doi: 10.1128/mcb.13.2.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murnane J P, Sabatier L, Marder B A, Morgan W F. EMBO J. 1994;13:4953–4962. doi: 10.1002/j.1460-2075.1994.tb06822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robertson E J. In: Teratocarcinomas and Embryonic Stem Cells: A Practical Approach. Robertson E J, editor. Oxford: Oxford Univ. Press; 1987. pp. 71–112. [Google Scholar]
- 22.Hanish J P, Yanowitz J L, De Lange T. Proc Natl Acad Sci USA. 1994;91:8861–8865. doi: 10.1073/pnas.91.19.8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murata S, Matsuzaki T, Takai S, Yaoita H, Noda M. Mutat Res. 1995;334:375–383. doi: 10.1016/0165-1161(95)90075-6. [DOI] [PubMed] [Google Scholar]
- 24.Brisebois J J, DuBow M S. Mutat Res. 1993;287:191–205. doi: 10.1016/0027-5107(93)90012-5. [DOI] [PubMed] [Google Scholar]
- 25.Soriano P, Montgomery C, Geske R, Bradley A. Cell. 1991;64:693–702. doi: 10.1016/0092-8674(91)90499-o. [DOI] [PubMed] [Google Scholar]
- 26.Donoho G, Jasin M, Berg P. Mol Cell Biol. 1998;18:4070–4078. doi: 10.1128/mcb.18.7.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elliott B, Richardson C, Winderbaum J, Nickoloff J A, Jasin M. Mol Cell Biol. 1998;18:93–101. doi: 10.1128/mcb.18.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richardson C, Moynahan M E, Jasin M. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rouet P, Smih F, Jasin M. Proc Natl Acad Sci USA. 1994;91:6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smih F, Rouet P, Romanienko P J, Jasin M. Nucleic Acids Res. 1995;23:5012–5019. doi: 10.1093/nar/23.24.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luria S E, Delbruck M. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Capizzi R L, Jameson J W. Mutat Res. 1972;17:147–148. doi: 10.1016/0027-5107(73)90265-0. [DOI] [PubMed] [Google Scholar]
- 33.Richard C W, III, Withers D A, Meeker T C, Maurer S, Evans G A, Myers R M, Cox D R. Am J Hum Genet. 1991;49:1189–1196. [PMC free article] [PubMed] [Google Scholar]
- 34.Barnett M A, Buckle J, Evans E P, Porter A C G, Rout D, Smith A G, Brown W R A. Nucleic Acids Res. 1993;21:27–36. doi: 10.1093/nar/21.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farr C, Fantes J, Goodfellow P, Cooke H. Proc Natl Acad Sci USA. 1991;88:7006–7010. doi: 10.1073/pnas.88.16.7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sprung C N, Sabatier L, Murnane J P. Exp Cell Res. 1999;247:29–37. doi: 10.1006/excr.1998.4293. [DOI] [PubMed] [Google Scholar]
- 37.Zijlmans J M J M, Martens U M, Poon S S S, Raap A K, Tanke H J, Ward R K, Lansdorp P M. Proc Natl Acad Sci USA. 1997;94:7423–7428. doi: 10.1073/pnas.94.14.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weng A, Magnuson T, Storb U. Development (Cambridge, UK) 1995;121:2853–2859. doi: 10.1242/dev.121.9.2853. [DOI] [PubMed] [Google Scholar]
- 39.Kleymenova E V, Yuan X, LaBate M E, Walker C L. Oncogene. 1998;16:713–720. doi: 10.1038/sj.onc.1201583. [DOI] [PubMed] [Google Scholar]
- 40.Ivanov E L, Sugawara N, White C I, Fabre F, Haber J E. Mol Cell Biol. 1994;14:3414–3425. doi: 10.1128/mcb.14.5.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee S E, Moore J K, Holmes A, Umezu K, Kolodner R D, Haber J E. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 42.Meuth M. In: Mobile DNA. Berg D E, Howe M M, editors. Washington, DC: Am. Soc. Microbiol.; 1989. pp. 833–860. [Google Scholar]
- 43.McClintock B. Genetics. 1941;41:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aladjem M I, Spike B T, Rodewald L W, Hope T J, Klemm M, Jaenisch R, Wahl G M. Curr Biol. 1998;8:145–155. doi: 10.1016/s0960-9822(98)70061-2. [DOI] [PubMed] [Google Scholar]